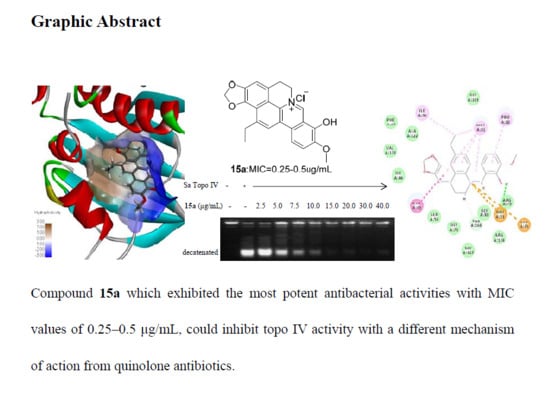
Evolution and Antibacterial Evaluation of 8-Hydroxy-cycloberberine Derivatives as a Novel Family of Antibacterial Agents Against MRSA
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Pharmacological Evaluation
2.2.1. SAR for Anti-Bacterial Activity
2.2.2. Time-kill Curve Study of 15a
2.2.3. Evaluation of Metabolic Stability and Acute Toxicity Assay of Compound 15a
2.2.4. Preliminary Mechanism of Compound 15a
2.2.5. Molecular Docking Study of Key Compound 15a
3. Experimental Section
3.1. Apparatus, Materials, and Analysis Reagents
3.2. Chemistry
3.2.1. Synthesis of 1,2-Dimethoxy-8-hydroxy-9-methoxycycloberberine Chloride (6)
3.2.2. Synthesis of 1,2,8-Trihydroxy-9-methoxycycloberberine Chloride (10)
3.2.3. General Procedure for the Synthesis of 13a–t
3.2.4. General Procedure for the Synthesis of 15a–c
3.3. Antimicrobial Assay
3.4. Time-kill Curves of 15a
3.5. Stability Assay of Key Compound in Liver Microsomes
3.6. Stability Assay of Key Compound in SD Rat Whole Blood
3.7. Acute Toxicity Assay
3.8. Enzymatic Assay
3.9. Molecular Docking Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References and Notes
- Antibiotic Resistance: The Global Threat (U.S.); Centers for Disease Control and Prevention: Atlanta, GA, USA. Available online: http://stacks.cdc.gov/view/cdc/31340 (accessed on 27 February 2015).
- Marr, A.K.; Gooderham, W.J.; Hancock, R.E. Antibacterial peptides for therapeutic use: Obstacles and realistic outlook. Curr. Opin. Pharmacol. 2006, 6, 468–472. [Google Scholar] [CrossRef] [PubMed]
- WHO. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Filice, G.A.; Nyman, J.A.; Lexau, C.; Lees, C.H.; Bockstedt, L.A.; ComoSabetti, K.; Lesher, L.J.; Lynfield, R. Excess costs and utilization associated with methicillin resistance for patients with Staphylococcus aureus infection. Infect. Control Hosp. Epidemiol. 2010, 31, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T.; Fischbach, M.A. Squashing superbugs—The race for new antibiotics. Sci. Am. 2009, 301, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Hanaki, H.; Ino, T.; Yabuta, K.; Oquri, T.; Tenover, F.C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 1997, 40, 135–136. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Aritaka, N.; Hanaki, H.; Kawasaki, S.; Hosoda, Y.; Hori, S.; Fukuchi, Y.; Kobayashi, I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 1997, 350, 1670–1673. [Google Scholar] [CrossRef]
- Auckland, C.; Teare, L.; Cooke, F.; Kaufmann, M.E.; Warner, M.; Jones, G.; Bamford, K.; Ayles, H.; Johnson, A.P. Linezolid-resistant enterococci: Report of the first isolates in the United Kingdom. J. Antimicrob. Chemother. 2002, 50, 743–746. [Google Scholar] [CrossRef] [PubMed]
