Derivatives of 3-Aminopyrazine-2-carboxamides: Synthesis, Antimicrobial Evaluation, and in Vitro Cytotoxicity
Abstract
1. Introduction
2. Results and Discussion
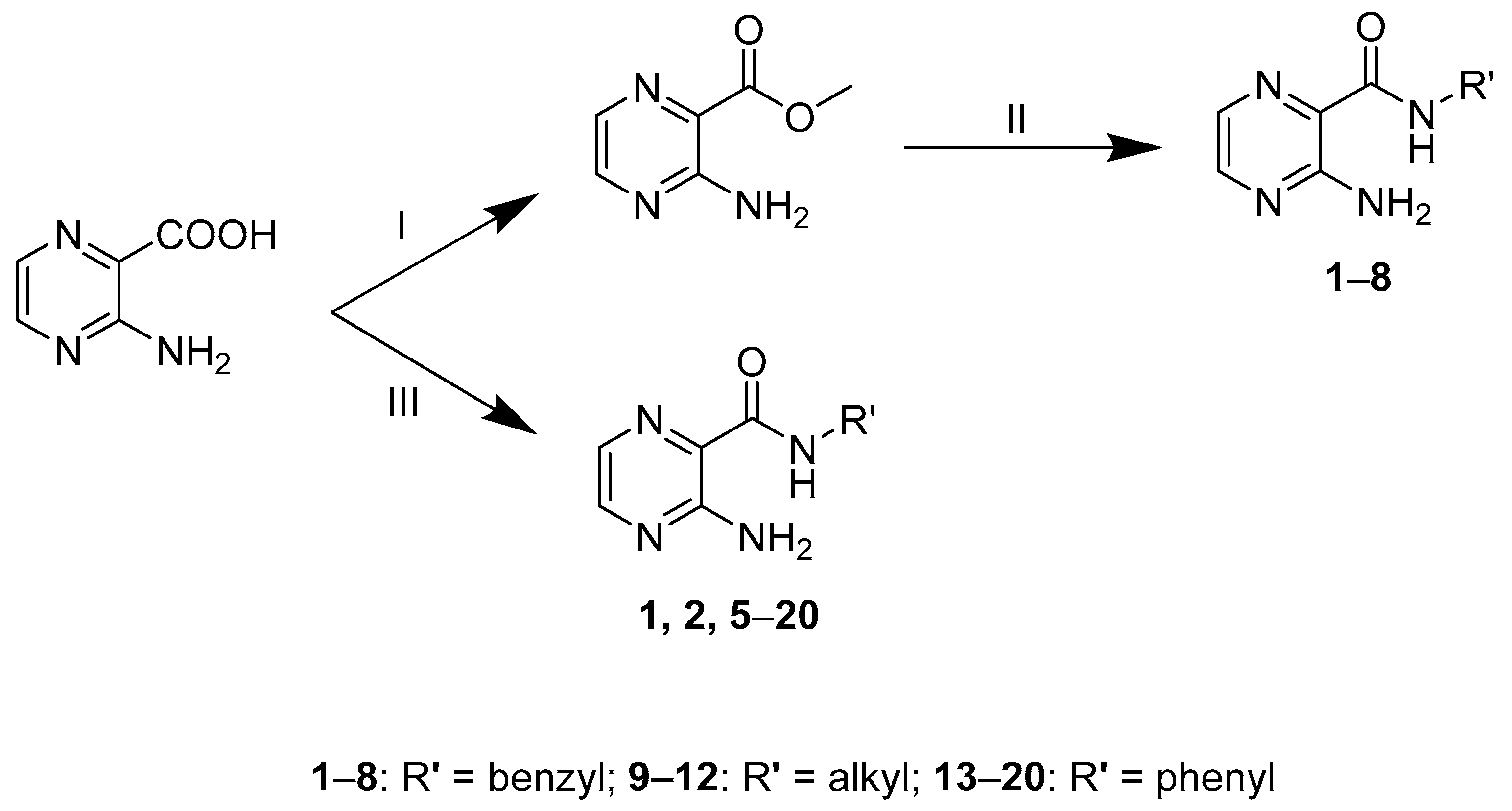
2.1. Synthesis
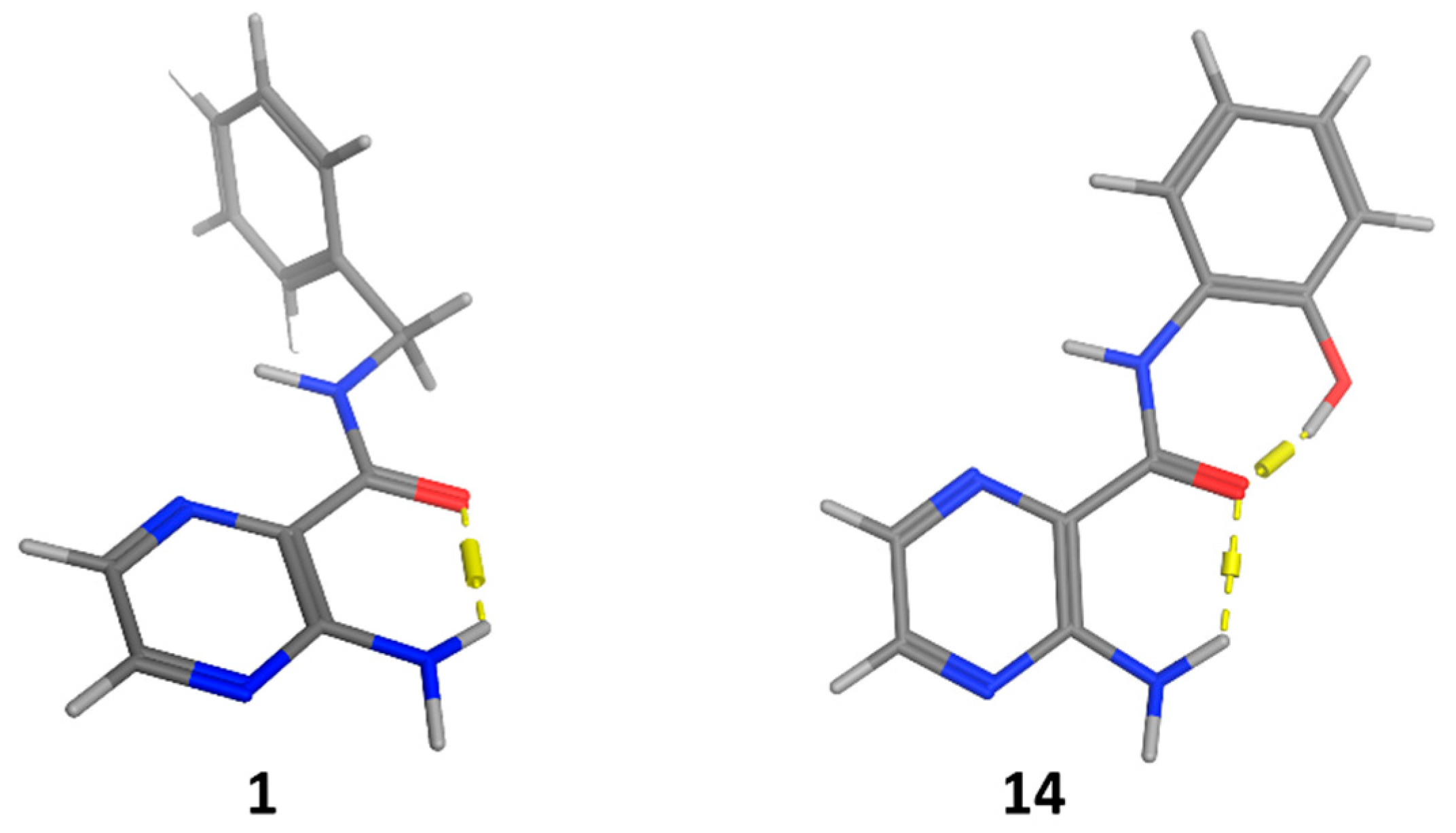
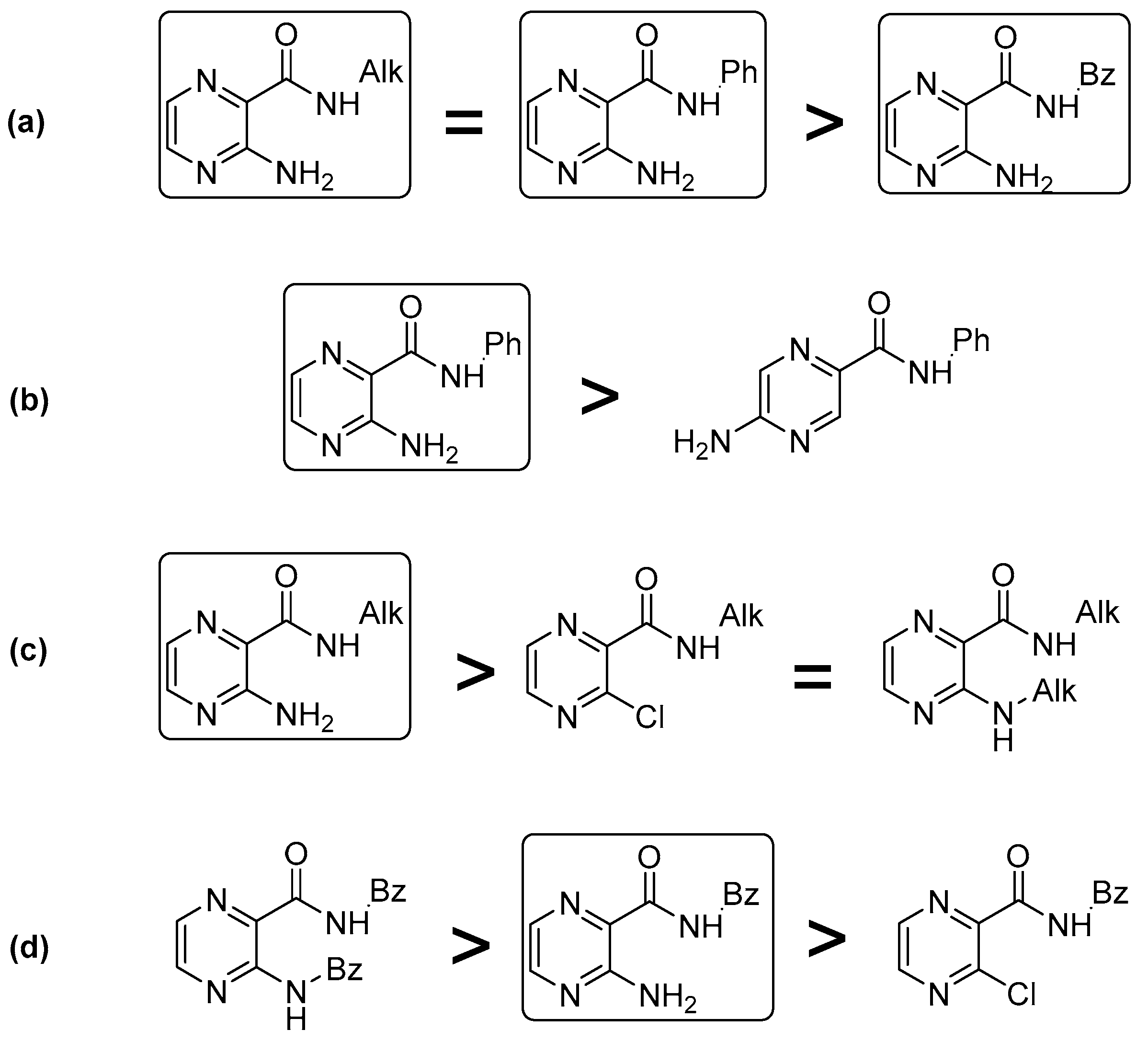
2.2. Predicted 3D Structure
2.3. Antimycobacterial Activity
2.4. Antibacterial and Antifungal Activity
2.5. Cytotoxicity
3. Experimental
3.1. General Information
3.2. Chemistry
3.2.1. General Procedure A
3.2.2. General Procedure B (Used for Compounds 1–2 and 5–20)
3.3. Analytical Data
3.4. Evaluation of Antimycobacterial Activity
3.4.1. Mycobacterium tuberculosis, Mycobacterium kansasii, and Mycobacterium avium
3.4.2. Mycobacterium smegmatis and Mycobacterium aurum
3.5. Evaluation of Antibacterial Activity
3.6. Evaluation of Antifungal Activity
3.7. Evaluation of Cytotoxicity
3.8. Determination of Lipophilicity by HPLC (Log k)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2018. WHO/CDC/TB/2018.20. Available online: http://www.who.int/tb/publications/global_report/en/ (accessed on 20 February 2019).
