The Analogs of Temporin-GHa Exhibit a Broader Spectrum of Antimicrobial Activity and a Stronger Antibiofilm Potential against Staphylococcus aureus
Abstract
1. Introduction
2. Results
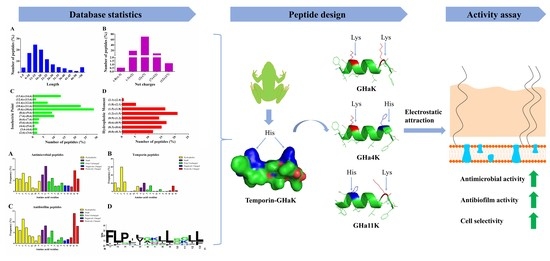
2.1. Statistical Analysis of Bioinformatics
2.2. Peptide Design and Physicochemical Properties
2.3. The Activity and Structure Prediction
2.4. Derived Peptides Showed Higher and Broader Spectrum Antibacterial Activity Than the Parent Peptide
2.5. Growth Inhibition Kinetics
2.6. Killing Kinetics of GHa and Its Derived Peptides
2.7. Effect of GHa and Its Derived Peptides on the Bacterial Membrane Permeability
2.8. Effects of GHa and Its Analogues on Bacterial Initial Attachment, Biofilm Formation, and Preformed Biofilms
2.9. Hemolytic Activity of Peptides
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Bioinformatics Statistical Analysis, Peptide Design and Physicochemical Properties
4.3. Activity and Structure Prediction
4.4. Synthesis of Peptides
4.5. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) Assays
4.6. Growth Curve Assay
4.7. Time-Killing Curves
4.8. Membrane Permeation by Fluorescence Spectroscopy
4.9. Antibiofilm Assay
4.9.1. Inhibition of bacterial initial attachment
4.9.2. Inhibition of biofilm growth
4.9.3. Preformed biofilm disruption
4.9.4. Biofilm metabolic activity assay (MTT assay)
4.10. Hemolytic Assays
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lin, C.; Lee, P.R. The Antibiotic Paradox: How the Misuse of Antibiotics Destroys Their Curative Powers (review). Perspect. Boil. Med. 2003, 46, 603–604. [Google Scholar]
- Molton, J.S.; Tambyah, P.A.; Ang, B.S.P.; Ling, M.L.; Fisher, D.A. The Global Spread of Healthcare-Associated Multidrug-Resistant Bacteria: A Perspective from Asia. Clin. Infect. Dis. 2013, 56, 1310–1318. [Google Scholar] [PubMed]
- Fridkin, S.K.; Steward, C.D.; Edwards, J.R.; Pryor, E.R.; McGowan, J.E., Jr.; Archibald, L.K.; Gaynes, R.P.; Tenover, F.C. Surveillance of antimicrobial use and antimicrobial resistance in United States hospitals: project ICARE phase 2. Project Intensive Care Antimicrobial Resistance Epidemiology (ICARE) hospitals. Clin. Infect. Dis. 1999, 29, 245–252. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef]
- Klevens, R.M.; Morrison, M.A.; Nadle, J.; Petit, S.; Gershman, K.; Ray, S.; Harrison, L.H.; Lynfield, R.; Dumyati, G.; Townes, J.M.; et al. Invasive Methicillin-Resistant Staphylococcus aureus Infections in the United States. JAMA 2007, 298, 1763. [Google Scholar] [CrossRef]
- Nguyen, D.B.; Lessa, F.C.; Belflower, R.; Mu, Y.; Wise, M.; Nadle, J.; Bamberg, W.M.; Petit, S.; Ray, S.M.; Harrison, L.H.; et al. Invasive methicillin-resistant Staphylococcus aureus infections among patients on chronic dialysis in the United States, 2005–2011. Clin. Infect. Dis. 2013, 57, 1393–1400. [Google Scholar] [CrossRef]
- Tiwari, H.K.; Sen, M.R. Emergence of vancomycin resistant Staphylococcus aureus (VRSA) from a tertiary care hospital from northern part of India. BMC Infect. Dis. 2006, 6, 156. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm Formation as Microbial Development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Wu, H.; Moser, C.; Wang, H.-Z.; Høiby, N.; Song, Z.-J. Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 2015, 7, 1–7. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Scott, M.G.; Hancock, R.E.W. Cationic antimicrobial peptides and their multifunctional role in the immune system. Crit. Rev. Immunol. 2000, 20, 24. [Google Scholar] [CrossRef]
- Li, L.; Sun, J.; Xia, S.; Tian, X.; Cheserek, M.J.; Le, G. Mechanism of antifungal activity of antimicrobial peptide APP, a cell-penetrating peptide derivative, against Candida albicans: intracellular DNA binding and cell cycle arrest. Appl. Microbiol. Biotechnol. 2016, 100, 3245–3253. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Luo, W.; Zhong, H.; Wang, M.; Song, Y.; Deng, S.; Zhang, Y. Molecular cloning and characterization of antimicrobial peptides from skin of Hylarana guentheri. Acta Biochim. et Biophys. Sin. 2017, 49, 450–457. [Google Scholar] [CrossRef] [PubMed]
- DBAASP. Available online: https://dbaasp.org/statistics (accessed on 10 January 2019).
