Simultaneously Quantitative Analysis of Naringin and Its Major Human Gut Microbial Metabolites Naringenin and 3-(4′-Hydroxyphenyl) Propanoic Acid via Stable Isotope Deuterium-Labeling Coupled with RRLC-MS/MS Method
Abstract
1. Introduction
2. Results and Discussion
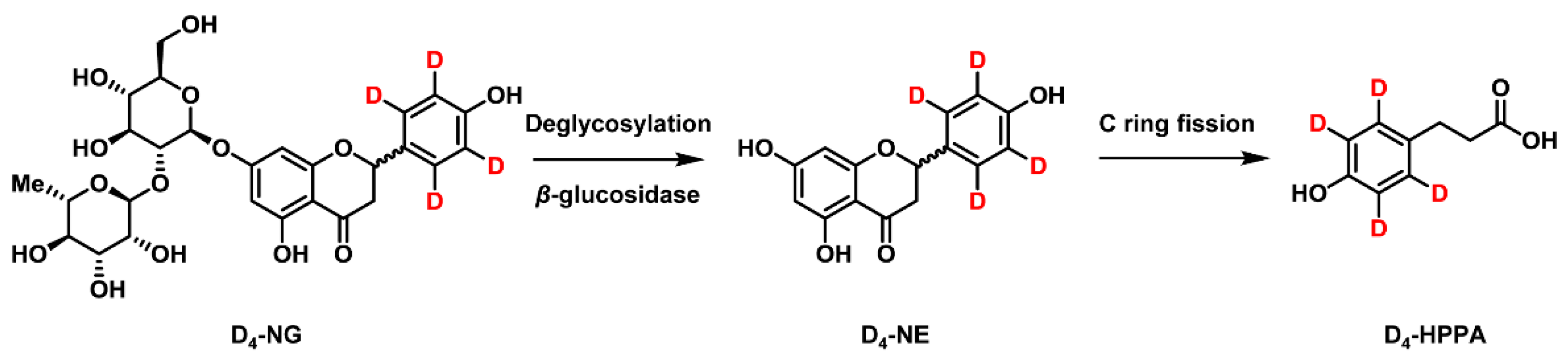
2.1. Synthesis of [2′,3′,5′,6′-D4]naringenin (D4-NE) and 3-(4′-Hydroxyphenyl)-[2′,3′,5′,6′-D4]propanoic Acid (D4-HPPA)
2.2. Method Development and Optimization
2.3. Method Validation
2.3.1. Specificity
2.3.2. Linearity
2.3.3. Precision and Accuracy
2.3.4. Extract Recovery
2.3.5. Matrix Effect
2.3.6. Stability
2.4. Simultaneously Quantitative Analysis of D4-NG, D4-NE, and D4-HPPA after Anaerobic Co-Incubation of D4-NG with Human Gut Microbiota Solution
3. Experimental Procedures
3.1. Chemicals and Reagents
3.2. NMR, Infra-Red Spectra and Melting Point Analysis
3.3. Synthesis of [2′,3′,5′,6′-D4]Naringenin
3.4. Synthesis of 3-(4′-Hydroxyphenyl)-[2′,3′,5′,6′-D4]Propanoic Acid
3.5. Preparation of Stock Solutions, Calibration Standards, and Quality Control Samples
3.6. Recruitment of Human Participants
3.7. Feces Sample Collection and Gut Microbiota Solution Preparation
3.8. Anaerobic Incubation of D4-NG with Gut Microbiota Solution
3.9. Sample Preparation
3.10. RRLC-MS/MS Conditions
3.11. Method Validation
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Esfahani, A.; Wong, J.M.; Truan, J.; Villa, C.R.; Mirrahimi, A.; Srichaikul, K.; Kendall, C.W. Health effects of mixed fruit and vegetable concentrates: A systematic review of the clinical interventions. J. Am. Coll. Nutr. 2011, 30, 285–294. [Google Scholar] [CrossRef] [PubMed]
- García-Lafuente, A.; Guillamón, E.; Villares, A.; Rostagno, M.A.; Martínez, J.A. Flavonoids as anti-inflammatory agents: Implications in cancer and cardiovascular disease. Inflamm. Res. 2009, 58, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Benavente-García, O.; Castillo, J. Update on uses and properties of citrus flavonoids: New findings in anticancer, cardiovascular, and anti-inflammatory activity. J. Agric. Food Chem. 2008, 56, 6185–6205. [Google Scholar] [CrossRef] [PubMed]
- Bacanlı, M.; Başaran, A.A.; Başaran, N. The antioxidant and antigenotoxic properties of citrus phenolics limonene and naringin. Food Chem. Toxicol. 2015, 81, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Amini, N.; Sarkaki, A.; Dianat, M.; Mard, S.A.; Ahanqarpour, A.; Badavi, M. Protective effects of naringin and trimetazidine on remote effect of acute renal injury on oxidative stress and myocardial injury through Nrf-2 regulation. Pharmacol. Rep. 2019, 71, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Xulu, S.; Oroma Owira, P.M. Naringin ameliorates atherogenic dyslipidemia but not hyperglycemia in rats with type 1 diabetes. J. Cardiovasc. Pharmacol. 2012, 59, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, K.; Sudhandiran, G. Protective effect of naringin on 3-nitropropionic acid-induced neurodegeneration through the modulation of matrix metalloproteinases and glial fibrillary acidic protein. Can. J. Physiol. Pharm. 2016, 94, 65–71. [Google Scholar] [CrossRef]
