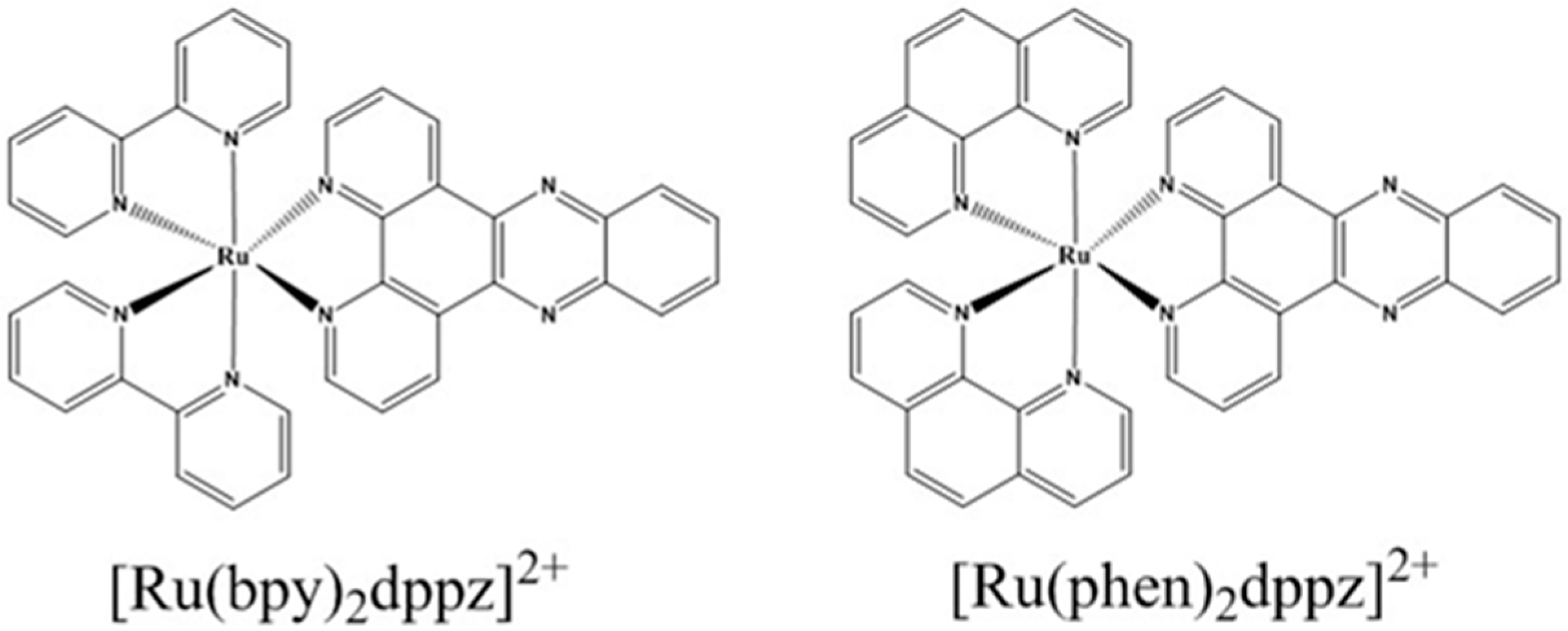
Recent Developments in the Interactions of Classic Intercalated Ruthenium Compounds: [Ru(bpy)2dppz]2+ and [Ru(phen)2dppz]2+ with a DNA Molecule
Abstract
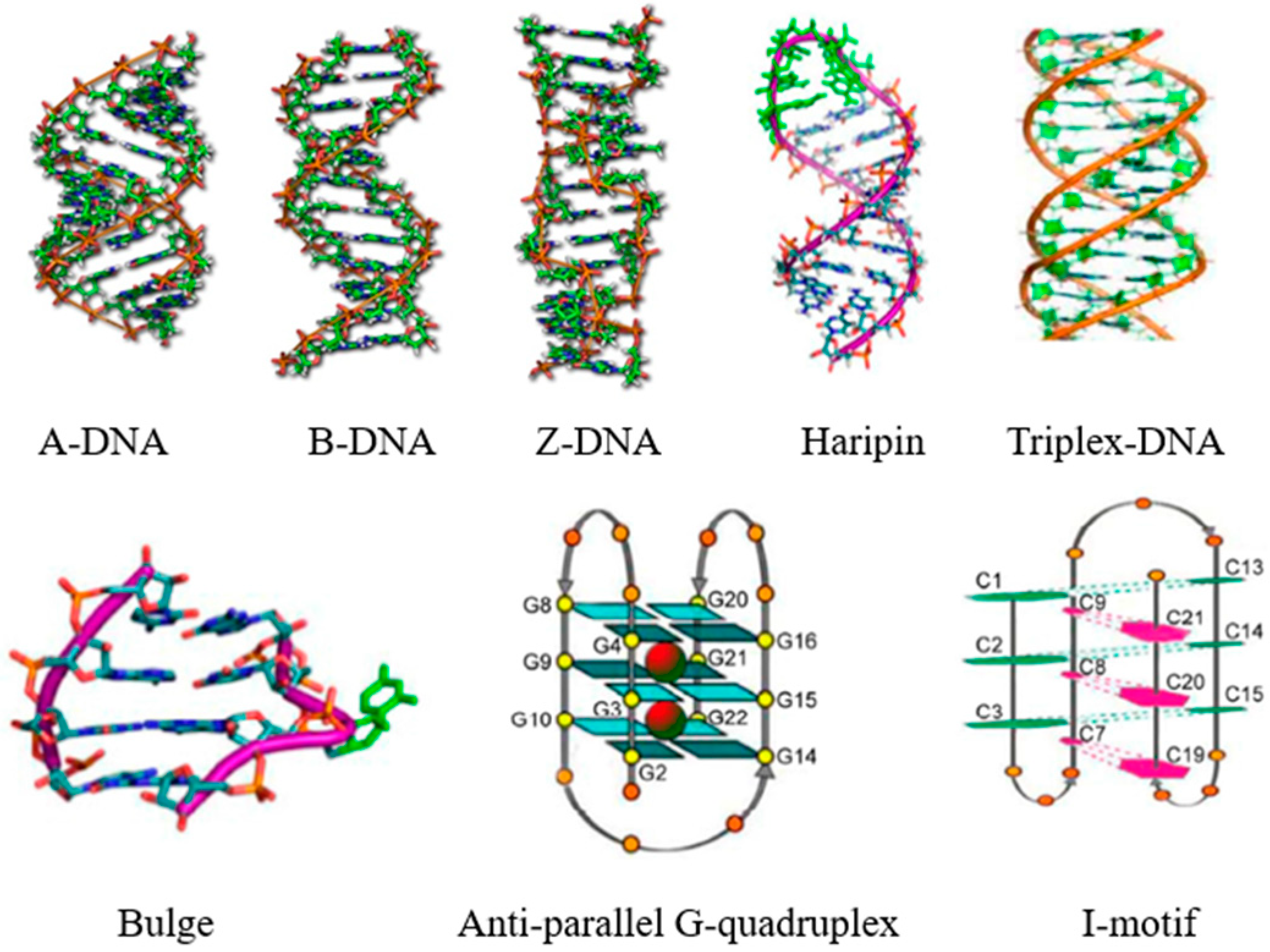
:1. Introduction
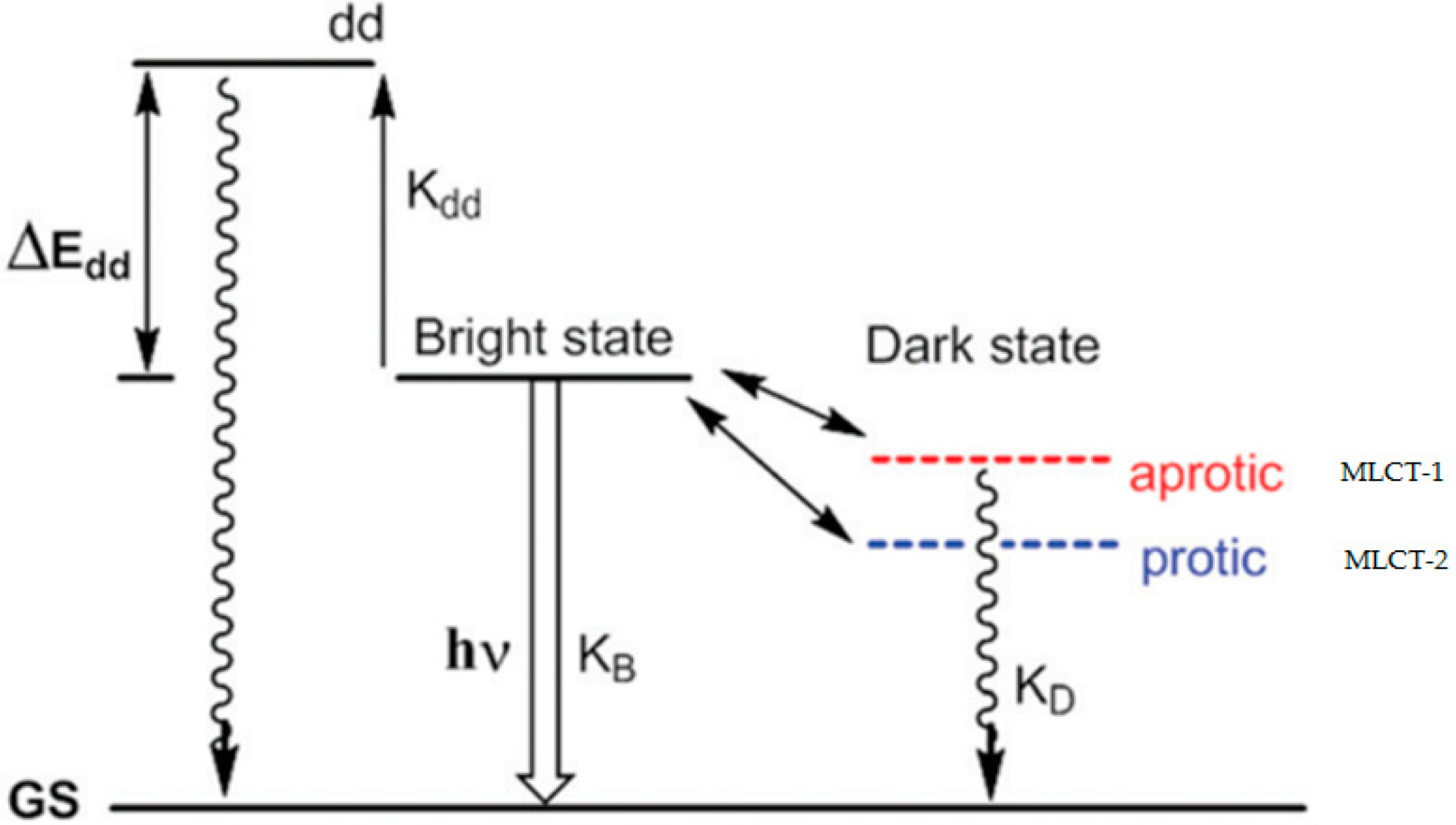
2. Mechanism of the Molecular Light Switch
3. Sensitive Luminescent Reporter of Nucleic Acid Structures
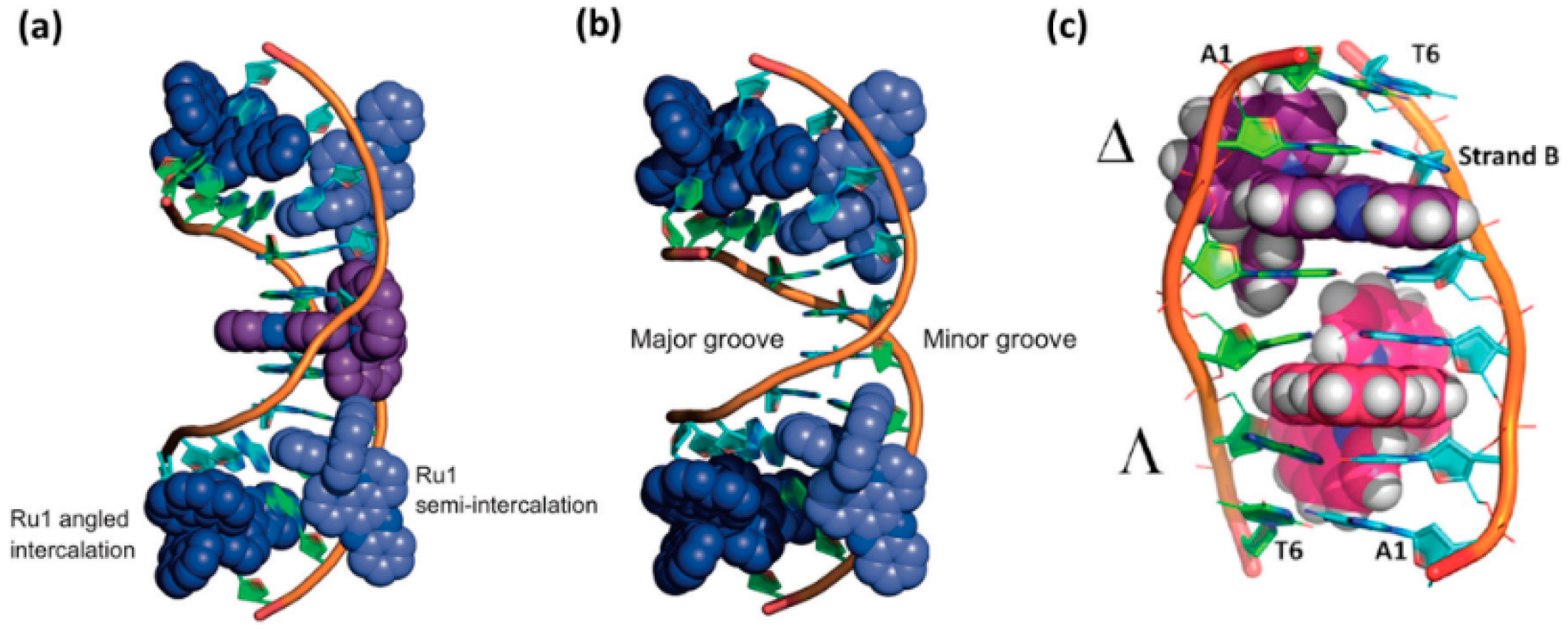
4. Binding Mode
5. Single Molecule Force Spectroscopy Study
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| dsDNA | Double strand DNA |
| bpy | Bipyridine |
| phen | Phenanthroline |
| dppz | Dipyridophenazine |
| SMFS | Single molecule force spectroscopy |
References
- Sherman, S.E.; Gibson, D.; Wang, A.H.; Lippard, S.J. X-ray structure of the major adduct of the anticancer drug cisplatin with DNA: Cis-[Pt(NH3)2(d(pGpG))]. Science 1985, 230, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Belmont, P.; Constant, J.F.; Demeunynck, M. Nucleic Acid Conformation Diversity: From Structure to Function and Regulation. Chem. Soc. Rev. 2001, 30, 70–81. [Google Scholar] [CrossRef]
- Martin, E. Nucleic acid crystallography: Current progress. Curr. Opin. Chem. Biol. 2004, 8, 580–591. [Google Scholar]
- Shi, S.; Yao, T.; Ji, L.N. DNA molecular light switch and biosensors based on ruthenium(II) polypyridyl complexes. Sci. China-Chem. 2014, 44, 460–470. [Google Scholar]
- Zeglis, B.M.; Pierre, V.C.; Barton, J.K. Metallo-intercalators and metallo-insertors. Chem. Commun. 2007, 44, 4565–4579. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.K.; Sadler, P.J. Metal complexes as DNA intercalators. Acc. Chem. Res. 2011, 44, 349–359. [Google Scholar] [CrossRef]
- Pages, B.J.; Ang, D.L.; Wright, E.P.; Aldrich-Wright, J.R. Metal complex interactions with DNA. Dalton Trans. 2015, 44, 3505–3526. [Google Scholar] [CrossRef]
- Keene, F.R.; Smith, J.A.; Collins, J.G. Metal complexes as structure-selective binding agents for nucleic acids. Coordin. Chem. Rev. 2009, 253, 2021–2035. [Google Scholar] [CrossRef]
- Gill, M.R.; Thomas, J.A. Ruthenium(II) polypyridyl complexes and DNA—From structural probes to cellular imaging and therapeutics. Chem. Soc. Rev. 2012, 41, 3179–3192. [Google Scholar] [CrossRef]
- Mari, C.; Pierroz, V.; Ferrari, S.; Gasser, G. Combination of Ru(ii) complexes and light: New frontiers in cancer therapy. Chem. Sci. 2015, 6, 2660–2686. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.E.; Barton, J.K.; Chambron, J.C.; Sauvage, J.P.; Turro, N.J. Molecular “Light Switch” for DNA: [Ru(bpy)2(dppz)]2+. J. Am. Chem. Soc. 1990, 112, 4960–4962. [Google Scholar] [CrossRef]
- Jenkins, Y.; Friedman, A.E.; Turro, N.J.; Barton, J.K. Characterization of dipyridophenazine complexes of ruthenium(II): The light switch effect as a function of nucleic acid sequence and conformation. Biochemistry 1992, 31, 10809–10816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, E.J.C.; Hu, D.; Hörmann, A.; Jonkman, A.M.; Arkin, M.R.; Stemp, E.D.A.; Barton, J.K.; Barbara, P.F. First observation of the key intermediate in the light-switch mechanism of [Ru(Phen)2dppz]2+. J. Am. Chem. Soc. 1997, 119, 11458–11467. [Google Scholar] [CrossRef]
- Hartshorn, R.M.; Barton, J.K. Novel dipyridophenazine complexes of ruthenium(II): Exploring luminescent reporters of DNA. J. Am. Chem. Soc. 1992, 114, 5919–5925. [Google Scholar] [CrossRef]
- Lim, M.H.; Song, H.; Olmon, E.D.; Dervan, E.E.; Barton, J.K. Sensitivity of [Ru(bpy)2dppz]2+ luminescence to DNA defects. Inorg. Chem. 2009, 48, 5392–5397. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Geng, X.; Zhao, J.; Yao, T.; Wang, C.; Yang, D.; Zheng, L.; Ji, L. Interaction of [Ru(bpy)2(dppz)]2+ with human telomeric DNA: Preferential binding to G-quadruplexes over i-motif. Biochimie 2010, 92, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Turro, C.; Bossmann, S.H.; Jenkins, Y.; Barton, J.K.; Turro, N.J. Proton transfer quenching of the MLCT excited state of Ru(phen)2dppz2+ in homogeneous solution and bound to DNA. J. Am. Chem. Soc. 1995, 117, 9026–9032. [Google Scholar] [CrossRef]
