Anthra[1,2-d][1,2,3]triazine-4,7,12(3H)-triones as a New Class of Antistaphylococcal Agents: Synthesis and Biological Evaluation
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Studies
2.2.1. Determination of MIC Values of Synthesized Agents against Reference Strains of Bacteria and Yeasts and Clinical Isolates of S. aureus
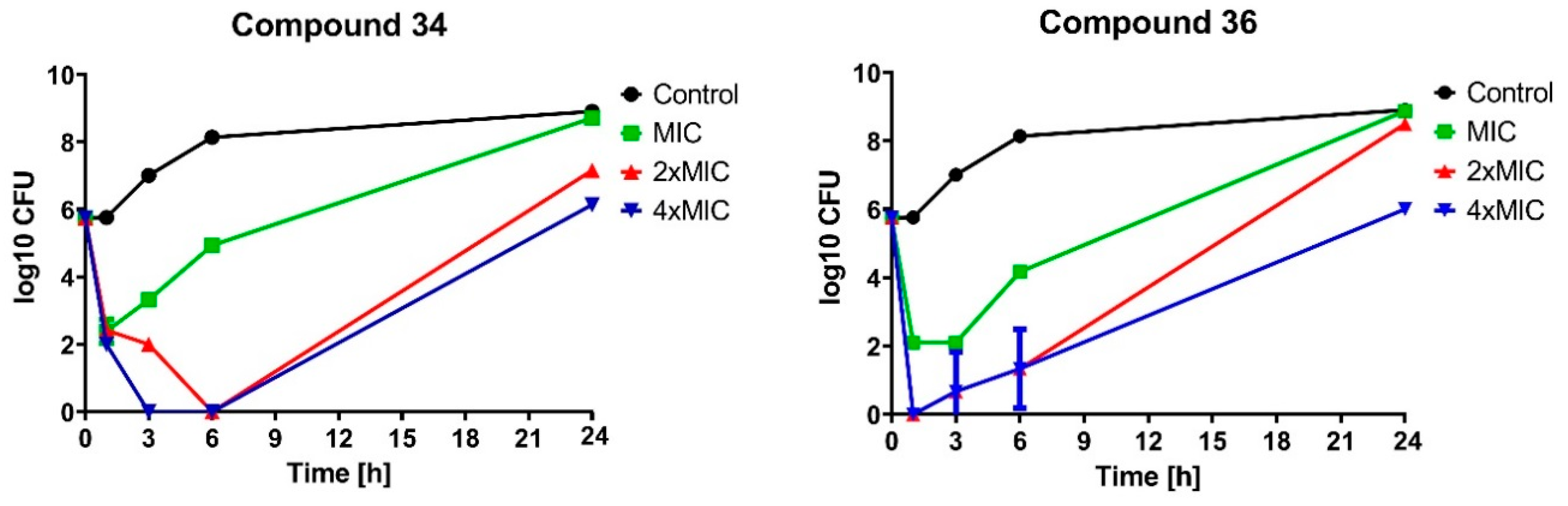
2.2.2. Bactericidal Effect of Selected Compounds
2.2.3. Activity of the Selected Compounds against Staphylococcal Biofilm
2.2.4. Influence of the Selected Compounds on Metabolic Activity
3. Materials and Methods
3.1. General Information
3.2. Chemistry
3.2.1. General Procedure for the Synthesis of Amides 6–13, 15–21
3.2.2. General Procedure for the Synthesis of Anthra[1,2-d][1,2,3]triazine-4,7,12(3H)-triones 22–38
3.3. Biology
3.3.1. Bacterial Strains and Media
3.3.2. Investigation of Antimicrobial Potential of Synthesized Agents
3.3.3. Time-Kill Assay
3.3.4. Biofilm Formation and Determination of Antibiofilm Activity
3.3.5. Influence of Investigated Agents on Enzymatic Activity of S. aureus
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hutchings, M.; Truman, A.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Frieri, M.; Kumar, K.; Boutin, A. Antibiotic resistance. J. Infect. Public Health 2017, 10, 369–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pristov, K.E.; Ghannoum, M.A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 2019, 25, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G.J. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiggli, B.J.; Frei, R.; Laffer, R.; Tschudin Sutter, S.; Widmer, A.F. Survival from methicillin-sensitive Staphylococcus aureus bloodstream infections over 20 years: A cohort of 1328 patients. Swiss Med. Wkly. 2017, 147, w14508. [Google Scholar] [CrossRef]
- Szweda, P.; Schielmann, M.; Kotlowski, R.; Gorczyca, G.; Zalewska, M.; Milewski, S. Peptidoglycan hydrolases-potential weapons against Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2012, 96, 1157–1174. [Google Scholar] [CrossRef] [Green Version]
- Grecka, K.; Kuś, P.M.; Okińczyc, P.; Worobo, R.W.; Walkusz, J.; Szweda, P. The anti-staphylococcal potential of ethanolic polish propolis extracts. Molecules 2019, 24, 1732. [Google Scholar] [CrossRef] [Green Version]
- Ansari, S.; Jha, R.K.; Mishra, S.K.; Tiwari, B.R.; Asaad, A.M. Recent advances in Staphylococcus aureus infection: Focus on vaccine development. Infect. Drug Resist. 2019, 12, 1243. [Google Scholar] [CrossRef] [Green Version]