- Ahmed, S.; Khan, H.; Aschner, M.; Hasan, M.M.; Hassan, S.T.S. Therapeutic potential of naringin in neurological disorders. Food Chem. Toxicol. 2019, 132, 110646. [Google Scholar] [CrossRef]
- Luo, Y.L.; Li, P.B.; Zhang, C.C.; Zheng, Y.F.; Wang, S.; Nie, Y.C.; Zhang, K.J.; Su, W.W. Effects of four antitussives on airway neurogenic inflammation in a guinea pig model of chronic cough induced by cigarette smoke exposure. Inflamm. Res. 2013, 62, 1053–1061. [Google Scholar] [CrossRef]
- Luo, Y.L.; Zhang, C.C.; Li, P.B.; Nie, Y.C.; Wu, H.; Shen, J.G.; Su, W.W. Naringin attenuates enhanced cough, airway hyperresponsiveness and airway inflammation in a guinea pig model of chronic bronchitis induced by cigarette smoke. Int. Immunopharmacol. 2012, 13, 301–307. [Google Scholar] [CrossRef]
- Gao, S.; Li, P.; Yang, H.; Fang, S.; Su, W. Antitussive effect of naringin on experimentally induced cough in Guinea pigs. Planta Med. 2011, 77, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wu, H.; Nie, Y.C.; Li, P.B.; Shen, J.G.; Su, W.W. Mucoactive effects of naringin in lipopolysaccharide-induced acute lung injury mice and beagle dogs. Environ. Toxicol. Pharmacol. 2014, 38, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Nie, Y.C.; Luo, Y.L.; Lin, F.; Zheng, Y.F.; Cheng, G.H.; Wu, H.; Zhang, K.J.; Su, W.W.; Shen, J.G.; et al. Protective effects of naringin against paraquat-induced acute lung injury and pulmonary fibrosis in mice. Food Chem. Toxicol. 2013, 58, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Gerritsen, J.; Smidt, H.; Rijkers, G.T.; de Vos, W.M. Intestinal microbiota in human health and disease: The impact of probiotics. Genes Nutr. 2011, 6, 209–240. [Google Scholar] [CrossRef] [PubMed]
- Koppel, N.; Rekdal, V.M.; Balskus, E.P. Chemical transformation of exnobiotics by the human gut microbiota. Science 2017, 356, 1246–1257. [Google Scholar] [CrossRef]
- Sousa, T.; Paterson, R.; Moore, V.; Carlsson, A.; Abrahamsson, B.; Basit, A.W. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int. J. Pharm. 2008, 363, 1–25. [Google Scholar] [CrossRef]
- Feng, T.; Wang, K.; Liu, F.; Ye, R.; Zhu, X.; Zhuang, H.; Xu, Z. Structural characterization and bioavailability of ternary nanoparticles consisting of amylose, α-linoleic acid and β-lactoglobulin complexed with naringin. Int. J. Biol. Macromol. 2017, 99, 365–374. [Google Scholar] [CrossRef]