- Boynton, A.N.; Marcélis, L.; Barton, J.K. [Ru(Me4phen)2dppz]2+, a light switch for DNA mismatches. J. Am. Chem. Soc. 2016, 138, 5020–5023. [Google Scholar] [CrossRef]
- Cadena, A.D.L.; Davydova, D.Y.; Tolstik, T.; Reichardt, C.; Shukla, S.; Akimov, D.; Heintzmann, R.; Popp, J.; Dietzek, B. Ultrafastin cellulophotoinduced dynamics processes of the paradigm molecular light switch [Ru(bpy)2dppz]2+. Sci. Rep. 2016, 6, 33547–33556. [Google Scholar] [CrossRef] [PubMed]
- Cadena, A.D.L.; Pascher, T.; Davydova, D.Y.; Akimov, D.; Herrmann, F.; Presselt, M.; Wächtler, M.; Dietzek, B. Intermolecular exciton–exciton annihilation in phospholipid vesicles doped with [Ru(bpy)2dppz]2+. Chem. Phys. Lett. 2016, 644, 56–61. [Google Scholar] [CrossRef]
- Li, G.; Sun, L.; Ji, L.; Chao, H. Ruthenium(ii) complexes with dppz: From molecular photoswitch to biological applications. Dalton Trans. 2016, 47, 13261–13276. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Walker, M.G.; Ramu, V.; Meijer, A.J.H.M.; Das, A. A ratiometric sensor for DNA based on a dual emission Ru(dppz) light-switch complex. Dalton Trans. 2017, 46, 6079–6086. [Google Scholar] [Green Version]
- Despax, S.; Jia, F.; Pfeffer, M.; Hébraud, P. Complexation of DNA with ruthenium organometallic compounds: The high complexation ratio limit. Phys. Chem. Chem. Phys. 2014, 16, 10491–10502. [Google Scholar] [CrossRef] [PubMed]
- Very, T.; Despax, S.; Hébraud, P.; Monari, A.; Assfeld, X. Spectral properties of polypyridyl ruthenium complexes intercalated in DNA: Theoretical insights into the surrounding effects of [Ru(dppz)(bpy)2]2+. Phys. Chem. Chem. Phys. 2012, 14, 12496–12504. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Kaiser, J.T.; Barton, J.K. Crystal structure of Δ-[Ru(bpy)2dppz]2+ bound to mismatched DNA reveals side-by-side metalloinsertion and intercalation. Nat. Chem. 2012, 4, 615–620. [Google Scholar] [CrossRef]
- Niyazi, H.; Hall, J.P.; O’Sullivan, K.; Winter, G.; Sorensen, T.; Kelly, J.M.; Cardin, C.J. Crystal structures of Λ-[Ru(phen)2dppz]2+ with oligonucleotides containing TA/TA and AT/AT steps show two intercalation modes. Nat. Chem. 2012, 4, 621–628. [Google Scholar] [CrossRef]
- Liao, G.; Chen, X.; Ji, L.; Chao, H. Visual specific luminescent probing of hybrid G-quadruplex DNA by a ruthenium polypyridyl complex. Chem. Commun. 2012, 48, 10781–10783. [Google Scholar] [CrossRef]
- Vladescu, I.D.; McCauley, M.J.; Nuñez, M.E.; Rouzina, I.; Williams, M.C. Quantifying force-dependent and zero-force DNA intercalation by single-molecule stretching. Nat. Methods 2007, 4, 517–522. [Google Scholar] [CrossRef]
- Brennaman, M.K.; Alstrum-Acevedo, J.H.; Fleming, C.N.; Paul, J.; Meyer, T.J.; Papanikolas, J.M. Turning the [Ru(bpy)2dppz]2+ light-switch on and off with temperature. J. Am. Chem. Soc. 2002, 124, 15094–15098. [Google Scholar] [CrossRef]
- Brennaman, M.K.; Meyer, T.J.; Papanikolas, J.M. [Ru(bpy)2dppz]2+ light-switch mechanism in protic solvents as studied through temperature-dependent lifetime measurements. J. Phys. Chem. A 2004, 108, 9938–9944. [Google Scholar] [CrossRef]
- Hu, L.; Bian, Z.; Li, H.; Han, S.; Yuan, Y.; Gao, L.; Xu, G. [Ru(bpy)2dppz]2+ electrochemiluminescence switch and its applications for DNA interaction study and label-free ATP aptasensor. Anal. Chem. 2009, 81, 9807–9811. [Google Scholar] [CrossRef] [PubMed]
- Marcelina, K.; Pascal, H.; Claude, S.; Christian, G.; Sebastien, H. DNA binding to an anticancer organo-ruthenium complex. J. Phys. Chem. B 2010, 114, 14041–14047. [Google Scholar]
- Sun, Y.; Collins, S.N.; Joyce, L.E.; Turro, C. Unusual photophysical properties of a ruthenium(II) Complex related to [Ru(bpy)2(dppz)]2+. Inorg. Chem. 2010, 49, 4257–4262. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Turro, C.; Friedman, L.A.; Barton, J.K.; Turro, N.J. Resonance raman investigation of Ru(phen)2(dppz)2+ and related complexes in water and in the presence of DNA. J. Phys. Chem. B 1997, 101, 6995–7000. [Google Scholar] [CrossRef]