- Diekema, D.J.; Pfaller, M.A.; Shortridge, D.; Zervos, M.; Jones, R.N. Twenty-Year trends in antimicrobial susceptibilities among Staphylococcus aureus from the SENTRY antimicrobial surveillance program. Open Forum Infect. Dis. 2019, 6 (Suppl. 1), S47–S53. [Google Scholar] [CrossRef]
- Kang, J.; Dietz, M.J.; Hughes, K.; Xing, M.; Li, B. Silver nanoparticles present high intracellular and extracellular killing against Staphylococcus aureus. J. Antimicrob. Chemother. 2019, 74, 1578–1585. [Google Scholar] [CrossRef]
- Mathur, H.; Field, D.; Rea, M.C.; Cotter, P.D.; Hil, C.; Ross, R.P. Fighting biofilms with lantibiotics and other groups of bacteriocins. npj Biofilms Microbiomes 2018, 4, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehman, S.M.; Mearns, G.; Rankin, D.; Cole, R.A.; Smrekar, F.; Branston, S.D.; Morales, S. Design and preclinical development of a phage product for the treatment of antibiotic-resistant Staphylococcus aureus Infections. Viruses 2019, 11, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauhan, D.; Hati, S.; Priyadarshini, R.; Sen, S. Transcriptome analysis predicts mode of action of benzimidazole molecules against Staphylococcus aureus UAMS-1. Drug. Dev. Res. 2019, 80, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Subedi, Y.P.; Alfindee, M.N.; Shrestha, J.P.; Chang, C.-W.T. Tuning the biological activity of cationic anthraquinone analogues specifically toward Staphylococcus aureus. Eur. J. Med. Chem. 2018, 157, 683–690. [Google Scholar] [CrossRef]
- Zvarych, V.I.; Stasevych, M.V.; Stanko, O.V.; Komarovskaya-Porokhnyavets, E.Z.; Poroikov, V.V.; Rudik, A.V.; Lagunin, A.A.; Vovk, M.V.; Novikov, V.P. Computerized prediction, synthesis, and antimicrobial activity of new amino-acid derivatives of 2-Chloro-N-(9,10-Dioxo-9,10-Dihydroanthracen-1-yl) acetamide. Pharm. Chem. J. 2014, 48, 584–588. [Google Scholar] [CrossRef]
- Stasevych, M.; Zvarych, V.; Lunin, V.; Halenova, T.; Savchuk, O.; Dudchak, O.; Vovk, M.; Novikov, V. Novel anthraquinone-based derivatives as potent inhibitors for receptor tyrosine kinases. Indian J. Pharm. Sci. 2015, 77, 634–637. [Google Scholar] [CrossRef] [Green Version]
- Stasevych, M.; Zvarych, V.; Lunin, V.; Kopak, N.; Komarovska-Porokhnyavets, O.; Deniz, N.G.; Sayil, C.; Ozyurek, M.; Guclu, K.; Vovk, M.; et al. Synthesis, investigation of antimicrobial and antioxidant activity of anthraquinonylhydrazones. Monatsh. Chem. 2018, 149, 1111–1119. [Google Scholar] [CrossRef]