- World Health Organisation. The End TB Strategy. Global Strategy and Targets for Tuberculosis Prevention, Care and Control after 2015; WHO: Geneva, Switzerland, 2016; Available online: http://www.who.int/tb/ post2015_ TBstrategy.pdf?ua=1 (accessed on 20 February 2019).
- Singh, P.; Mishra, A.K.; Malonia, S.K.; Chauhan, D.S.; Sharma, V.D.; Venkatesan, K.; Katoch, V.M. The Paradox of Pyrazinamide: An Update on the Molecular Mechanisms of Pyrazinamide Resistance in Mycobacteria. J. Commun. Dis. 2006, 38, 288–298. [Google Scholar]
- Tripathi, R.P.; Tewari, N.; Dwivedi, N.; Tiwari, V.K. Fighting tuberculosis: An old disease with new challenges. Med. Res. Rev. 2005, 25, 93–131. [Google Scholar] [CrossRef]
- Zhang, Y.; Mitchison, D. The curious characteristics of pyrazinamide: A review. Int. J. Tuberc. Lung Dis. 2003, 7, 6–21. [Google Scholar]
- Zimhony, O.; Cox, J.S.; Welch, J.T.; Vilcheze, C.; Jacobs, W.R., Jr. Pyrazinamide inhibits the eukaryotic-like fatty acid synthetase I (FASI) of Mycobacterium tuberculosis. Nat. Med. 2000, 6, 1043–1047. [Google Scholar] [CrossRef] [PubMed]
- Zimhony, O.; Vilcheze, C.; Arai, M.; Welch, J.T.; Jacobs, W.R., Jr. Pyrazinoic acid and its n-propyl ester inhibit fatty acid synthase type I in replicating tubercle bacilli. Antimicrob. Agents Chemother. 2007, 51, 752–754. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.L.; Chen, J.Z.; Feng, J.; Cui, P.; Zhang, S.; Weng, X.H.; Zhang, W.; Zhang, Y. Aspartate decarboxylase (PanD) as a new target of pyrazinamide in Mycobacterium tuberculosis. Emerg. Microbes Infect. 2014, 3, e58. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Shibayama, K.; Rimbara, E.; Mori, S. Biochemical Characterization of Quinolinic Acid Phosphoribosyltransferase from Mycobacterium tuberculosis H37Rv and Inhibition of Its Activity by Pyrazinamide. PLoS ONE 2014, 9, e100062. [Google Scholar] [CrossRef]
- Shi, W.; Zhang, X.; Jiang, X.; Yuan, H.; Lee, J.S.; Barry, C.E., 3rd; Wang, H.; Zhang, W.; Zhang, Y. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 2011, 333, 1630–1632. [Google Scholar] [CrossRef]
- Yang, J.; Liu, Y.; Bi, J.; Cai, Q.; Liao, X.; Li, W.; Guo, C.; Zhang, Q.; Lin, T.; Zhao, Y.; et al. Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide. Mol. Microbiol. 2015, 95, 791–803. [Google Scholar] [CrossRef]
- Dillon, N.A.; Peterson, N.D.; Feaga, H.A.; Keiler, K.C.; Baughn, A.D. Anti-tubercular Activity of Pyrazinamide is Independent of trans-Translation and RpsA. Sci. Rep. 2017, 7, 6135. [Google Scholar] [CrossRef] [PubMed]
- Simoes, M.F.; Valente, E.; Gomez, M.J.R.; Anes, E.; Constantino, L. Lipophilic pyrazinoic acid amide and ester prodrugs Stability, activation and activity against M. tuberculosis. Eur. J. Pharm. Sci. 2009, 37, 257–263. [Google Scholar] [CrossRef]