- Marty, F.M.; Yeh, W.W.; Wennersten, C.B.; Venkataraman, L.; Albano, E.; Alyea, E.P.; Gold, H.S.; Baden, L.R.; Pillai, S.K. Emergence of a clinical daptomycin-resistant Staphylococcus aureus isolate during treatment of methicillin-resistant Staphylococcus aureus bacteremia and osteomyelitis. J. Clin. Microbiol. 2006, 44, 595–597. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.C.; Basuino, L.; Diep, B.; Hamilton, S.; Chatterjee, C.C.; Chambers, H.F. Ceftobiprole- and ceftaroline-resistant methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2015, 59, 2960–2963. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Liu, Y.X.; Xiao, C.L.; Wang, Y.X.; Li, Y.H.; Yang, Y.H.; Li, Y.B.; Bi, C.W.; Gao, L.M.; Jiang, J.D.; Song, D.Q. Synthesis, structure-activity relationship and in vitro anti-mycobacterial evaluation of 13-n-octylberberine derivatives. Eur. J. Med. Chem. 2012, 52, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.G.; Hu, X.X.; Li, C.R.; Li, Y.H.; Wang, Y.X.; Jiang, J.D.; Bi, C.W.; Tang, S.; You, X.F.; Song, D.Q. Design, synthesis and biological evaluation of monobactams as antibacterial agents against gram-negative bacteria. Eur. J. Med. Chem. 2016, 110, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.B.; Zhao, W.L.; Wang, Y.X.; Zhang, C.X.; Jiang, J.D.; Bi, C.W.; Tang, S.; Chen, R.X.; Shao, R.G.; Song, D.Q. Discovery, synthesis and biological evaluation of cycloprotoberberine derivatives as potential antitumor agents. Eur. J. Med. Chem. 2013, 68, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.Y.; Hu, X.X.; Tang, S.; Liu, X.J.; Wang, Y.X.; Deng, H.B.; You, X.F.; Jiang, J.D.; Li, Y.H.; Song, D.Q. Discovery and development of 8-substituted cycloberberine derivatives as novel antibacterial agents against MRSA. ACS Med. Chem. Lett. 2018, 9, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.Y.; Wang, Y.X.; Tang, S.; Hu, X.X.; Zeng, Q.X.; Pang, J.; Yang, Y.S.; You, X.F.; Song, D.Q. Synthesis and antibacterial evaluation of 13-substituted cycloberberine derivatives as a novel class of anti-MRSA agents. Eur. J. Med. Chem. 2018, 157, 877–886. [Google Scholar] [CrossRef] [PubMed]
- National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; pp. M07–M10. [Google Scholar]
- Lu, X.; Yang, X.Y.; Li, X.; Lu, Y.; Ren, Z.T.; Zhao, L.Y.; Hu, X.X.; Jiang, J.D.; You, X.F. In vitro activity of sodium new houttuyfonate alone and in combination with oxacillin or netilmicin against methicillin-resistant Staphylococcus aureus. PLoS ONE 2013, 8, e68053. [Google Scholar] [CrossRef] [PubMed]
- Plech, T.; Kaproń, B.; Paneth, A.; Kosikowska, U.; Malm, A.; Strzelczyk, A.; Stączek, P.; Świątek, Ł.; Rajtar, B.; Polz-Dacewicz, M. Search for factors affecting antibacterial activity and toxicity of 1,2,4-triazole-ciprofloxacin hybrids. Eur. J. Med. Chem. 2015, 97, 94–103. [Google Scholar] [CrossRef] [PubMed]