- Kumar, R.; Singh, A.D.; Singh, J.; Singh, H.; Roy, R.K.; Chaudhary, A. 1,2,3-Triazine scaffold as a potent biologically active moiety: A mini review. Mini Rev. Med. Chem. 2014, 14, 72–83. [Google Scholar] [CrossRef]
- Kumar, R.; Kumar, N.; Roy, R.K.; Singh, A. Triazines—A comprehensive review of their synthesis and diverse biological importance. Curr. Med. Drug Res. 2017, 1, 173. [Google Scholar]
- Kumar, K.S.; Adepu, R.; Sandra, S.; Rambabu, D.; Krishna, G.R.; Reddy, C.M.; Misra, P.; Pal, M. Cu-mediated N-arylation of 1,2,3-triazin-4-ones: Synthesis of fused triazinone derivatives as potential inhibitors of chorismate mutase. Bioorganic. Med. Chem. Lett. 2012, 22, 1146–1150. [Google Scholar] [CrossRef]
- Cascioferro, S.; Parrino, B.; Spanò, V.; Carbone, A.; Montalbano, A.; Barraja, P.; Diana, P.; Cirrincione, G. Synthesis and antitumor activities of 1,2,3-triazines and their benzo- and heterofused derivatives. Eur. J. Med. Chem. 2017, 142, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Hamama, W.S.; El-Bana, G.G.; Shaaban, S.; Habib, O.M.O.; Zoorob, H.H. Advances in the domain of 4-amino-3-mercapto-1,2,4-triazine-5-ones. RSC Adv. 2016, 6, 24010–24049. [Google Scholar] [CrossRef]
- Srinivasa Rao, D.; Pavan Kumar, G.V.; Pooja, B.; Harika, G.; Anil Kumar, Y.; Sadasiva Rao, G. An extensive review on 1,2,3 and 1,2,4-triazines scaffold-valuable lead molecules with potent and diverse pharmacological activities. Der Chem. Sin. 2016, 7, 101–130. [Google Scholar]
- Khalid, Z.; Ahmad, H.A.; Munawar, M.A.; Khan, M.; Gul, S. 1,2,3-Benzotriazin-4(3H)-ones: Synthesis, reactions and applications. Heterocycles 2017, 94, 3–54. [Google Scholar] [CrossRef]
- Stasevych, M.; Zvarych, V.; Lunin, V.; Deniz, N.G.; Gokmen, Z.; Akgun, O.; Ulukaya, E.; Poroikov, V.; Gloriozova, T.; Novikov, V. Computer-aided prediction and cytotoxicity evaluation of some dithiocarbamates of 9,10-anthracenedione as new anticancer agents. SAR QSAR Environ. Res. 2017, 28, 355–366. [Google Scholar] [CrossRef]
- Stasevych, M.V.; Zvarych, V.I.; Lunin, V.V.; Khomyak, S.V.; Vovk, M.V.; Novikov, V.P. Synthesis of pyrazole and tetrazole derivatives of 9,10-anthraquinonylhydrazones. Chem. Heterocycl. Compd. 2017, 53, 942–944. [Google Scholar] [CrossRef]
- Stasevych, M.V.; Zvarych, V.I.; Stan’ko, O.V.; Vovk, M.V.; Novikov, V.P. Synthesis of 2-(N-benzoylimino)-N-(9,10-dioxo-9,10-dihydroanthracen-1-yl) thiazoles. Chem. Heterocycl. Compd. 2014, 49, 1831–1833. [Google Scholar] [CrossRef]