- Semelkova, L.; Konecna, K.; Paterova, P.; Kubicek, V.; Kunes, J.; Novakova, L.; Marek, J.; Naesens, L.; Pesko, M.; Kralova, K.; et al. Synthesis and Biological Evaluation of N-Alkyl-3-(alkylamino)-pyrazine-2-carboxamides. Molecules 2015, 20, 8687–8711. [Google Scholar] [CrossRef] [PubMed]
- Semelkova, L.; Jandourek, O.; Konecna, K.; Paterova, P.; Navratilova, L.; Trejtnar, F.; Kubíček, V.; Kuneš, J.; Doležal, M.; Zitko, J. 3-Substituted N-Benzylpyrazine-2-carboxamide Derivatives: Synthesis, Antimycobacterial and Antibacterial Evaluation. Molecules 2017, 22, 495. [Google Scholar] [CrossRef] [PubMed]
- Zitko, J.; Franco, F.; Paterova, P. Synthesis and anti-infective evaluation of 5-amino-N-phenylpyrazine-2-carboxamides. Ceska Slov. Farm. 2015, 64, 19–24. [Google Scholar] [PubMed]
- Kajino, M.; Morimoto, S.; Inaba, A.; Nagaya, H. Preparation and Formulation of Quinazoline Derivatives as Allergy Inhibitors. WO 9914203, 25 March 1999. [Google Scholar]
- Luo, H.; Shi, J.; Lu, L.; Wu, F.; Zhou, M.; Hou, X.; Zhang, W.; Ding, Z.; Li, R. Molecular dynamics-based self-organizing molecular field analysis on 3-amino-6-arylpyrazines as the ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase inhibitors. Med. Chem. Res. 2014, 23, 747–758. [Google Scholar] [CrossRef]
- Bouz, G.; Juhas, M.; Niklova, P.; Jandourek, O.; Paterova, P.; Janousek, J.; Tůmová, L.; Kovalíková, Z.; Kastner, P.; Doležal, M.; et al. Ureidopyrazine Derivatives: Synthesis and Biological Evaluation as Anti-infectives and Abiotic Elicitors. Molecules 2017, 22, 1797. [Google Scholar] [CrossRef] [PubMed]
- Eric, V.; Bradley, M. Amide Bond Formation: Beyond the Myth of Coupling Reagents. Chem. Soc. Rev. 2009, 38, 606–631. [Google Scholar]
- Shalaeva, M.; Caron, G.; Abramov, Y.A.; O’Connell, T.N.; Plummer, M.S.; Yalamanchi, G.; Farley, K.A.; Goetz, G.H.; Philippe, L.; Shapiro, M.J. Integrating Intramolecular Hydrogen Bonding (IMHB) Considerations in Drug Discovery Using Delta logP As a Tool. J. Med. Chem. 2013, 56, 4870–4879. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, T.A.; Brown, A.J.; Bell, I.A.W.; Cockroft, S.L. The Limit of Intramolecular H-Bonding. JACS 2016, 138, 15114–15117. [Google Scholar] [CrossRef]
- Nagy, P.I. Competing Intramolecular vs. Intermolecular Hydrogen Bonds in Solution. Int. J. Mol. Sci. 2014, 15, 19562–19633. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, C.H.; Tsai, T.F.; Hsueh, P.R. Characteristics of skin and soft tissue infection caused by non-tuberculous mycobacteria in Taiwan. Int. J. Tuberc. Lung Dis. 2011, 15, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Bhakta, S. An integrated surrogate model for screening of drugs against Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2012, 67, 1380–1391. [Google Scholar] [CrossRef]
- Servusova, B.; Paterova, P.; Mandikova, J.; Kubicek, V.; Kucera, R.; Kunes, J.; Doležal, M.; Zitko, J. Alkylamino derivatives of pyrazinamide: Synthesis and antimycobacterial evaluation. Bioorg. Med. Chem. Lett. 2014, 24, 450–453. [Google Scholar] [CrossRef] [PubMed]