- Chen, T.; Su, W.; Yan, Z.; Wu, H.; Zeng, X.; Peng, W.; Gan, L.; Zhang, Y.; Yao, H. Identification of naringin metabolites mediated by human intestinal microbes with stable isotope-labeling method and UFLC-Q-TOF-MS/MS. J. Pharm. Biomed. Anal. 2018, 161, 262–272. [Google Scholar] [CrossRef]
- Kumar, S.; Tiku, A.B. Biochemical and molecular mechanisms of radioprotective effects of naringenin, a phytochemical from citrus fruits. J. Agric. Food Chem. 2016, 64, 1676–1685. [Google Scholar] [CrossRef]
- Zeng, W.; Jin, L.; Zhang, F.; Zhang, C.; Liang, W. Naringenin as a potential immunomodulator in therapeutics. Pharmacol. Res. 2018, 135, 122–126. [Google Scholar] [CrossRef]
- Chen, C.; Wei, Y.Z.; He, X.M.; Li, D.D.; Wang, G.Q.; Li, J.J.; Zhang, F. Naringenin produces neuroprotection against LPS-induced dopamine neurotoxicity via the inhibition of microglial NLRP3 inflammasome activation. Front. Immunol. 2019, 10, 936. [Google Scholar] [CrossRef] [PubMed]
- Steed, A.L.; Christophi, G.P.; Kaiko, G.E.; Sun, L.; Goodwin, V.M.; Jain, U.; Esaulova, E.; Artyomov, M.N.; Morales, D.J.; Holtzman, M.J.; et al. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 2017, 357, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Blaut, M.; Schoefer, L.; Braune, A. Transformation of flavonoids by intestinal microorganisms. Int. J. Vitam. Nutr. Res. 2003, 73, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.Y.; Shao, L.; Zhang, W.; Wang, C.Z.; Zhou, H.H.; Huang, W.H.; Yuan, C.S. Metabolic analysis of Panax notoginseng saponins with gut gutmicrobiota-mediated biotransformation by HPLC-DAD-Q-TOF-MS/MS. J. Pharm. Biomed. Anal. 2018, 150, 199–207. [Google Scholar] [CrossRef]
- Pierce, C.L.; Williams, T.L.; Moura, H.; Pirkle, J.L.; Cox, N.J.; Stevens, J.; Donis, R.O.; Barr, J.R. Quantification of immunoreactive viral influenza proteins by immunoaffinity capture and isotope-dilution liquid chromatography-tandem mass spectrometry. Anal. Chem. 2011, 83, 4729–4737. [Google Scholar] [CrossRef]
- Chen, Q.S.; Wu, J.; Zhang, Y.; Lin, J.M. Qualitative and quantitative analysis of tumor cell metabolism via stable isotope labeling assisted microfluidic chip electrospray ionization mass spectrometry. Anal. Chem. 2012, 84, 1695–1701. [Google Scholar] [CrossRef]
- Liu, J.; Tang, M.; Lai, H.; Dong, Y.; Xie, C.; Ye, H.; Ma, L.; Qiu, N.; Li, Y.; Cai, L.; et al. Identification of metabolites of honokiol in rat urine using 13C stable isotope labeling and liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry. J. Chromatogr. A 2013, 1295, 48–56. [Google Scholar] [CrossRef]
- Wei, G.J.; Sheen, J.F.; Lu, W.C.; Hwang, L.S.; Ho, C.T.; Lin, C.I. Identification of sinensetin metabolites in rat urine by an isotope-labeling method and ultrahigh-performance liquid chromatography-electrospray ionization mass spectrometry. J. Agric. Food Chem. 2013, 61, 5016–5021. [Google Scholar] [CrossRef]