- Coates, C.G.; Mcgarvey, J.J.; Callaghan, P.L.; Coletti, M.; Hamilton, J.G. Probing the interaction of [Ru(phen)2(dppz)]2+ with single-stranded DNA what degree of protection is required for operation of the “light-switch effect”? J. Phys. Chem. B 2001, 105, 730–735. [Google Scholar] [CrossRef]
- Olofsson, J.; Wilhelmsson, L.M.; Lincoln, P. Effects of methyl substitution on radiative and solvent quenching rate constants of [Ru(phen)2dppz]2+ in polyol solvents and bound to DNA. J. Am. Chem. Soc. 2004, 126, 15458–15465. [Google Scholar] [CrossRef]
- Pourtois, G.; Beljonne, D.; Moucheron, C.; Schumm, S.; Mesmaeker, A.K.D.M.; Lazzaroni, R.; Brédas, J. Photophysical properties of ruthenium(II) polyazaaromatic compounds: a theoretical insight. J. Am. Chem. Soc. 2004, 126, 683–692. [Google Scholar] [CrossRef]
- Fantacci, S.; De, A.F.; Sgamellotti, A.; Marrone, A.; Re, N. Photophysical properties of [Ru(phen)2(dppz)]2+ intercalated into DNA: An integrated Car-Parrinello and TDDFT study. J. Am. Chem. Soc. 2005, 127, 14144–14145. [Google Scholar] [CrossRef]
- Lutterman, D.A.; Chouai, A.; Liu, Y.; Sun, Y.; Stewart, C.D.; Dunbar, K.R.; Turro, C. Intercalation is not required for DNA light-switch behavior. J. Am. Chem. Soc. 2008, 130, 1163–1170. [Google Scholar] [CrossRef]
- Holmlin, R.E.; Stemp, E.D.; Barton, J.K. Ru(phen)2dppz2+ Luminescence: Dependence on DNA sequences and groove-binding agents. Inorg. Chem. 1998, 37, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Mcconnell, A.J.; Hee, L.M.; Olmon, E.D.; Hang, S.; Dervan, E.E.; Barton, J.K. Luminescent properties of ruthenium(II) complexes with sterically expansive ligands bound to DNA defects. Inorg. Chem. 2012, 51, 12511–12520. [Google Scholar] [CrossRef]
- Almaqwashi, A.A.; Paramanathan, T.; Rouzina, I.; Williams, M.C. Mechanisms of small molecule-DNA interactions probed by single-molecule force spectroscopy. Nucleic Acids Res. 2016, 44, 3971–3988. [Google Scholar] [CrossRef]
- Shi, S.; Zhao, J.; Geng, X.T.; Yao, T.M.; Huang, H.L.; Liu, T.L.; Zheng, L.F.; Li, Z.H.; Yang, D.J.; Ji, L.N. Molecular “light switch” for G-quadruplexes and i-motif of human telomeric DNA: [Ru(phen)2(dppz)]2+. Dalton Trans. 2010, 39, 2490–2493. [Google Scholar] [CrossRef]
- Lincoln, P.; Nordén, B. DNA binding geometries of ruthenium(II) complexes with 1,10-phenanthroline and 2,2‘-bipyridine ligands studied with linear dichroism spectroscopy. Borderline cases of intercalation. J. Phys. Chem. B 1998, 102, 9583–9594. [Google Scholar] [CrossRef]
- Dupureur, C.M.; Barton, J.K. Use of selective deuteration and 1H NMR in demonstrating major groove binding of Δ-[Ru(phen)2dppz]2+ to d(GTCGAC)2. J. Am. Chem. Soc. 1994, 116, 10286–10287. [Google Scholar] [CrossRef]
- Collins, J.G.; Sleeman, A.D.; Aldrich-Wright, J.R.; Greguric, I.; Hambley, T.W. A H-1 NMR study of the DNA binding of ruthenium(II) polypyridyl complexes. Inorg. Chem. 1998, 37, 3133–3141. [Google Scholar] [CrossRef]
- Hiort, C.; Lincoln, P.; Norden, B. DNA binding of Δ- and Λ-[Ru(phen)2DPPZ]2+. J. Am. Chem. Soc. 1993, 115, 3448–3454. [Google Scholar] [CrossRef]
- Tuite, E.; Lincoln, P.; Nordén, B. Photophysical evidence that Δ- and Λ-[Ru(phen)2(dppz)]2+ intercalate DNA from the minor groove. J. Am. Chem. Soc. 1997, 119, 239–240. [Google Scholar] [CrossRef]
- Hall, J.P.; Kyra, O.S.; Abeer, N.; Smith, J.A.; Kelly, J.M.; Cardin, C.J. Structure determination of an intercalating ruthenium dipyridophenazine complex which kinks DNA by semiintercalation of a tetraazaphenanthrene ligand. Proc. Natl. Acad. Sci. USA 2011, 108, 17610–17614. [Google Scholar] [CrossRef]
- Hall, J.P.; Sanchez-Weatherby, J.; Alberti, C.; Quimper, C.H.; O’Sullivan, K.; Brazier, J.A.; Winter, G.; Sorensen, T.; Kelly, J.M.; Cardin, D.J. Controlled dehydration of a ruthenium complex-DNA crystal induces reversible DNA kinking. J. Am. Chem. Soc. 2014, 136, 17505–17512. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.P.; Cook, D.; Morte, S.R.; McIntyre, P.; Buchner, K.; Beer, H.; Cardin, D.J.; Brazier, J.A.; Winter, G.; Kelly, J.M.; et al. X-ray crystal structure of rac-[Ru(phen)2dppz]2+ with d(ATGCAT)2 shows enantiomer orientations and water ordering. J. Am. Chem. Soc. 2013, 135, 12652–12659. [Google Scholar] [CrossRef] [PubMed]