- O’Dowd, H.; Dean, E.; Shannon, K.R.; Chandupatla, V.D.; Juntyma, J.E.; Ye, Z.Q.; Jones, S.M.; O’Brien, C.F.; Nicolau, D.P.; Tessier, P.R.; et al. Discovery and characterization of a water-soluble prodrug of a dual inhibitor of bacterial DNA gyrase and topoisomerase IV. ACS Med. Chem. Lett. 2015, 6, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Badshah, S.L.; Ullah, A. New developments in non-quinolone-based antibiotics for the inhibition of bacterial gyrase and topoisomerase IV. Eur. J. Med. Chem. 2018, 152, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Champoux, J. DNA topoisomerases: Structure, function and mechanism. J. Annu. Rev. Biochem. 2001, 70, 369–413. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.W.; Goetz, M.A.; Smith, S.K.; Zink, D.L.; Polishook, J.; Onishi, R.; Salowe, S.; Wiltsie, J.; Allocco, J.; Sigmund, J.; et al. Discovery of kibdelomycin, a potent new class of bacterial type II topoisomerase inhibitor by chemical-genetic profiling in Staphylococcus aureus. Chem. Biol. 2011, 18, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Patel, S.; Sharma, N.; Soisson, S.M.; Kishii, R.; Takei, M.; Fukuda, Y.; Lumb, K.J.; Singh, S.B. Structures of kibdelomycin bound to Staphylococcus aureus GyrB and ParE showed a novel U-shaped binding mode. ACS Chem. Biol. 2014, 9, 2023–2031. [Google Scholar] [CrossRef] [PubMed]
- Verma, P. Methods for determining bactericidal activity and antimicrobial interactions synergy testing, time-kill curves, and population analysis. In Antimicrobial Susceptibility Testing Protocols; Schwalbe, R., Steele-Moore, L., Goodwin, A.C., Eds.; CRC Press: Roca Raton, FL, USA, 2017; pp. 275–298. [Google Scholar]
- Wang, B.; Wang, K.; Meng, P.; Hu, Y.; Yang, F.; Liu, K.; Lei, Z.; Chen, B.; Tian, Y. Design, synthesis, and evaluation of carboxyl-modified oseltamivir derivatives with improved lipophilicity as neuraminidase inhibitors. Bioorg. Med. Chem. Lett. 2018, 28, 3477–3482. [Google Scholar] [CrossRef] [PubMed]
- Bielenica, A.; Stefańska, J.; Stępień, K.; Napiórkowska, A.; Augustynowicz-Kopeć, E.; Sanna, G.; Madeddu, S.; Boi, S.; Giliberti, G.; Wrzosek, M.; et al. Synthesis, cytotoxicity and antimicrobial activity of thiourea derivatives incorporating 3-(trifluoromethyl)phenyl moiety. Eur. J. Med. Chem. 2015, 101, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Sharma, M.; Shakeel, E.; Jamal, Q.M.; Kamal, M.A.; Sayeed, U.; Khan, M.K.; Siddiqui, M.H.; Arif, J.M.; Akhtar, S. Molecular interaction and computational analytical studies of pinocembrin for its antiangiogenic potential targeting VEGFR-2: A persuader of metastasis. Med. Chem. 2018, 14, 626–640. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 6, 10, 13a–t and 15a–c are available from the authors. |