- Gao, D.; Chen, X.; Yang, X.; Wu, Q.; Jin, F.; Wen, H.; Jiang, Y.; Liu, H. Stable isotope labeling strategy for curcumin metabolite study in human liver microsomes by liquid chromatography-tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2015, 26, 686–694. [Google Scholar] [CrossRef]
- Luo, P.; Dai, W.; Yin, P.; Zeng, Z.; Kong, H.; Zhou, L.; Wang, X.; Chen, S.; Lu, X.; Xu, G. Multiple reaction monitoring-ion pair finder: A systematic approach to transform nontargeted mode to pseudotargeted mode for metabolomics study based on liquid chromatography-mass spectrometry. Anal. Chem. 2015, 87, 5050–5055. [Google Scholar] [CrossRef]
- Kim, K.Y.; Joo, H.J.; Kwon, H.W.; Kim, H.; Hancock, W.S.; Paik, Y.K. Development of a method to quantitate nematode pheromone for study of small-molecule metabolism in Caenorhabditis elegans. Anal. Chem. 2013, 85, 2681–2688. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Luo, Y.; Liu, M.; Chen, S.; Wang, S.; Nie, Y.; Cheng, G.; Su, W.; Zhang, K. Human intestinal microbial metabolism of naringin. Eur. J. Drug Metab. Pharmacokinet. 2015, 40, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Girón, A.; Queipo-Ortuño, M.I.; Boto-Ordóñez, M.; Muñoz-González, I.; Sánchez-Patán, F.; Monagas, M.; Martín-Álvarez, P.J.; Murri, M.; Tinahones, F.J.; Andrés-Lacueva, C.; et al. Comparative study of microbial-derived phenolic metabolites in human feces after intake of gin, red wine, and dealcoholized red wine. J. Agric. Food Chem. 2013, 61, 3909–3915. [Google Scholar] [CrossRef] [PubMed]
- Stojančević, M.; Bojić, G.; Salami, H.A.; Mikov, M. The Influence of Intestinal Tract and Probiotics on the Fate of Orally Administered Drugs. Curr. Issues Mol. Biol. 2014, 16, 55–68. [Google Scholar] [PubMed]
- Michlmayr, H.; Kneifel, W. β-Glucosidase activities of lactic acid bacteria: Mechanisms, impact on fermented food and human health. Fems Microbiol. Lett. 2014, 352, 1–10. [Google Scholar] [CrossRef]
- Winter, J.; Moore, L.H.; Dowell, V.R.; Bokkenheuser, V.D. C-ring cleavage of flavonoids by human intestinal bacteria. Appl. Environ. Microbiol. 1989, 55, 1203–1208. [Google Scholar]
- Jin, J.S.; Hattori, M. Isolation and characterization of a human intestinal bacterium Eggerthella sp. CAT– capable of cleaving the C-ring of (+)-catechin and (-)epicatechin, followed by p-dehydroxylation of the B-ring. Biol. Pharm. Bull. 2012, 35, 2252–2256. [Google Scholar] [CrossRef]
- Braune, A.; Blaut, M. Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes 2016, 7, 216–234. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Smith, C.C.R.; Snowberg, L.K.; Gregory Caporaso, J.; Knight, R.; Bolnick, D.I. Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. Isme J. 2015, 9, 2515–2526. [Google Scholar] [CrossRef]