- Zvarych, V.I.; Stasevych, M.V.; Stan’ko, O.V.; Musyanovych, R.Y.; Novikov, V.P. Amino acid derivatives of 2-chloro-N-(9,10-dioxy-9,10-dihydroanthracen-1-yl) acetamide. Rus. J. Org. Chem. 2014, 50, 306–307. [Google Scholar] [CrossRef]
- Zvarych, V.I.; Stasevych, M.V.; Lunin, V.V.; Vovk, M.V.; Novikov, V.P. Synthesis of 9,10-Anthracenedione Diethyldithiocarbamates. Rus. J. Gen. Chem. 2016, 86, 2699–2701. [Google Scholar] [CrossRef]
- Stasevych, M.; Zvarych, V.; Musyanovych, R.; Novikov, V.; Vovk, M. Synthesis of N-benzoyl-N’-(9,10-dioxo-9,10-dihydroanthracene-1-yl)-thioureas and quantum-chemical analysis of the reaction passing. Chem. Chem. Technol. 2014, 8, 135–140. [Google Scholar] [CrossRef]
- Rubin, J.E.; Ball, K.R.; Chirino-Trejo, M. Antimicrobial susceptibility of Staphylococcus aureus and Staphylococcus pseudintermedius isolated from various animals. Can. Vet. J. 2011, 52, 153–157. [Google Scholar] [PubMed]
- Atshan, S.S.; Nor Shamsudin, M.; Lung, L.T.; Sekawi, Z.; Pei Pei, C.; Karunanidhi, A.; Jeevajothi Nathan, J.; Mateg Ali, A.; Ghaznavi-Rad, E.; Abduljaleel, S.A.; et al. Genotypically different clones of Staphylococcus aureus are diverse in the antimicrobial susceptibility patterns and biofilm formations. Biomed Res. Int. 2013, 2013, 515712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bondock, S.; Rabie, R.; Etman, H.A.; Fadda, A.A. Synthesis and antimicrobial activity of some new heterocycles incorporating antipyrine moiety. Eur J. Med. Chem. 2008, 43, 2122–2129. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, J.; Mohan, S.; Roy, J.J. Synthesis of some 3-substituted amino-4,5-tetramethylene thieno[2,3-d] [1,2,3]-triazin-4 (3H)-ones as potential antimicrobial agents. Eur J. Med. Chem. 2010, 45, 4365–4369. [Google Scholar] [CrossRef] [PubMed]
- El-Gohary, N.S.; Hawas, S.S.; Gabr, M.T.; Shaaban, M.I.; El-Ashmawy, M.B. New series of fused pyrazolopyridines: Synthesis, molecular modeling, antimicrobial, antiquorum-sensing and antitumor activities. Bioorganic Chem. 2019, 92, 103109. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Long, S.; Rakesh, K.P.; Zha, G.F. Structure-activity relationships (SAR) of triazine derivatives: Promising antimicrobial agents. Eur. J. Med. Chem. 2019, 23, 111804. [Google Scholar] [CrossRef] [PubMed]