- Boer, D.R.; Wu, L.; Lincoln, P.; Coll, M. Thread insertion of a bis(dipyridophenazine) diruthenium complex into the DNA double helix by the extrusion of AT base pairs and cross-linking of DNA duplexes. Angew. Chem. 2014, 53, 1949–1952. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.P.; Keane, P.M.; Beer, H.; Buchner, K.; Winter, G.; Sorensen, T.L.; Cardin, D.J.; Brazier, J.A.; Cardin, C.J. Delta chirality ruthenium ‘light-switch’ complexes can bind in the minor groove of DNA with five different binding modes. Nucleic Acids Res. 2016, 44, 9472–9482. [Google Scholar] [CrossRef] [PubMed]
- Cardin, C.J.; Kelly, J.M.; Quinn, S.J. Photochemically active DNA-intercalating ruthenium and related complexes—Insights by combining crystallography and transient spectroscopy. Chem. Sci. 2017, 8, 4705–4723. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.D.; Kim, M.S.; Lincoln, P.; Tuite, E.; Norden, B. Binding mode of [Ruthenium(II) (1,10-phenanthroline)2L]2+ with Poly(dT*dA-dT) triplex. Ligand size effect on third-strand stabilization. Biochemistry 1997, 36, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.N.; Zhu, Z.Y.; Tan, L.F. Binding differences of two homochiral [Ru(bpy)2dppz]2+ complexes with poly(U)·poly(A)*poly(U) triplex RNA. Inorg. Chem. 2017, 56, 7312–7315. [Google Scholar] [CrossRef] [PubMed]
- Lipfert, J.; Klijnhout, S.; Dekker, N.H. Torsional sensing of small-molecule binding using magnetic tweezers. Nucleic Acids Res. 2010, 38, 7122–7132. [Google Scholar] [CrossRef] [Green Version]
- Coury, J.E.; Anderson, J.R.; McFail-Isom, L.; Williams, L.D.; Bottomley, L.A. Scanning force microscopy of small ligand-nucleic acid complexes: Tris(o-phenanthroline) ruthenium(II) as a test for a new assay. J. Am. Chem. Soc. 1997, 119, 3792–3796. [Google Scholar] [CrossRef]
- Ros, R.; Eckel, R.; Bartels, F.; Sischka, A.; Baumgarth, B.; Wilking, S.D.; Pühler, A.; Sewald, N.; Becker, A.; Anselmetti, D. Single molecule force spectroscopy on ligand–DNA complexes: From molecular binding mechanisms to biosensor applications. J. Biotechnol. 2004, 112, 5–12. [Google Scholar] [CrossRef]
- Husale, S.; Grange, W.; Hegner, M. DNA mechanics affected by small DNA interacting ligands. Single Mol. 2002, 3, 91–96. [Google Scholar] [CrossRef]
- Gunther, K.; Mertig, M.; Seidel, R. Mechanical and structural properties of YOYO-1 complexed DNA. Nucleic Acids Res. 2010, 38, 6526–6532. [Google Scholar] [CrossRef] [Green Version]
- McCauley, M.J.; Williams, M.C. Mechanisms of DNA binding determined in optical tweezers experiments. Biopolymers 2006, 85, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Mihailovic, A.; Vladescu, I.; McCauley, M.; Ly, E.; Williams, M.C.; Spain, E.M.; Nuñez, M.E. Exploring the interaction of ruthenium (II) polypyridyl complexes with DNA using single-molecule techniques. Langmuir 2006, 22, 4699–4709. [Google Scholar] [CrossRef] [PubMed]
- Thayaparan, P.; Fredrik, W.; Mccauley, M.J.; Ioulia, R.; Per, L.; Williams, M.C. Mechanically manipulating the DNA threading intercalation rate. J. Am. Chem. Soc. 2008, 130, 3752–3753. [Google Scholar]
- Clark, A.G.; Naufer, M.N.; Westerlund, F.; Lincoln, P.; Rouzina, I.; Paramanathan, T.; Williams, M.C. Reshaping the energy landscape transforms the mechanism and binding kinetics of DNA threading intercalation. Biochemistry 2017, 57, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Kleinwächter, V.; Koudelka, J. Thermal denaturation of deoxyribonucleic acid-acridine orange complexes. Biochim. Biophys. Acta 1964, 91, 539–540. [Google Scholar] [CrossRef]
- Müller, W.; Crothers, D.M. Studies of the binding of actinomycin and related compounds to DNA. J. Mol. Biol. 1968, 35, 251–290. [Google Scholar] [CrossRef]
- Wu, L.S.; Reymer, A.; Persson, C.; Kazimierczuk, K.; Brown, T.; Lincoln, P.; Nordén, B.; Billeter, M. Initial DNA interactions of the binuclear threading intercalator Λ,Λ-[μ-bidppz(bipy)4Ru2]4+: An NMR study with [d(CGCGAATTCGCG)]2. Chem-Eur. J. 2013, 19, 5401–5410. [Google Scholar] [CrossRef]