- Servusova-Vanaskova, B.; Jandourek, O.; Paterova, P.; Kordulakova, J.; Plevakova, M.; Kubicek, V.; Kucera, R.; Garaj, V.; Naesens, L.; Kunes, J.; et al. Alkylamino derivatives of N-benzylpyrazine-2-carboxamide: Synthesis and antimycobacterial evaluation. MedChemComm 2015, 6, 1311–1317. [Google Scholar] [CrossRef]
- Zitko, J.; Servusova, B.; Janoutova, A.; Paterova, P.; Mandikova, J.; Garaj, V.; Vejsová, M.; Marek, J.; Doležal, M. Synthesis and antimycobacterial evaluation of 5-alkylamino-N-phenylpyrazine-2-carboxamides. Bioorg. Med. Chem. 2015, 23, 174–183. [Google Scholar] [CrossRef]
- Servusova-Vanaskova, B.; Paterova, P.; Garaj, V.; Mandikova, J.; Kunes, J.; Naesens, L.; Jilek, P.; Dolezal, M.; Zitko, J. Synthesis and Antimicrobial Evaluation of 6-Alkylamino-N-phenylpyrazine-2-carboxamides. Chem. Biol. Drug Des 2015, in press. [Google Scholar] [CrossRef]
- El Bouazzi, O.; Hammi, S.; Bourkadi, J.E.; Tebaa, A.; Tanani, D.S.; Soulaymani-Bencheikh, R.; Badrane, N.; Bengueddour, R. First line anti-tuberculosis induced hepatotoxicity: Incidence and risk factors. Pan Afr. Med. J. 2016, 25, 167. [Google Scholar] [CrossRef]
- Tostmann, A.; Boeree, M.J.; Peters, W.H.M.; Roelofs, H.M.J.; Aarnoutse, R.E.; van der Ven, A.J.A.M.; Dekhuijzen, P.N. Isoniazid and its toxic metabolite hydrazine induce in vitro pyrazinamide toxicity. Int. J. Antimicrob. Agents 2008, 31, 577–580. [Google Scholar] [CrossRef]
- Ellingson, R.C.; Henry, R.L.; McDonald, F.G. Pyrazine Chemistry. I. Derivatives of 3-Aminopyrazinoic Acid. JACS 1945, 67, 1711–1713. [Google Scholar] [CrossRef]
- Dermer, O.C.; King, J. N-BENZYLAMIDES AS DERIVATIVES FOR IDENTIFYING THE ACYL GROUP IN ESTERS1,2. JOC 1943, 8, 168–173. [Google Scholar] [CrossRef]
- Clark, J.; Neath, G.; Smith, C. Heterocyclic Studies. 7. Action of Methoxyamine and Methylhydrazines on 4-Hydroxypteridine and Its Methyl Derivatives. J. Chem. Soc. C-Org. 1969, 1297–1301. [Google Scholar] [CrossRef]
- EUCAST DISCUSSION DOCUMENT E.Dis 5.1. Determination of Minimum Inhibitory Concentrations (MICs) of Antibacterial Agents by Broth Dilution. Clin. Microbiol. Infect. 2003, 9, 1–7. [Google Scholar]
- EUCAST DEFINITIVE DOCUMENT E.DEF 7.3.1. Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Yeasts; EUCAST: Växjö, Sweden, 2017; pp. 1–21. [Google Scholar]
- EUCAST DEFINITIVE DOCUMENT E.DEF 9.3.1. Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Conidia Forming Moulds; EUCAST: Växjö, Sweden, 2017; pp. 1–23. [Google Scholar]
Sample Availability: Samples of title compounds are available from the authors. |

| Cpd. | Yield [%] | |
|---|---|---|
| Procedure A | Procedure B | |
| 1 | 29 | 75 |
| 2 | 27 | 91 |
| 3 | 69 | - |
| 4 | 57 | - |
| 5 | 39 | 26 |
| 6 | 45 | 79 |
| 7 | 18 | 74 |