- Gebreyohannes, G.; Nyerere, A.; Bii, C.; Sbhatu, D.B. Challenges of intervention, treatment, and antibiotic resistance of biofilm-forming microorganisms. Heliyon 2019, 5, e02192. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Armarego, W.L.F.; Chai, C. Purification of Laboratory Chemicals, 5th ed.; Elsevier: Oxford, UK, 2003. [Google Scholar]
- Hargreaves, T.; Eyles, H.G.; Peters, A.T. New intermediates and dyes. Part XI. Novel nucleophilic substitution and group elimination in the 2-carbamoylanthraquinone series. J. Chem. Soc. C Org. 1968, 19, 2431–2435. [Google Scholar] [CrossRef]
- Samoilova, A.A.; Gladkova, V.V. Compatibility of bicomponent mixtures of disperse dyes. Tekstil’naya Promyshlennost 1981, 8, 57–59. [Google Scholar]
- Clinical Laboratory Standard Institute. Performance Standards for Antimicrobial Susceptibility Testing; Seventeenth Informational Supplement; CLSI document M100–S17; CLSI: Wayne, PA, USA, 2007. [Google Scholar]
- Rex, J.H. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard—Third Edition; Clinical & Laboratory Standards Institute: Wayne, PA, USA, 2008; CLSI Document M27-A3. [Google Scholar]
- Walencka, E.; Sadowska, B.; Rozalska, S.; Hryniewicz, W.; Rozalska, B. Lysostaphin as a potential therapeutic agent for staphylococcal biofilm eradication. Pol. J. Microbiol. 2005, 54, 191–200. [Google Scholar] [PubMed]
- Kairo, S.K.; Bedwell, J.; Tyler, P.C.; Carter, A.; Corbel, M.J. Development of a tetrazolium salt assay for rapid determination of viability of BCG vaccines. Vaccine 1999, 17, 2423–2428. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 5–38 are available from the authors. |


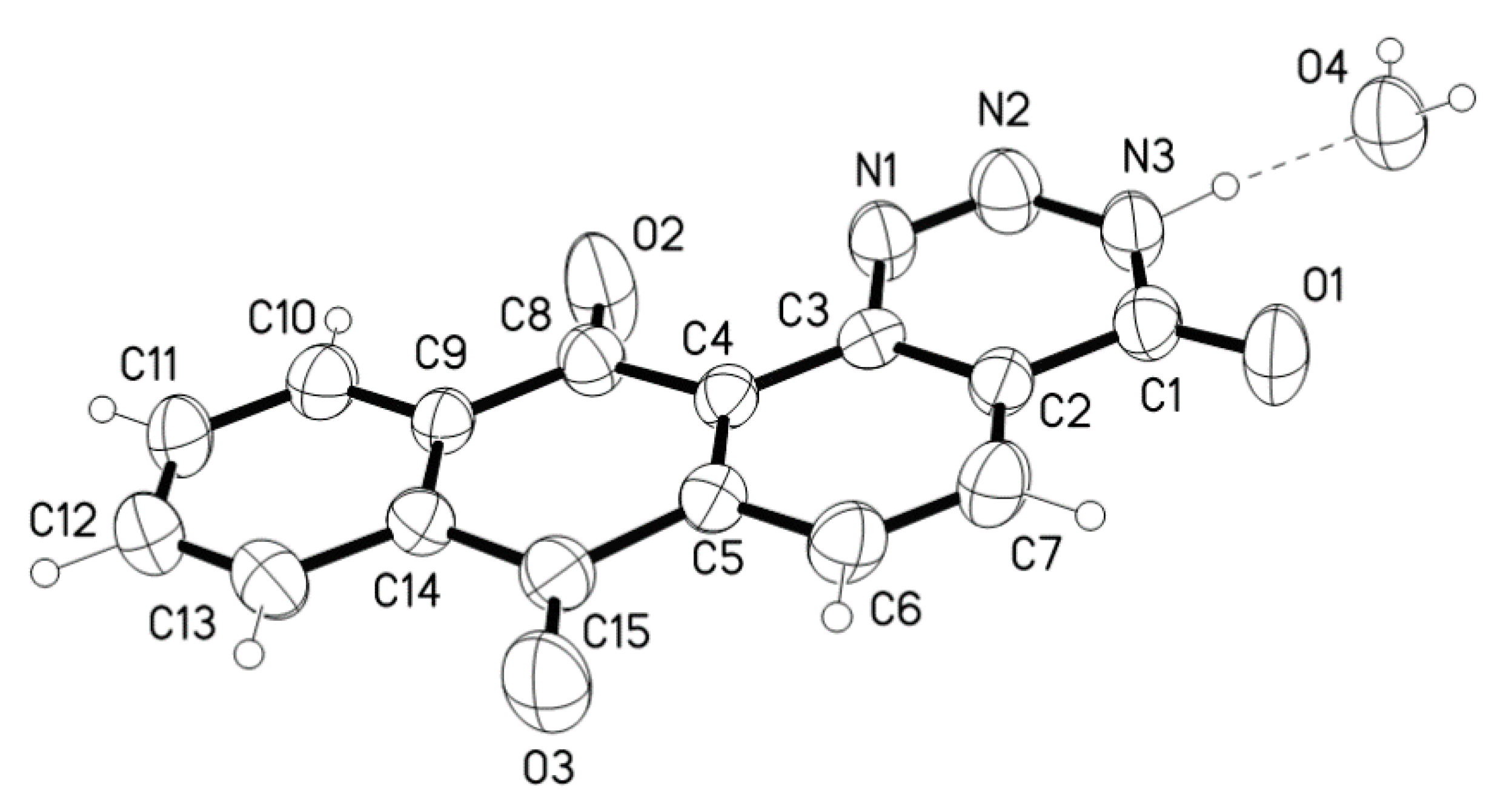


| D-H ···A | D-H(Å) | D···A(Å) | D-H-A(°) |
|---|---|---|---|
| N3-H3···O4 | 1.01(3) | 2.732(3) | 175 (3) |
| O4-H41··· O1_$1 * | 0.92(5) | 2.844(3) | 166 (4) |
| O4-H42 ···O2_$2 * | 0.91(5) | 2.867(4) | 142 (3) |
| O4-H42 ···N1_$2 * | 0.91(5) | 3.097(3) | 143 (3) |
| Agent | Pathogen | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| S. aureus ATCC 25923 | S. aureus ATCC 29213 | S. epidermidis ATCC 12228 | E. coli ATCC 25922 | P. aeruginosa ATCC 27853 | C. albicans ATCC 10231 | C. albicans SC 5314 | C. glabrata DSM11226 | C. krusei DSM6128 | |
| 22 | 4.0 | 4.0 | 8.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 23 | 2.0 | 2.0 | 1.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 24 | 0.5 | 0.5 | 0.5 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | 32.0 |