- Walter, K. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc. Natl. Acad. Sci. USA 1975, 72, 4876–4880. [Google Scholar]
- Lincoln, P.; Nordén, B. Binuclear ruthenium(II) phenanthroline compounds with extreme binding affinity for DNA. Chem. Commun. 1996, 18, 2145–2146. [Google Scholar] [CrossRef]
- Mazzitelli, C.L.; Chu, Y.; Reczek, J.J.; Iverson, B.L.; Brodbelt, J.S. Screening of threading bis-intercalators binding to duplex DNA by electrospray ionization tandem mass spectrometry. J. Am. Chem. Soc. Mass Spec. 2007, 18, 311–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihmels, H.; Faulhaber, K.; Vedaldi, D.; Dall’Acqua, F.; Viola, G. Intercalation of organic dye molecules into double-stranded DNA. Part 2: The annelated quinolizinium ion as a structural motif in DNA intercalators. Photochem. Photobiol. 2005, 81, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Neto, B.A.D.; Lapis, A.A.M. Recent developments in the chemistry of deoxyribonucleic acid (DNA) intercalators: Principles, design, synthesis, applications and trends. Molecules 2009, 14, 1725–1746. [Google Scholar] [CrossRef] [PubMed]
- Thayaparan, P.; Ioana, V.; Mccauley, M.J.; Ioulia, R.; Williams, M.C. Force spectroscopy reveals the DNA structural dynamics that govern the slow binding of Actinomycin D. Nucleic Acids Res. 2012, 40, 4925–4932. [Google Scholar] [Green Version]
- Mameren, J.V.; Peterman, E.J.G.; Wuite, G.J.L. See me, feel me: Methods to concurrently visualize and manipulate single DNA molecules and associated proteins. Nucleic Acids Res. 2008, 36, 4381–4389. [Google Scholar] [CrossRef] [PubMed]
- Mccauley, M.J.; Williams, M.C. Measuring DNA-protein binding affinity on a single molecule using optical tweezers. Methods Mol. Biol. 2011, 749, 305–315. [Google Scholar] [PubMed]
- Selvam, S.; Yu, Z.; Mao, H. Exploded view of higher order G-quadruplex structures through click-chemistry assisted single-molecule mechanical unfolding. Nucleic Acids Res. 2016, 44, 45–55. [Google Scholar] [CrossRef]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 6, 491–505. [Google Scholar] [CrossRef]
- Ritzefeld, M.; Walhorn, V.; Anselmetti, D.; Sewald, N. Analysis of DNA interactions using single-molecule force spectroscopy. Amino Acids 2013, 44, 1457–1475. [Google Scholar] [CrossRef]
- Pi, J.; Jin, H.; Jiang, J.H.; Yang, F.; Cai, H.H.; Yang, P.H.; Cai, J.Y.; Chen, Z.W. Single molecule force spectroscopy for in-situ probing oridonin inhibited ROS-mediated EGF-EGFR interactions in living KYSE-150 cells. Pharmacol. Res. 2017, 119, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.M.; Liu, J.; Chen, J.C.; Tan, C.P.; Shi, S.; Zheng, K.C.; Ji, L.N. Synthesis, characterization, DNA-binding and spectral properties of complexes [Ru(L)4(dppz)]2+ (L = Im and MeIm). J. Inorg. Biochem. 2008, 102, 330–341. [Google Scholar] [CrossRef] [PubMed]
- Almaqwashi, A.A.; Thayaparan, P.; Per, L.; Ioulia, R.; Fredrik, W.; Williams, M.C. Strong DNA deformation required for extremely slow DNA threading intercalation by a binuclear ruthenium complex. Nucleic Acids Res. 2014, 42, 11634–11641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahira, M.; McCauley, M.J.; Almaqwashi, A.A.; Lincoln, P.; Westerlund, F.; Rouzina, I.; Williams, M.C. A ruthenium dimer complex with a flexible linker slowly threads between DNA bases in two distinct steps. Nucleic Acids Res. 2015, 43, 8856–8867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, Y.; Ji, L.N. Synthesis, DNA-binding and DNA-mediated luminescence quenching of Ru(II) polypyridine complexes. Coordin. Chem. Rev. 1999, 185, 711–733. [Google Scholar] [CrossRef]
- Liu, J.G.; Ye, B.H.; Li, H.; Ji, L.N.; Li, R.H.; Zhou, J.Y. Synthesis, characterization and DNA-binding properties of novel dipyridophenazine complex of ruthenium (II): [Ru(IP)2(DPPZ)]2+. J. Inorg. Biochem. 1999, 73, 117–122. [Google Scholar] [CrossRef]
- Önfelt, B.; Lincoln, P.; Nordén, B. A molecular staple for DNA: Threading bis-intercalating [Ru(phen)2dppz]2+ dimer. J. Am. Chem. Soc. 1999, 121, 10846–10847. [Google Scholar] [CrossRef]
