PEG-OligoRNA Hybridization of mRNA for Developing Sterically Stable Lipid Nanoparticles toward In Vivo Administration
Abstract
1. Introduction
2. Results
2.1. Hybridization of mRNA with PEG-OligoRNAs
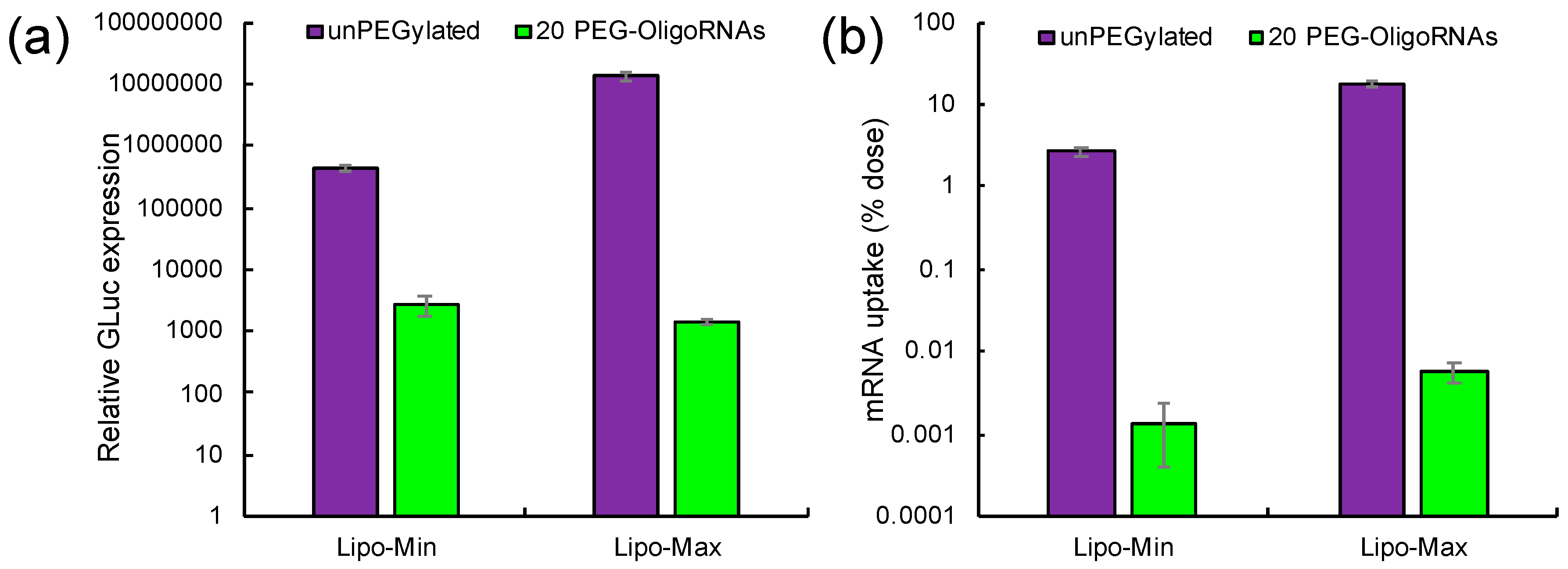
2.2. Translation Activity of mRNA after Hybridization with PEG-OligoRNAs
2.3. Characterization of Lipofectamine LTX-based LNPs Loading PEG-OligoRNAs/mRNA
2.4. Transmission Electron Microscopic Observation of Lipofectamine LTX-based LNPs
2.5. Loading of PEG-OligoRNA/mRNA to DOTAP/Chol liposome
2.6. Introduction of Lipofectamine LTX-Based LNPs to Cultured Cells
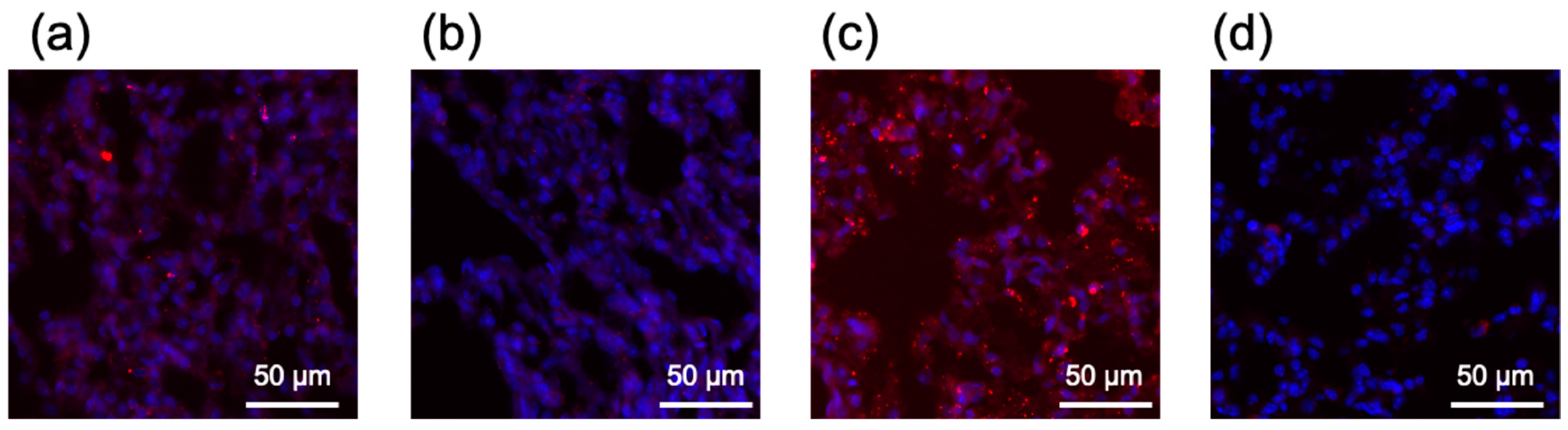
2.7. In Vivo Behavior of Lipofectamine LTX-Based LNPs after Systemic Delivery
3. Discussion
4. Materials and Methods
4.1. Preparation of mRNA
4.2. Hybridization of mRNA with PEG-OligoRNAs
4.3. Cell Free Translation
4.4. Preparation of Lipofectamine LTX/mRNA LNPs
4.5. Characterization of LNPs
4.6. Transmission Electron Microscopic Observation
4.7. Preparation of DOTAP/Chol LNPs
4.8. mRNA Introduction to Cultured Cells
4.9. Systemic Injection of LNPs to Mice
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sahin, U.; Kariko, K.; Tureci, O. mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef]
- Hajj, K.A.; Whitehead, K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017, 2, 17056. [Google Scholar] [CrossRef]
- Kranz, L.M.; Diken, M.; Haas, H.; Kreiter, S.; Loquai, C.; Reuter, K.C.; Meng, M.; Fritz, D.; Vascotto, F.; Hefesha, H.; et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 2016, 534, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. [Google Scholar] [CrossRef]
- Yin, H.; Song, C.Q.; Suresh, S.; Wu, Q.; Walsh, S.; Rhym, L.H.; Mintzer, E.; Bolukbasi, M.F.; Zhu, L.J.; Kauffman, K.; et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017, 35, 1179–1187. [Google Scholar] [CrossRef]
- Huebner, S.; Battersby, B.J.; Grimm, R.; Cevc, G. Lipid-DNA Complex Formation: Reorganization and Rupture of Lipid Vesicles in the Presence of DNA As Observed by Cryoelectron Microscopy. Biophys. J. 1999, 76, 3158–3166. [Google Scholar] [CrossRef]
- Fenton, O.S.; Kauffman, K.J.; Kaczmarek, J.C.; McClellan, R.L.; Jhunjhunwala, S.; Tibbitt, M.W.; Zeng, M.D.; Appel, E.A.; Dorkin, J.R.; Mir, F.F.; et al. Synthesis and Biological Evaluation of Ionizable Lipid Materials for the In Vivo Delivery of Messenger RNA to B Lymphocytes. Adv. Mater. 2017, 29, 1606944. [Google Scholar] [CrossRef] [PubMed]