| 25 | 4.0 | 8.0 | 8.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 26 | 2.0 | 4.0 | 2.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 27 | 4.0 | 8.0 | 8.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 28 | 1.0 | 2.0 | 2.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 29 | 2.0 | 16.0 | 2.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 30 | 2.0 | 2.0 | 2.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 31 | 8.0 | 4.0 | 8.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 32 | 0.25 | 0.25 | 0.125 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 33 | 0.25 | 0.25 | 0.25 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 34 | 0.25 | 0.5 | 0.25 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 35 | 4.0 | 8.0 | 8.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 36 | 0.25 | 0.5 | 0.125 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 37 | 0.5 | 1.0 | 0.125 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 |
| 38 | 1.0 | 2.0 | 2.0 | >32.0 | >32.0 | >32.0 | >32.0 | >32.0 | 32.0 |
| Ampicillin | 0.25 | 8.0 | 16 | >32.0 | >32.0 | ND | ND | ND | ND |
| Gentamicin | 0.125 | 0.5 | 0.5 | 2.0 | 2.0 | ND | ND | ND | ND |
| Fusidic acid | 0.5 | 0.25 | 0.125 | >32.0 | >32.0 | ND | ND | ND | ND |
| Linezolid | 2.0 | 1.0 | 1.0 | >32.0 | >32.0 | ND | ND | ND | ND |
| Daptomycin | 1.0 | 1.0 | 2.0 | >32.0 | >32.0 | ND | ND | ND | ND |
| Oxacillin | 0.25 | 0.125 | 0.125 | >32.0 | >32.0 | ND | ND | ND | ND |
| Levofloxacin | 0.125 | 0.25 | 0.125 | >32.0 | >32.0 | ND | ND | ND | ND |
| Agent | Pathogens—Clinical Isolates of S. aureus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | MRSA1 | MRSA2 | MRSA3 | MRSA4 | |
| 23 | 4.0 | 8.0 | 4.0 | 8.0 | 8.0 | 8.0 | 8.0 | 8.0 | 8.0 | 2.0 | 8.0 | 8.0 |
| 24 | 1.0 | 1.0 | 0.5 | 0.25 | 0.25 | 1.0 | 0.5 | 0.5 | 1.0 | 1.0 | 0.5 | 0.5 |
| 26 | 2.0 | 2.0 | 1.0 | 1.0 | 1.0 | 4.0 | 1.0 | 2.0 | 4.0 | 2.0 | 1.0 | 2.0 |
| 32 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.5 | 0.25 | 0.125 | 0.25 | 0.25 | 0.25 | 0.25 |
| 33 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.25 | 0.5 |
| 34 | 0.25 | 0.5 | 0.25 | 0.25 | 0.5 | 0.25 | 0.25 | 0.25 | 0.5 | 0.125 | 0.25 | 0.25 |
| 36 | 0.25 | 0.5 | 0.25 | 0.5 | 1.0 | 0.5 | 0.25 | 0.5 | 0.5 | 1.0 | 0.25 | 0.5 |
| 37 | 0.5 | 0.5 | 0.5 | 1.0 | 1.0 | 0.5 | 1.0 | 0.5 | 1.0 | 1.0 | 1.0 | 2.0 |
| Compound | Enzyme | ||||
|---|---|---|---|---|---|
| Alkaline Phosphatase | Esterase (C4) | Esterase Lipase (C8) | Acid Phosphatase | Naphthol-AS-BI-Phosphohydrolase | |
| Control | 5 | 5 | 5 | 5 | 5 |