- Ossipov, D.; Pradeepkumar, P.I.; Holmer, M.; Chattopadhyaya, J. Synthesis of [Ru(phen)2dppz]2+-tethered oligo-DNA and studies on the metallointercalation mode into the DNA duplex. J. Am. Chem. Soc. 2001, 123, 3551–3562. [Google Scholar] [CrossRef]
- Ossipov, D.; Gohil, S.; Chattopadhyaya, J. Synthesis of the DNA−[Ru(tpy)(dppz)(CH3CN)]2+ conjugates and their photo cross-linking studies with the complementary DNA strand. J. Am. Chem. Soc. 2002, 124, 13416–13433. [Google Scholar] [CrossRef]
- Metcalfe, C.; Adams, H.; Haq, I.; Thomas, J.A. A ruthenium dipyridophenazine complex that binds preferentially to GC sequences. Chem. Commun. 2003, 10, 1152–1153. [Google Scholar] [CrossRef]
- Nair, R.B.; Teng, E.S.; Kirkland, S.L.; Murphy, C.J. Synthesis and DNA-binding properties of [Ru(NH3)4dppz]2+. Inorg. Chem. 1998, 37, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.B.; Yeung, L.K.; Murphy, C.J. Synthesis and solvent-dependent properties of Ru(acac)2dppz. Inorg. Chem. 1999, 38, 2536–2538. [Google Scholar] [CrossRef]
- Liu, J.G.; Zhang, Q.L.; Shi, X.F.; Ji, L.N. Interaction of [Ru(dmp)2(dppz)]2+ and [Ru(dmb)2(dppz)]2+ with DNA: Effects of the ancillary ligands on the DNA-binding behaviors. Inorg. Chem. 2001, 40, 5045–5050. [Google Scholar] [CrossRef] [PubMed]
- Greguric, A.; Greguric, I.D.; Hambley, T.W.; Aldrich-Wright, J.R.; Collins, J.G. Minor groove intercalation of Δ-[Ru(Me2phen)2dppz]2+ to the hexanucleotide d(GTCGAC)2. Dalton Trans. 2002, 6, 849–855. [Google Scholar] [CrossRef]
- Rüba, E.; Hart, J.R.; Barton, J.K. [Ru(bpy)2(L)]Cl2: Luminescent metal complexes that bind DNA base mismatches. Inorg. Chem. 2004, 43, 4570–4578. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chouai, A.; Degtyareva, N.N.; Lutterman, D.A.; Dunbar, K.R.; Turro, C. Chemical control of the DNA light switch: Cycling the switch on and off. J. Am. Chem. Soc. 2005, 127, 10796–10797. [Google Scholar] [CrossRef]
- Hall, J.P.; Beer, H.; Buchner, K.; Cardin, D.J.; Cardin, C.J. The structural effect of methyl substitution on the binding of polypyridyl Ru–dppz complexes to DNA. Organometallics 2015, 34, 2481–2486. [Google Scholar] [CrossRef]
- Önfelt, B.; Lincoln, P.; Nordén, B. Enantioselective DNA threading dynamics by phenazine-linked [Ru(phen)2dppz]2+ dimers. J. Am. Chem. Soc. 2001, 123, 3630–3637. [Google Scholar] [CrossRef]
- Puckett, C.A.; Barton, J.K. Methods to explore cellular uptake of ruthenium complexes. J. Am. Chem. Soc. 2007, 129, 46–47. [Google Scholar] [CrossRef]
- Puckett, C.A.; Barton, J.K. Mechanism of cellular uptake of a ruthenium polypyridyl complex. Biochemistry 2008, 47, 11711–11716. [Google Scholar] [CrossRef]
- Pierroz, V.; Joshi, T.; Leonidova, A.; Mari, C.; Schur, J.; Ott, I.; Spiccia, L.; Ferrari, S.; Gasser, G. Molecular and cellular characterization of the biological effects of ruthenium(II) complexes incorporating 2-pyridyl-2-pyrimidine-4-carboxylic acid. J. Am. Chem. Soc. 2012, 134, 20376–20387. [Google Scholar] [CrossRef] [PubMed]
- Rajendiran, V.; Palaniandavar, M.; Periasamy, V.; Akbarsha, M. [Ru(phen)2(dppz)]2+ as an efficient optical probe for staining nuclear components. J. Inorg. Biochem. 2010, 104, 217–220. [Google Scholar] [CrossRef] [PubMed]
- Gill, M.R.; Garcia-Lara, J.; Foster, S.J.; Smythe, C.; Battaglia, G.; Thomas, J.A. A ruthenium(II) polypyridyl complex for direct imaging of DNA structure in living cells. Nat. Chem. 2009, 1, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Schatzschneider, U.; Niesel, J.; Ott, I.; Gust, R.; Alborzinia, H.; Wölfl, S. Cellular uptake, cytotoxicity, and metabolic profiling of human cancer cells treated with ruthenium(II) polypyridyl complexes [Ru(bpy)2(N--N)]Cl2 with N--N=bpy, phen, dpq, dppz, and dppn. ChemMedChem 2008, 3, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Gill, M.R.; Derrat, H.; Smythe, C.G.; Battaglia, G.; Thomas, J.A. Ruthenium(II) metallo-intercalators: DNA imaging and cytotoxicity. ChemBioChem 2011, 12, 877–880. [Google Scholar] [CrossRef]