- APD3. Available online: http://aps.unmc.edu/AP/main.php (accessed on 10 January 2019).
- WebLogo. Available online: https://weblogo.berkeley.edu/ (accessed on 10 January 2019).
- Lata, S.; Mishra, N.K.; Raghava, G.P. AntiBP2: improved version of antibacterial peptide prediction. BMC Bioinform. 2010, 11, S19. [Google Scholar] [CrossRef] [PubMed]
- Saporito, P.; Vang Mouritzen, M.; Løbner-Olesen, A.; Jenssen, H. LL-37 fragments have antimicrobial activity against Staphylococcus epidermidis biofilms and wound healing potential in HaCaT cell line. J. Pept. Sci. 2018, 24, e3080. [Google Scholar] [CrossRef]
- CAMPR3. Available online: http://www.camp.bicnirrh.res.in/prediction.php (accessed on 12 January 2019).
- Heliquest. Available online: http://heliquest.ipmc.cnrs.fr/ (accessed on 12 January 2019).
- PEP-FOLD. Available online: http://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD/ (accessed on 12 January 2019).
- Zhu, X.; Dong, N.; Wang, Z.; Ma, Z.; Zhang, L.; Ma, Q.; Shan, A. Design of imperfectly amphipathic α-helical antimicrobial peptides with enhanced cell selectivity. Acta Biomater. 2014, 10, 244–257. [Google Scholar] [CrossRef]
- Yuan, Y.; Zai, Y.; Xi, X.; Ma, C.; Wang, L.; Zhou, M.; Shaw, C.; Chen, T. A novel membrane-disruptive antimicrobial peptide from frog skin secretion against cystic fibrosis isolates and evaluation of anti-MRSA effect using Galleria mellonella model. Biochim. et Biophys. Acta (BBA)-Gen. Subj. 2019, 1863, 849–856. [Google Scholar] [CrossRef]
- Mishra, B.; Wang, X.; Lushnikova, T.; Zhang, Y.; Golla, R.M.; Narayana, J.L.; Wang, C.; McGuire, T.R.; Wang, G. Antibacterial, antifungal, anticancer activities and structural bioinformatics analysis of six naturally occurring temporins. Peptides 2018, 106, 9–20. [Google Scholar] [CrossRef]
- Zhang, S.-K.; Song, J.-W.; Gong, F.; Li, S.-B.; Chang, H.-Y.; Xie, H.-M.; Gao, H.-W.; Tan, Y.-X.; Ji, S.-P. Design of an α-helical antimicrobial peptide with improved cell-selective and potent anti-biofilm activity. Sci. Rep. 2016, 6, 27394. [Google Scholar] [CrossRef]
- Zaet, A.; Dartevelle, P.; Daouad, F.; Ehlinger, C.; Quilès, F.; Francius, G.; Boehler, C.; Bergthold, C.; Frisch, B.; Prévost, G.; et al. D-Cateslytin, a new antimicrobial peptide with therapeutic potential. Sci. Rep. 2017, 7, 15199. [Google Scholar] [CrossRef]
- Mangoni, M.L.; Maisetta, G.; Di Luca, M.; Gaddi, L.M.H.; Esin, S.; Florio, W.; Brancatisano, F.L.; Barra, D.; Campa, M.; Batoni, G. Comparative analysis of the bactericidal activities of amphibian peptide analogues against multidrug-resistant nosocomial bacterial strains. Antimicrob. Agents Chemother. 2008, 52, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Ong, Z.Y.; Wiradharma, N.; Yang, Y.Y. Strategies employed in the design and optimization of synthetic antimicrobial peptide amphiphiles with enhanced therapeutic potentials. Adv. Drug Deliv. Rev. 2014, 78, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Sikora, K.; Jaskiewicz, M.; Neubauer, D.; Bauer, M.; Bartoszewska, S.; Barańska-Rybak, W.; Kamysz, W. Counter-ion effect on antistaphylococcal activity and cytotoxicity of selected antimicrobial peptides. Amino Acids 2018, 50, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Grassi, L.; Maisetta, G.; Maccari, G.; Esin, S.; Batoni, G. Analogs of the Frog-skin Antimicrobial Peptide Temporin 1Tb Exhibit a Wider Spectrum of Activity and a Stronger Antibiofilm Potential as Compared to the Parental Peptide. Front. Chem. 2017, 5, 1173. [Google Scholar] [CrossRef] [PubMed]