| 32 | 4 | 5 | 4 | 5 | 5 |
| 33 | 3 | 4 | 4 | 5 | 5 |
| 34 | 3 | 3 | 4 | 5 | 5 |
| 36 | 4 | 2 | 4 | 5 | 5 |
| No. | Code of the Strain | Material | Ward of Hospital | Antibiogram—Susceptibility Profile 1 |
|---|---|---|---|---|
| 1 | 4471313 | Nasal swab | Intensive care | Pen.—R, Met.—S, Clin.—S, Ery.—S |
| 2 | 4475564 | Nasal swab | Internal | Pen.—R, Met.—S, Clin.—R, Ery.—R |
| 3 | 4476206 | Sputum | Internal | Pen.—R, Met.—S, Clin.—R, Ery.—R |
| 4 | 4475131 | Pus | Internal | Pen.—R, Met.—S, Clin.– R, Ery.—R |
| 5 | 4466686 | Sputum | Surgical | Pen.—R, Met.—S, Clin.—R, Ery.—R |
| 6 | 4466380 | Wound | Surgical | Pen.—R, Met.—S, Clin.—S, Ery.—S |
| 7 | 4466896 | Nasal swab | Internal | Pen.—R, Met.—S, Clin.—S, Ery.—S |
| 8 | 4468792 | Pharyngeal swab | Pediatrics | Pen.—R, Met.—S, Clin.—S, Ery.—S |
| 9-MRSA | 9572250 | Wound | Internal | Pen.—R, Met.—R, Clin.—R, Ery.—R |
| 10-MRSA | 8007171 | Wound | Laryngology | Pen.—R, Met.—R, Clin.—R, Ery.—R |
| 11-MRSA | 45300223 | Blood | Pediatrics | Pen.—R, Met—R, Clin.—R, Ery.—R |
| 12-MRSA | 9935169 | Wound | Dispensary | Pen.—R, Met.—R, Clin.—R, Ery.—R |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zvarych, V.; Stasevych, M.; Novikov, V.; Rusanov, E.; Vovk, M.; Szweda, P.; Grecka, K.; Milewski, S. Anthra[1,2-d][1,2,3]triazine-4,7,12(3H)-triones as a New Class of Antistaphylococcal Agents: Synthesis and Biological Evaluation. Molecules 2019, 24, 4581. https://doi.org/10.3390/molecules24244581
Zvarych V, Stasevych M, Novikov V, Rusanov E, Vovk M, Szweda P, Grecka K, Milewski S. Anthra[1,2-d][1,2,3]triazine-4,7,12(3H)-triones as a New Class of Antistaphylococcal Agents: Synthesis and Biological Evaluation. Molecules. 2019; 24(24):4581. https://doi.org/10.3390/molecules24244581
Chicago/Turabian StyleZvarych, Viktor, Maryna Stasevych, Volodymyr Novikov, Eduard Rusanov, Mykhailo Vovk, Piotr Szweda, Katarzyna Grecka, and Slawomir Milewski. 2019. "Anthra[1,2-d][1,2,3]triazine-4,7,12(3H)-triones as a New Class of Antistaphylococcal Agents: Synthesis and Biological Evaluation" Molecules 24, no. 24: 4581. https://doi.org/10.3390/molecules24244581
APA StyleZvarych, V., Stasevych, M., Novikov, V., Rusanov, E., Vovk, M., Szweda, P., Grecka, K., & Milewski, S. (2019). Anthra[1,2-d][1,2,3]triazine-4,7,12(3H)-triones as a New Class of Antistaphylococcal Agents: Synthesis and Biological Evaluation. Molecules, 24(24), 4581. https://doi.org/10.3390/molecules24244581