- Yu, K.U.; Jang, I.S.; Kang, K.H.; Sung, C.K.; Kim, D.H. Metabolism of saikosaponin c and naringin by human intestinal bacteria. Arch. Pharmacal Res. 1997, 20, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Day, A.J.; DuPont, M.S.; Ridley, S.; Rhodes, M.; Rhodes, M.J.C.; Morgan, M.R.A.; Williamson, G. Deglycosylation of flavonoid and isoflavonoid glycosides by human small intestine and liver β-glucosidase activity. Febs Lett. 1998, 436, 71–75. [Google Scholar] [CrossRef]
- Rechner, A. Colonic metabolism of dietary polyphenols: Influence of structure on microbial fermentation products. Free Radic. Biol. Med. 2004, 36, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sakurai, T.; Chen, X.; Sun, H.; Wang, Z.; Sun, Q.; Sun, W.; Cao, H. Hydrolysis of flavanone glycosides and degradation of the corresponding aglycones from dried immature Citrus fruit by human fecal flora in vitro. Planta Med. 2008, 74, 1751–1755. [Google Scholar] [CrossRef]
- Pereira-Caro, G.; Borges, G.; Ky, I.; Ribas, A.; Calani, L.; Del Rio, D.; Clifford, M.N.; Roberts, S.A.; Crozier, A. In vitro colonic catabolism of orange juice (poly)phenols. Mol. Nutr. Food Res. 2015, 59, 465–475. [Google Scholar] [CrossRef]
- Pereira-Caro, G.; Fernández-Quirós, B.; Ludwig, I.A.; Pradas, I.; Crozier, A.; Moreno-Rojas, J.M. Catabolism of citrus flavanones by the probiotics Bifidobacterium longum and Lactobacillus rhamnosus. Eur. J. Nutr. 2018, 57, 231–242. [Google Scholar] [CrossRef]
- Konieczka, P.; Namieśnik, J. Estimating uncertainty in analytical procedures based on chromatographic techniques. J. Chromatogr. A 2010, 1217, 882–891. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |





| Compound | Retention Time (Min) | MRM Transition | Dwell Time (ms) | Fragmentor Voltage (V) | Collision Energy (V) |
|---|---|---|---|---|---|
| D4-NG | 1.6 | 583.2→275.1 | 250 | 215 | 34 |
| D4-NE | 2.3 | 275.0→151.0 | 250 | 125 | 12 |
| D4-HPPA | 1.7 | 169.1→125.0 | 250 | 70 | 7 |
| D5-BA | 2.2 | 126.0→82.1 | 250 | 75 | 8 |
| Compound | Concentration (ng mL−1) | Intra-Day (n = 6) | Inter-Day (n = 18) | Recovery (%, n = 6) | ||
|---|---|---|---|---|---|---|
| RSD (%) | RE (%) | RSD (%) | RE (%) | Mean ± SD | ||
| D4-NG | 10 | 0.91 | 7.98 | 1.58 | 6.73 | 57.60 ± 1.34 |
| 20 | 3.00 | −2.92 | 3.25 | −6.25 | 57.78 ± 0.80 | |
| 200 | 3.58 | 6.08 | 8.20 | −4.04 | 56.22 ± 0.89 | |
| 1500 | 2.24 | 4.98 | 3.55 | 9.81 | 58.02 ± 1.43 | |
| D4-NE | 5 | 2.51 | −2.47 | 4.16 | 0.10 | 53.97 ± 1.56 |
| 10 | 3.49 | 4.54 | 4.38 | −5.15 | 54.69 ± 1.78 | |
| 100 | 1.63 | 2.50 | 6.41 | −4.78 | 54.97 ± 0.85 | |
| 750 | 1.25 | −1.46 | 5.57 | −0.25 | 58.50 ± 1.63 | |
| D4-HPPA | 2.5 | 9.90 | 5.51 | 11.39 | −2.44 | 49.53 ± 5.23 |
| 7.5 | 4.75 | 8.73 | 7.40 | 3.71 | 59.16 ± 0.65 | |
| 50 | 5.91 | 8.69 | 4.46 | 8.30 | 61.71 ± 0.91 | |
| 375 | 1.45 | −9.42 | 2.24 | −8.90 | 70.69 ± 0.83 | |
| Matrix Resource | D4-NG Concentration (ng mL−1) | D4-NE Concentration (ng mL−1) | D4-HPPA Concentration (ng mL−1) | |||