- Sang, M.; Wu, Q.; Xi, X.; Ma, C.; Wang, L.; Zhou, M.; Burrows, J.F.; Chen, T. Identification and target-modifications of temporin-PE: A novel antimicrobial peptide in the defensive skin secretions of the edible frog, Pelophylax kl. esculentus. Biochem. Biophys. Res. Commun. 2018, 495, 2539–2546. [Google Scholar] [CrossRef]
- Abbassi, F.; Raja, Z.; Oury, B.; Gazanion, E.; Piesse, C.; Sereno, D.; Nicolas, P.; Foulon, T.; Ladram, A. Antibacterial and leishmanicidal activities of temporin-SHd, a 17-residue long membrane-damaging peptide. Biochimie 2013, 95, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Mecozzi, S.; West, A.P.; Dougherty, D.A. Cation-pi interactions in aromatics of biological and medicinal interest: electrostatic potential surfaces as a useful qualitative guide. Proc. Natl. Acad. Sci. 1996, 93, 10566–10571. [Google Scholar] [CrossRef]
- Dougherty, D.A. Cation-π interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science 1996, 271, 163–168. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Sugishita, K.-I.; Harada, M.; Fujii, N.; Miyajima, K. Interactions of an antimicrobial peptide, magainin 2, with outer and inner membranes of Gram-negative bacteria. Biochim. et Biophys. Acta (BBA)-Bioenerg. 1997, 1327, 119–130. [Google Scholar] [CrossRef]
- Papo, N.; Shai, Y.; Humphries, J.D.; Schofield, N.R.; Mostafavi-Pour, Z.; Green, L.J.; Mould, A.P.; Garratt, A.N. A Molecular Mechanism for Lipopolysaccharide Protection of Gram-negative Bacteria from Antimicrobial Peptides. J. Boil. Chem. 2005, 280, 10378–10387. [Google Scholar] [CrossRef]
- Rosenfeld, Y.; Barra, D.; Simmaco, M.; Shai, Y.; Mangoni, M.L. A Synergism between Temporins toward Gram-negative Bacteria Overcomes Resistance Imposed by the Lipopolysaccharide Protective Layer. J. Boil. Chem. 2006, 281, 28565–28574. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Rinaldi, A.C.; Di Giulio, A.; Simmaco, M.; Kinnunen, P.K. Interactions of the antimicrobial peptides temporins with model biomembranes. Comparison of temporins B and L. Biochemistry 2002, 41, 4425–4436. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.D.; Palmer, M. The action mechanism of daptomycin. Bioorganic Med. Chem. 2016, 24, 6253–6268. [Google Scholar]
- Falanga, A.; Valiante, S.; Galdiero, E.; Franci, G.; Scudiero, O.; Morelli, G.; Galdiero, S. Dimerization in tailoring uptake efficacy of the HSV-1 derived membranotropic peptide gH625. Sci. Rep. 2017, 7, 9434. [Google Scholar] [CrossRef] [PubMed]
- Dean, S.N.; Bishop, B.M.; Van Hoek, M.L. Natural and synthetic cathelicidin peptides with anti-microbial and anti-biofilm activity against Staphylococcus aureus. BMC Microbiol. 2011, 11, 114. [Google Scholar] [CrossRef]