- Joshi, T.; Pierroz, V.; Ferrari, S.; Gasser, G. Bis(dipyridophenazine)(2-(2′-pyridyl)pyrimidine-4-carboxylic acid)ruthenium(II) Hexafluorophosphate: A Lesson in Stubbornness. Chemmedchem 2014, 9, 1419–1427. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Zhang, P.Y.; Yu, B.L.; Chen, Y.; Wang, J.Q.; Ji, L.N.; Chao, H. Targeting nucleus DNA with a cyclometalated dipyridophenazineruthenium(II) complex. J. Med. Chem. 2014, 57, 8971–8983. [Google Scholar] [CrossRef]
- Huang, H.; Yu, B.; Zhang, P.; Huang, J.; Chen, Y.; Gasser, G.; Ji, L.; Chao, H. Highly charged ruthenium(II) polypyridyl complexes as lysosome-localized photosensitizers for two-photon photodynamic therapy. Angew. Chem. 2015, 127, 14255–14258. [Google Scholar] [CrossRef]
- Marcélis, L.; Ghesquière, J.; Garnir, K.; Mesmaeker, K.D.; Moucheron, C. Photo-oxidizing RuII complexes and light: Targeting biomolecules via photoadditions. Coordin. Chem. Rev. 2012, 256, 1569–1582. [Google Scholar] [CrossRef]
- Shi, G.; Monro, S.; Hennigar, R.; Colpitts, J.; Fong, J.; Kasimova, K.; Yin, H.; Decoste, R.; Spencer, C.; Chamberlain, L. Ru(II) dyads derived from α-oligothiophenes: A new class of potent and versatile photosensitizers for PDT. Coordin. Chem. Rev. 2015, 282, 127–138. [Google Scholar] [CrossRef]
- Yu, H.J.; Chao, H.; Jiang, L.; Li, L.Y.; Huang, S.M.; Ji, L.N. Single oxygen-mediated DNA photocleavage of a di-bithiazolyl ruthenium(II) complex [Ru(btz)(dppz)]. Inorg. Chem. Commun. 2008, 11, 553–556. [Google Scholar] [CrossRef]
- Foxon, S.P.; Alamiry, M.A.H.; Walker, M.G.; Meijer, A.J.H.M.; Sazanovich, I.V.; Weinstein, J.A.; Thomas, J.A. Photophysical properties and singlet oxygen production by ruthenium(II) complexes of benzo[i]dipyrido[3,2-a:2′,3′-c]phenazine: Spectroscopic and TD-DFT study. J. Phys. Chem. A 2009, 113, 12754–12762. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Stephenson, M.; Gibson, J.; Sampson, E.; Shi, G.; Sainuddin, T.; Monro, S.; McFarland, S.A. In vitro multiwavelength PDT with 3IL states: Teaching old molecules new tricks. Inorg. Chem. 2014, 53, 4548–4559. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, S.P.; Li, S.; Korn, R.; Xie, X.; Meggers, E. Solid-phase synthesis of tris-heteroleptic ruthenium(II) complexes and application to acetylcholinesterase inhibition. Inorg. Chem. 2008, 47, 5030–5032. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.P.; Torres, V.; Jain, D.; Martí, A.A. Sensing amyloid-β aggregation using luminescent dipyridophenazine ruthenium(II) complexes. J. Am. Chem. Soc. 2011, 133, 11121–11123. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.P.; Mehmet, O.; Christina, K.; Rajeev, P.; Martí, A.A. Unraveling the photoluminescence response of light-switching ruthenium(II) complexes bound to amyloid-β. J. Am. Chem. Soc. 2013, 135, 10810–10816. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.P.; Kilpatrick, K.; Segatori, L.; Martí, A.A. Detection of α-synuclein amyloidogenic aggregates in vitro and in cells using light-switching dipyridophenazine ruthenium(II) Complexes. J. Am. Chem. Soc. 2012, 134, 20776–20782. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Fang, X.; Bai, C. Signaling aptamer/protein binding by a molecular light switch complex. Anal. Chem. 2004, 76, 5230–5235. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, Y.; Zhou, C.; Fang, X. Aptamer-based ATP assay using a luminescent light switching complex. Anal. Chem. 2005, 77, 3542–3546. [Google Scholar] [CrossRef]
- Choi, M.S.; Yoon, M.; Baeg, J.O.; Kim, J. Label-free dual assay of DNA sequences and potassium ions using an aptamer probe and a molecular light switch complex. Chem. Commun. 2009, 47, 7419–7421. [Google Scholar] [CrossRef]