| Code | R1/R2 | R3 | MSSA c | MSSE c | S.s c | S.h c | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATCC 29213 | 15 b | 16-1 b | ATCC 12228 | 16-5 b | ATCC 15305 | ATCC 35982 | ||||||
| CBBR | - | - | >64 | >64 | >64 | >64 | >64 | >64 | >64 | |||
| 1 | - | - | 0.5 | 4 | 1 | 0.5 | 0.5 | >64 | 1 | |||
| 6 | - | - | >64 | >64 | >64 | >64 | >64 | >64 | >64 | |||
| 10 | - | - | 32 | 16 | 32 | 8 | 16 | 32 | 32 | |||
| 13a | H | H | 8 | 16 | 8 | 16 | 8 | 8 | 8 | |||
| 13b | H | p-F | >64 | >64 | >64 | >64 | >64 | >64 | >64 | |||
| 13c | H | 2′,4′-(F)2 | 8 | 8 | 8 | 8 | 8 | 16 | 8 | |||
| 13d | H | 2′,6′-(F)2 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | |||
| 13e | H | p-Cl | 8 | 8 | 4 | 8 | 4 | 16 | 8 | |||
| 13f | H | p-Br | 4 | 4 | 2 | 4 | 2 | 4 | 4 | |||
| 13g | H | o-CH3 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | |||
| 13h | H | m-CH3 | 4 | 4 | 4 | 4 | 2 | 16 | 8 | |||
| 13i | H | p-CH3 | 8 | 8 | 4 | 8 | 4 | 16 | 4 | |||
| 13j | H | 3′,5′-(CH3)2 | 32 | 32 | 32 | 32 | 16 | 64 | 32 | |||
| 13k | H | p-tBu | 32 | 32 | 16 | 16 | 8 | 32 | 32 | |||
| 13l | H | p-OCF3 | 32 | 32 | 32 | 32 | 32 | 64 | 64 | |||
| 13m | H | o-CN | >64 | >64 | >64 | >64 | >64 | >64 | >64 | |||
| 13n | H | m-CN | >64 | >64 | >64 | 16 | 32 | >64 | 64 | |||
| 13o | H | p-CN | 8 | 4 | 8 | 8 | 16 | 16 | 16 | |||
| 13p | H | o-NO2 | 8 | 8 | 4 | 8 | 4 | 16 | 8 | |||
| 13q | H | m-NO2 | >64 | >64 | 64 | 64 | 32 | >64 | 64 | |||
| 13r | H | p-NO2 | 16 | 8 | 16 | 4 | 4 | 32 | 8 | |||
| 13s | CH3 | H | 16 | 8 | 16 | 32 | 16 | 64 | 32 | |||
| 13t | CO2CH3 | H | 64 | 64 | >64 | 64 | 32 | >64 | 64 | |||
| 15a | CH2CH3 | - | 0.5 | 0.5 | 0.5 | 0.5 | 0.25 | 1 | 0.5 | |||
| 15b | (CH2)2CH3 | - | 2 | 2 | 2 | 2 | 1 | 8 | 2 | |||
| 15c | (CH2)3CH3 | - | 8 | 4 | 4 | 8 | 8 | 16 | 4 | |||
| Lev | 0.125 | 0.125 | 0.25 | 0.126 | 0.125 | 1 | 0.125 | |||||
| Code | MRSA | VISA | MRSE | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13-18 | 13-23 | 16-34 | ATCC 43300 | ATCC-B AA-1708 a | ATCC -BAA-976 b | ATCC 700698 | ATCC 700699 | 13-3 | 16-5 | |||
| CBBR | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | ||
| 1 | 1 | 1 | 0.5 | 0.5 | 0.5 | 1 | 1 | 0.5 | >64 | 0.5 | ||
| 6 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | ||
| 10 | 32 | 64 | 16 | 32 | 16 | 64 | 32 | 32 | 32 | 16 | ||
| 13a | 16 | 16 | 8 | 16 | 8 | 16 | 8 | 8 | 8 | 16 | ||
| 13b | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | ||
| 13c | 16 | 8 | 8 | 8 | 4 | 8 | 8 | 8 | 8 | 8 | ||
| 13d | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | 32 | ||
| 13e | 8 | 8 | 4 | 8 | 8 | 4 | 4 | 8 | 64 | 4 | ||
| 13f | 4 | 4 | 4 | 4 | 1 | 2 | 2 | 1 | 16 | 2 | ||
| 13g | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | 64 | 64 | ||