- Costerton, J.; Montanaro, L.; Arciola, C. Bacterial Communications in Implant Infections: A Target for an Intelligence War. Int. J. Artif. Organs 2007, 30, 757–763. [Google Scholar] [CrossRef]
- Jefferson, K.K.; Goldmann, D.A.; Pier, G.B. Use of Confocal Microscopy to Analyze the Rate of Vancomycin Penetration through Staphylococcus Aureus Biofilms. Antimicrob. Agents Chemother. 2005, 49, 2467–2473. [Google Scholar] [CrossRef]
- Kharidia, R.; Liang, J.F. The activity of a small lytic peptide PTP-7 on Staphylococcus aureus biofilms. J. Microbiol. 2011, 49, 663–668. [Google Scholar] [CrossRef]
- Hoffman, L.R.; D’Argenio, D.A.; MacCoss, M.J.; Zhang, Z.; Jones, R.A.; Miller, S.I. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 2005, 436, 1171–1175. [Google Scholar] [CrossRef]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Rational design of α-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar] [CrossRef]
- Ko, S.J.; Kang, N.H.; Kim, M.K.; Park, J.; Park, E.; Park, G.H.; Kang, T.W.; Na, D.E.; Park, J.B.; Yi, Y.E.; et al. Antibacterial and anti-biofilm activity, and mechanism of action of pleurocidin against drug resistant Staphylococcus aureus. Microb. Pathog. 2019, 127, 70–78. [Google Scholar] [CrossRef] [PubMed]
- dPABBs. Available online: http://ab-openlab.csir.res.in/abp/antibiofilm/index.php (accessed on 10 January 2019).
- ExPASy. Available online: https://www.expasy.org (accessed on 12 January 2019).
- Wang, Y.; Wang, X.; Jiang, W.; Wang, K.; Luo, J.; Li, W.; Zhou, X.; Zhang, L. Antimicrobial peptide GH12 suppresses cariogenic virulence factors of Streptococcus mutans. J. Oral Microbiol. 2018, 10, 1442089. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.; Shah, R.; Bhave, M.; Palombo, E.A. Inhibitory activity of yarrow essential oil on Listeria planktonic cells and biofilms. Food Control. 2013, 29, 125–130. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |






| Peptide | Sequence | MW b | μH a | Amphip-Athicity a | Charge a | PI a | BI b | GRAVY b |
|---|---|---|---|---|---|---|---|---|
| GHa | FLQHIIGALGHLF | 1464.76 | 1.71 | 0.32 | 1 | 7.67 | −1.49 | 1.315 |
| GHaK | FLQKIIGALGKLF | 1446.83 | 1.78 | 0.66 | 2 | 10.7 | −1.35 | 1.208 |
| GHa4K | FLQKIIGALGHLF | 1455.79 | 1.74 | 0.49 | 1.5 | 9.87 | −1.42 | 1.262 |
| GHa11K | FLQHIIGALGKLF | 1455.79 | 1.76 | 0.49 | 1.5 | 9.87 | −1.42 | 1.262 |
| Species | Strains | (μM) (μg/mL) | |||||
|---|---|---|---|---|---|---|---|
| GHa | GHaK | GHa4K | GHa11K | Kanamycin | |||
| Gram+ | SA | MIC | 12.5 (18.3) | 1.6 (2.3) | 3.1 (4.5) | 1.6 (2.3) | 6.2 (3.6) |
| MBC | 25 (36.6) | 1.6 (2.3) | 3.1 (4.5) | 1.6 (2.3) | |||
| SM | MIC | 25 (36.6) | 3.1 (4.5) | 6.2 (9.1) | 6.2 (9.1) | 6.2 (3.6) | |
| MBC | 50 (73.2) | 6.2 (9) | 6.2 (9.1) | 6.2 (9.1) | |||
| BS | MIC | >100 (>146.5) | 12.5 (18.1) | >100 (>145.6) | 25 (36.4) | >100 (>58.2) | |
| MBC | >100 (>146.5) | 25 (36.2) | >100 (>145.6) | 50 (72.8) | |||
| MRSA | MIC | 100 (146.5) | 6.2 (9) | 6.2 (9.1) | 6.2 (9.1) | >100 (>58.2) | |