| 8 | 16 | 55 |
| Structure | No. | R’ | log P* | ClogP * | MIC [µg/mL (µM)] | |
|---|---|---|---|---|---|---|
| Mtb | M. smegmatis | |||||
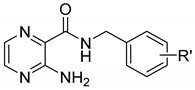 | 1 | H | 0.57 | 1.89 | ˃100(438.1) | ˃500(2191) |
| 2 | 2-CH3 | 1.06 | 2.34 | ˃100(412.7) | ˃500(2063.7) | |
| 3 | 4-CH3 | 1.06 | 2.39 | 100(412.7) | ˃500(2063.7) | |
| 4 | 4-OCH3 | 0.44 | 1.80 | 100(387.2) | ˃500(1935.8) | |
| 5 | 2,4-diOCH3 | 0.32 | 1.90 | ˃100(346.8) | ˃500(1734.2) | |
| 6 | 3,4-diCl | 1.69 | 3.20 | ˃100(336.5) | ˃500(1682.7) | |
| 7 | 3-CF3 | 1.49 | 2.77 | 50(168.8) | 250(843.9) | |
| 8 | 4-CF3 | 1.49 | 2.77 | 50(168.8) | ˃500(1687.8) | |
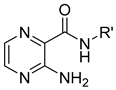 | 9 | C5H11 | 0.50 | 2.04 | ˃100(480.1) | ˃500(2400.7) |
| 10 | C6H13 | 0.91 | 2.57 | 100(449.9) | 250(1124.7) | |
| 11 | C7H15 | 1.33 | 3.10 | 50(211.6) | 62.5(264.5) | |
| 12 | C8H17 | 1.75 | 3.63 | 25 (99.9) | 62.5(249.7) | |
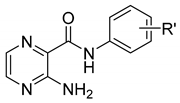 | 13 | H | 0.50 | 1.71 | ˃100(466.8) | ˃500(2333.9) |
| 14 | 2-OH | 0.11 | 1.04 | ˃100(434.3) | 125(542.9) | |
| 15 | 2,5-diCH3 | 1.48 | 2.06 | ˃100(412.7) | ˃500(2063.7) | |
| 16 | 4-C2H5 | 1.41 | 2.74 | ˃100(412.7) | ˃500(2063.7) | |
| 17 | 2,4-diOCH3 | 0.25 | 1.04 | 12.5(45.6) | ˃500(1823) | |
| 18 | 2,4-diF | 0.82 | 1.44 | ˃100(399.7) | ˃500(1998.3) | |
| 19 | 3,4-diCl | 1.62 | 3.06 | 50(176.6) | ˃500(1766.1) | |
| 20 | 4-CF3 | 1.42 | 2.65 | 50(176.6) | 31.25(110.7) | |
| PZA | 100 | ˃100 | ||||
| INH | 0.1–0.2 | 7.81–15.62 | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouz, G.; Semelková, L.; Janďourek, O.; Konečná, K.; Paterová, P.; Navrátilová, L.; Kubíček, V.; Kuneš, J.; Doležal, M.; Zitko, J. Derivatives of 3-Aminopyrazine-2-carboxamides: Synthesis, Antimicrobial Evaluation, and in Vitro Cytotoxicity. Molecules 2019, 24, 1212. https://doi.org/10.3390/molecules24071212
Bouz G, Semelková L, Janďourek O, Konečná K, Paterová P, Navrátilová L, Kubíček V, Kuneš J, Doležal M, Zitko J. Derivatives of 3-Aminopyrazine-2-carboxamides: Synthesis, Antimicrobial Evaluation, and in Vitro Cytotoxicity. Molecules. 2019; 24(7):1212. https://doi.org/10.3390/molecules24071212
Chicago/Turabian StyleBouz, Ghada, Lucia Semelková, Ondřej Janďourek, Klára Konečná, Pavla Paterová, Lucie Navrátilová, Vladimír Kubíček, Jiří Kuneš, Martin Doležal, and Jan Zitko. 2019. "Derivatives of 3-Aminopyrazine-2-carboxamides: Synthesis, Antimicrobial Evaluation, and in Vitro Cytotoxicity" Molecules 24, no. 7: 1212. https://doi.org/10.3390/molecules24071212
APA StyleBouz, G., Semelková, L., Janďourek, O., Konečná, K., Paterová, P., Navrátilová, L., Kubíček, V., Kuneš, J., Doležal, M., & Zitko, J. (2019). Derivatives of 3-Aminopyrazine-2-carboxamides: Synthesis, Antimicrobial Evaluation, and in Vitro Cytotoxicity. Molecules, 24(7), 1212. https://doi.org/10.3390/molecules24071212