| 13h | 8 | 8 | 4 | 8 | 4 | 8 | 4 | 4 | 4 | 4 | ||
| 13i | 8 | 8 | 8 | 16 | 2 | 4 | 4 | 4 | 2 | 2 | ||
| 13j | 32 | 32 | 16 | 32 | 16 | 32 | 32 | 32 | 16 | 16 | ||
| 13k | 16 | 32 | 16 | 32 | 8 | 16 | 32 | 32 | 8 | 8 | ||
| 13l | 32 | 32 | 32 | 64 | 32 | 64 | 64 | 32 | 32 | 32 | ||
| 13m | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | ||
| 13n | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | 64 | 32 | ||
| 13o | 8 | 8 | 8 | 4 | 8 | 16 | 4 | 8 | 16 | 8 | ||
| 13p | 16 | 8 | 4 | 8 | 4 | 8 | 8 | 4 | 32 | 8 | ||
| 13q | >64 | >64 | >64 | >64 | >64 | 64 | >64 | 64 | 64 | 32 | ||
| 13r | 32 | 8 | 8 | 64 | 4 | 32 | 4 | 4 | >64 | 16 | ||
| 13s | 16 | 8 | 16 | 16 | 16 | 32 | 32 | 32 | 32 | 32 | ||
| 13t | 64 | 64 | >64 | 64 | 64 | 64 | 64 | 64 | 64 | 32 | ||
| 15a | 2 | 0.5 | 0.25 | 0.5 | 0.25 | 0.5 | 0.5 | 0.25 | 1 | 0.25 | ||
| 15b | 4 | 4 | 2 | 4 | 2 | 2 | 4 | 2 | 4 | 2 | ||
| 15c | 4 | 8 | 8 | 4 | 8 | 4 | 4 | 4 | 16 | 8 | ||
| Lev | 32 | 64 | 64 | 32 | 16 | 0.125 | 32 | 8 | 32 | 8 | ||
| Code | Species | T1/2 (min) | CL Int (mic) (μL/min/mg) | Remaining Ratio (%) (T = 60 min) |
|---|---|---|---|---|
| 15a | Human | 105.3 | 13.2 | 70.6 |
| Rat | 50.9 | 27.3 | 47.9 | |
| Mouse | 18.9 | 73.4 | 10.4 | |
| Tes | Human | 12.9 | 107.2 | 3.9 |
| Rat | 0.6 | 2349.8 | 0 | |
| Mouse | 5.5 | 253.9 | 0 |
| Time (h) | 0 | 0.5 | 1 | 3 | 7 | 24 |
|---|---|---|---|---|---|---|
| 15a Rr (%) | 100 | 98.9 | 98.0 | 95.8 | 93.1 | 85.4 |
| Ems Rr (%) | 100 | 37.5 | 9.1 | 0 | 0 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.-S.; Wei, W.; Hu, X.-X.; Tang, S.; Pang, J.; You, X.-F.; Fan, T.-Y.; Wang, Y.-X.; Song, D.-Q. Evolution and Antibacterial Evaluation of 8-Hydroxy-cycloberberine Derivatives as a Novel Family of Antibacterial Agents Against MRSA. Molecules 2019, 24, 984. https://doi.org/10.3390/molecules24050984
Yang Y-S, Wei W, Hu X-X, Tang S, Pang J, You X-F, Fan T-Y, Wang Y-X, Song D-Q. Evolution and Antibacterial Evaluation of 8-Hydroxy-cycloberberine Derivatives as a Novel Family of Antibacterial Agents Against MRSA. Molecules. 2019; 24(5):984. https://doi.org/10.3390/molecules24050984
Chicago/Turabian StyleYang, Yuan-Shuai, Wei Wei, Xin-Xin Hu, Sheng Tang, Jing Pang, Xue-Fu You, Tian-Yun Fan, Yan-Xiang Wang, and Dan-Qing Song. 2019. "Evolution and Antibacterial Evaluation of 8-Hydroxy-cycloberberine Derivatives as a Novel Family of Antibacterial Agents Against MRSA" Molecules 24, no. 5: 984. https://doi.org/10.3390/molecules24050984
APA StyleYang, Y.-S., Wei, W., Hu, X.-X., Tang, S., Pang, J., You, X.-F., Fan, T.-Y., Wang, Y.-X., & Song, D.-Q. (2019). Evolution and Antibacterial Evaluation of 8-Hydroxy-cycloberberine Derivatives as a Novel Family of Antibacterial Agents Against MRSA. Molecules, 24(5), 984. https://doi.org/10.3390/molecules24050984