| MBC | >100 (>146.5) | 6.2 (9) | 6.2 (9.1) | 6.2 (9.1) | |||
| MRSA-2 | MIC | >100 (>146.5) | 6.2 (9) | 12.5 (18.2) | 6.2 (9.1) | >100 (>58.2) | |
| MBC | >100 (>146.5) | 12.5 (18.1) | 12.5 (18.2) | 12.5 (18.2) | |||
| Gram- | EC | MIC | 25 (36.6) | 6.2 (9) | 12.5 (18.2) | 12.5 (18.2) | 12.5 (7.3) |
| MBC | 50 (73.2) | 6.2 (9) | 12.5 (18.2) | 12.5 (18.2) | |||
| D31 | MIC | >100 (>146.5) | 12.5 (18.1) | >100 (>145.6) | 50 (72.8) | 25 (14.6) | |
| MBC | >100 (>146.5) | 12.5 (18.1) | >100 (>145.6) | 50 (72.8) | |||
| PAO1 | MIC | >100 (>146.5) | 12.5 (18.1) | >100 (>145.6) | >100 (>145.6) | 25 (14.6) | |
| MBC | >100 (>146.5) | 50 (72.3) | >100 (>145.6) | >100 (>145.6) | |||
| PA | MIC | >100 (>146.5) | 6.2 (9) | 6.2 (9.1) | 50 (72.8) | >100 (>58.2) | |
| MBC | >100 (>146.5) | 12.5 (18.1) | 6.2 (9.1) | >100 (>145.6) | |||
| Fungi | CA | MIC | 50 (73.2) | 25 (36.2) | >100 (>145.6) | 50 (72.8) | >100 (>58.2) |
| MBC | >100 (>146.5) | 50 (72.3) | >100 (>145.6) | >100 (>145.6) | |||
| Peptide | (μM) (μg/mL) | |||
|---|---|---|---|---|
| MBIC50 | MBIC90 | MBEC50 | MBEC90 | |
| GHa | 6.2 (9.2) | 12.5 (18.3) | 50 (73.2) | 100 (146.5) |
| GHaK | 0.4 (0.6) | 1.6 (2.3) | 3.1 (4.5) | 12.5 (18.1) |
| GHa4K | 0.8 (1.1) | 3.1 (4.5) | 12.5 (18.2) | 25 (36.4) |
| GHa11K | 0.8 (1.1) | 1.6 (2.3) | 3.1 (4.5) | 12.5 (18.2) |
| MHC a (μM) (μg/mL) | HL50 b (μM) (μg/mL) | CSI c | TI d | ||
|---|---|---|---|---|---|
| GHa | no bacterial | 20(29.3) | 115(168.4) | 9.2 | 0.5 |
| with S. aureus | 29(42.5) | 125(183.1) | |||
| GHaK | no bacterial | 16(23.1) | 66(95.5) | 42.3 | 2.8 |
| with S. aureus | 22(31.8) | 105(151.9) | |||
| GHa4K | no bacterial | 26(37.8) | 69(100.4) | 22.3 | 3.7 |
| with S. aureus | 70(101.9) | 156(227.1) | |||
| GHa11K | no bacterial | 15(21.8) | 40(58.2) | 25.6 | 2.3 |
| with S. aureus | 65(94.6) | 149(216.9) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Z.; Wei, H.; Meng, J.; Cheng, T.; Song, Y.; Wang, M.; Zhang, Y. The Analogs of Temporin-GHa Exhibit a Broader Spectrum of Antimicrobial Activity and a Stronger Antibiofilm Potential against Staphylococcus aureus. Molecules 2019, 24, 4173. https://doi.org/10.3390/molecules24224173
Xie Z, Wei H, Meng J, Cheng T, Song Y, Wang M, Zhang Y. The Analogs of Temporin-GHa Exhibit a Broader Spectrum of Antimicrobial Activity and a Stronger Antibiofilm Potential against Staphylococcus aureus. Molecules. 2019; 24(22):4173. https://doi.org/10.3390/molecules24224173
Chicago/Turabian StyleXie, Zhipeng, Hanqi Wei, Jiahui Meng, Tong Cheng, Yanting Song, Manchuriga Wang, and Yingxia Zhang. 2019. "The Analogs of Temporin-GHa Exhibit a Broader Spectrum of Antimicrobial Activity and a Stronger Antibiofilm Potential against Staphylococcus aureus" Molecules 24, no. 22: 4173. https://doi.org/10.3390/molecules24224173
APA StyleXie, Z., Wei, H., Meng, J., Cheng, T., Song, Y., Wang, M., & Zhang, Y. (2019). The Analogs of Temporin-GHa Exhibit a Broader Spectrum of Antimicrobial Activity and a Stronger Antibiofilm Potential against Staphylococcus aureus. Molecules, 24(22), 4173. https://doi.org/10.3390/molecules24224173