|---|---|---|---|---|---|---|
| 20 | 1500 | 10 | 750 | 7.5 | 375 | |
| 1 | 100.8 ± 3.77 | 103.8 ± 8.60 | 99.41 ± 3.24 | 104.0 ± 9.11 | 99.86 ± 1.35 | 98.12 ± 7.43 |
| 2 | 96.53 ± 4.76 | 101.9 ± 5.90 | 98.44 ± 4.99 | 103.7 ± 5.60 | 99.27 ± 6.15 | 104.2 ± 4.69 |
| 3 | 98.28 ± 1.71 | 103.5 ± 3.41 | 100.3 ± 2.30 | 102.4 ± 3.88 | 100.4 ± 2.50 | 104.3 ± 3.26 |
| 4 | 100.8 ± 1.18 | 106.6 ± 3.92 | 100.6 ± 3.83 | 107.1 ± 3.57 | 101.1 ± 1.20 | 100.7 ± 3.23 |
| 5 | 94.73 ± 8.36 | 101.8 ± 4.52 | 97.50 ± 5.19 | 102.9 ± 6.48 | 98.25 ± 5.53 | 105.0 ± 4.29 |
| 6 | 96.42 ± 4.04 | 104.5 ± 7.40 | 96.33 ± 3.42 | 105.8 ± 11.19 | 100.4 ± 2.02 | 103.6 ± 2.51 |
| RSD (%) | 2.66 | 1.74 | 1.70 | 1.70 | 1.09 | 2.66 |
| Compound | Concentration (ng mL−1) | Freeze and Thaw (3 Cycles) | Short-Term (24 h at 25 °C) | Long-Term (3 Months at −80 °C) |
|---|---|---|---|---|
| D4-NG | 20 | 97.82 ± 1.28 | 103.5 ± 0.67 | 107.3 ± 2.51 |
| 1500 | 99.36 ± 2.62 | 106.6 ± 1.96 | 96.54 ± 3.16 | |
| D4-NE | 10 | 96.17 ± 1.33 | 99.88 ± 1.24 | 104.8 ± 1.86 |
| 750 | 102.6 ± 0.29 | 96.38 ± 2.33 | 101.3 ± 2.33 | |
| D4-HPPA | 7.5 | 106.4 ± 1.37 | 105.7 ± 1.52 | 106.5 ± 3.12 |
| 375 | 97.85 ± 2.54 | 106.8 ± 3.01 | 94.84 ± 2.86 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.; Wu, H.; He, Y.; Pan, W.; Yan, Z.; Liao, Y.; Peng, W.; Gan, L.; Zhang, Y.; Su, W.; et al. Simultaneously Quantitative Analysis of Naringin and Its Major Human Gut Microbial Metabolites Naringenin and 3-(4′-Hydroxyphenyl) Propanoic Acid via Stable Isotope Deuterium-Labeling Coupled with RRLC-MS/MS Method. Molecules 2019, 24, 4287. https://doi.org/10.3390/molecules24234287
Chen T, Wu H, He Y, Pan W, Yan Z, Liao Y, Peng W, Gan L, Zhang Y, Su W, et al. Simultaneously Quantitative Analysis of Naringin and Its Major Human Gut Microbial Metabolites Naringenin and 3-(4′-Hydroxyphenyl) Propanoic Acid via Stable Isotope Deuterium-Labeling Coupled with RRLC-MS/MS Method. Molecules. 2019; 24(23):4287. https://doi.org/10.3390/molecules24234287
Chicago/Turabian StyleChen, Taobin, Hao Wu, Yan He, Wenjun Pan, Zenghao Yan, Yan Liao, Wei Peng, Li Gan, Yaohui Zhang, Weiwei Su, and et al. 2019. "Simultaneously Quantitative Analysis of Naringin and Its Major Human Gut Microbial Metabolites Naringenin and 3-(4′-Hydroxyphenyl) Propanoic Acid via Stable Isotope Deuterium-Labeling Coupled with RRLC-MS/MS Method" Molecules 24, no. 23: 4287. https://doi.org/10.3390/molecules24234287
APA StyleChen, T., Wu, H., He, Y., Pan, W., Yan, Z., Liao, Y., Peng, W., Gan, L., Zhang, Y., Su, W., & Yao, H. (2019). Simultaneously Quantitative Analysis of Naringin and Its Major Human Gut Microbial Metabolites Naringenin and 3-(4′-Hydroxyphenyl) Propanoic Acid via Stable Isotope Deuterium-Labeling Coupled with RRLC-MS/MS Method. Molecules, 24(23), 4287. https://doi.org/10.3390/molecules24234287