- Zhao, D.; Chan, W.H.; He, Z.; Qiu, T. Quantum dot-ruthenium complex dyads: Recognition of double-strand DNA through dual-color fluorescence detection. Anal. Chem. 2009, 81, 3537–3543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhao, D.; Ding, H.G.; Huang, Y.X.; Zhong, H.Z.; Xie, H.Y. Sensitive single-color fluorescence “off–on” switch system for dsDNA detection based on quantum dots-ruthenium assembling dyads. Biosens. Bioelectron. 2014, 56, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.Z.; Chao, X.J.; Huang, C.H.; Li, Y. Delivering the cell-impermeable DNA ‘light-switching’ Ru(ii) complexes preferentially into live-cell nucleus via an unprecedented ion-pairing method. Chem. Sci. 2016, 7, 4016–4023. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yu, T.; Guo, L.; Xie, J.; Shao, N.; He, Z. In vitro selection of DNA aptamer against abrin toxin and aptamer-based abrin direct detection. Biosens. Bioelectron. 2007, 22, 2456–2463. [Google Scholar] [CrossRef] [PubMed]



| Nucleic Acid | [Ru(byp)2dppz]2+ | [Ru(phen)2dppz]2+ | ||||||
|---|---|---|---|---|---|---|---|---|
| τ (ns) | % | λmax (nm) | RI | τ (ns) | % | λmax (nm) | RI | |
| poly[d(GC)]·poly[d(GC)] | 220 | 60 | 610 | 0.29 | 290 | 60 | 606 | 0.61 |
| 70 | 40 | 70 | 40 | |||||
| poly(dG)·poly(dC) | 260 | 30 | 610 | 0.29 | 400 | 40 | 607 | 0.74 |
| 70 | 70 | 90 | 60 | |||||
| poly[d(AT)]·poly[d(AT)] | 320 | 20 | 624 | 0.17 | 740 | 40 | 620 | 0.75 |
| 90 | 80 | 120 | 60 | |||||
| poly(dA)·poly(dT) | 340 | 40 | 626 | 0.23 | 840 | 60 | 621 | 1.39 |
| 80 | 60 | 110 | 40 | |||||
| poly[d(G-m5C)]·poly[d(d(G-m5C)] | 240 | 40 | 606 | 0.25 | 360 | 40 | 606 | 0.51 |
| 60 | 60 | 90 | 60 | |||||
| Z-poly[d(GC)]·poly[d(GC)] | 220 | 60 | 608 | 0.28 | 270 | 60 | 608 | 0.60 |
| 70 | 40 | 70 | 40 | |||||
| Calf thymus DNA Z conditions | 330 | 40 | 621 | 0.21 | 750 | 40 | 616 | 0.80 |
| 80 | 60 | 120 | 60 | |||||
| poly[r(AU)]·polyp[r(AU)] | 400 | 10 | 626 | 0.0057 | 490 | 20 | 620 | 0.10 |
| 50 | 90 | 80 | 80 | |||||
| poly(rG)·poly(dC) | 540 | 10 | 620 | 0.0067 | 520 | 30 | 616 | 0.04 |
| 70 | 90 | 80 | 70 | |||||
| poly(dT)·poly(dA)·poly(dT) | 430 | 70 | 621 | 0.60 | 530 | 60 | 621 | 1.45 |
| 170 | 30 | 170 | 40 | |||||
| tRNA | 300 | 30 | 624 | 0.06 | 300 | 30 | 620 | 0.18 |
| 60 | 70 | 70 | 70 | |||||
| Intercalator | Binding Constant K (× 10−6 M−1) | Binding Site Size n (Base Pairs) | Binding Equilibrium Elongation ΔXeq (nm/bp) |
|---|---|---|---|
| Ethidium | 0.036 ± 0.005 | 2.01 | |
| 10 | 2 | ||
| 0.46 ± 0.05 | 2.3 ± 0.1 | 0.25 ± 0.03 | |
| 0.13 ± 0.04 | 1.9 ± 0.1 | ||
| 0.145 | |||
| Daunomycin | 0.066 ± 0.024 | 3.04 | |
| AFP | 2.48 | 2 | |
| [Ru(phen)3]2+ | 0.0088 ± 0.0003 | 3.0 ± 0.2 | |
| 0.0016 ± 0.0002 | 3.0 ± 0.1 | 0.28 ± 0.01 | |
| [Ru(phen)2dppz]2+ | 0.15 ± 0.07 | 2.2 ± 0.4 | |
| 3.2 ± 0.1 (10 pN) | 3.0 ± 0.5 (10 pN) | ||
| 0.90 ± 0.10 | 2.9 ± 0.1 | 0.38 ± 0.02 | |
| [Ru(bpy)2dppz]2+ | 0.15 ± 0.07 | 2.2 ± 0.4 | |
| 3.2 ± 0.1 (10 pN) | 3.0 ± 0.5 (10 pN) | ||
| Oxazole Yellow (YO) | 0.578 ± 0.080 | 0.233 ± 0.013 | |
| 0.29 ± 0.09 | 3.8 ± 1.0 | 0.31 ± 0.03 | |
| Psoralen | 0.088 ± 0.024 | 1.43 ± 0.13 | |
| SYTOX Orange (SxO) | 2.4 ± 0.5 | 3.0 ± 0.4 | 0.30 ± 0.02 |
| SYTOX Green (SxG) | 14 ± 3 | 2.6 ± 0.6 | 0.27 ± 0.02 |
| SGold (SbG) | 7.8 ± 3.3 | 3.2 ± 0.5 | 0.30 ± 0.01 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, F.; Wang, S.; Man, Y.; Kumar, P.; Liu, B. Recent Developments in the Interactions of Classic Intercalated Ruthenium Compounds: [Ru(bpy)2dppz]2+ and [Ru(phen)2dppz]2+ with a DNA Molecule. Molecules 2019, 24, 769. https://doi.org/10.3390/molecules24040769
Jia F, Wang S, Man Y, Kumar P, Liu B. Recent Developments in the Interactions of Classic Intercalated Ruthenium Compounds: [Ru(bpy)2dppz]2+ and [Ru(phen)2dppz]2+ with a DNA Molecule. Molecules. 2019; 24(4):769. https://doi.org/10.3390/molecules24040769
Chicago/Turabian StyleJia, Fuchao, Shuo Wang, Yan Man, Parveen Kumar, and Bo Liu. 2019. "Recent Developments in the Interactions of Classic Intercalated Ruthenium Compounds: [Ru(bpy)2dppz]2+ and [Ru(phen)2dppz]2+ with a DNA Molecule" Molecules 24, no. 4: 769. https://doi.org/10.3390/molecules24040769
APA StyleJia, F., Wang, S., Man, Y., Kumar, P., & Liu, B. (2019). Recent Developments in the Interactions of Classic Intercalated Ruthenium Compounds: [Ru(bpy)2dppz]2+ and [Ru(phen)2dppz]2+ with a DNA Molecule. Molecules, 24(4), 769. https://doi.org/10.3390/molecules24040769