- Robinson, E.; MacDonald, K.D.; Slaughter, K.; McKinney, M.; Patel, S.; Sun, C.; Sahay, G. Lipid Nanoparticle-Delivered Chemically Modified mRNA Restores Chloride Secretion in Cystic Fibrosis. Mol. Ther. 2018, 26, 2034–2046. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, J.C.; Patel, A.K.; Kauffman, K.J.; Fenton, O.S.; Webber, M.J.; Heartlein, M.W.; DeRosa, F.; Anderson, D.G. Polymer-Lipid Nanoparticles for Systemic Delivery of mRNA to the Lungs. Angew. Chem. 2016, 55, 13808–13812. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Nakatani, T.; Furihata, T.; Tange, K.; Nakai, Y.; Yoshioka, H.; Harashima, H.; Akita, H. In Vivo Introduction of mRNA Encapsulated in Lipid Nanoparticles to Brain Neuronal Cells and Astrocytes via Intracerebroventricular Administration. Mol. Pharm. 2018, 15, 2060–2067. [Google Scholar] [CrossRef]
- Immordino, M.L.; Dosio, F.; Cattel, L. Stealth liposomes: Review of the basic science, rationale, and clinical applications, existing and potential. Int. J. Nanomed. 2006, 1, 297–315. [Google Scholar]
- Campbell, F.; Bos, F.L.; Sieber, S.; Arias-Alpizar, G.; Koch, B.E.; Huwyler, J.; Kros, A.; Bussmann, J. Directing Nanoparticle Biodistribution through Evasion and Exploitation of Stab2-Dependent Nanoparticle Uptake. ACS Nano 2018, 12, 2138–2150. [Google Scholar] [CrossRef]
- Kariko, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Kormann, M.S.; Hasenpusch, G.; Aneja, M.K.; Nica, G.; Flemmer, A.W.; Herber-Jonat, S.; Huppmann, M.; Mays, L.E.; Illenyi, M.; Schams, A.; et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nature Biotech. 2011, 29, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Andries, O.; Mc Cafferty, S.; De Smedt, S.C.; Weiss, R.; Sanders, N.N.; Kitada, T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 2015, 217, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Kataoka, K.; Itaka, K. Screening of mRNA Chemical Modification to Maximize Protein Expression with Reduced Immunogenicity. Pharmaceutics 2015, 7, 137–151. [Google Scholar] [CrossRef]
- Li, B.; Luo, X.; Dong, Y. Effects of Chemically Modified Messenger RNA on Protein Expression. Bioconjugate Chem 2016, 27, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Yoshinaga, N.; Yanagihara, K.; Yuba, E.; Kataoka, K.; Itaka, K. Designing immunostimulatory double stranded messenger RNA with maintained translational activity through hybridization with poly A sequences for effective vaccination. Biomaterials 2018, 150, 162–170. [Google Scholar] [CrossRef]
- Yoshinaga, N.; Uchida, S.; Naito, M.; Osada, K.; Cabral, H.; Kataoka, K. Induced packaging of mRNA into polyplex micelles by regulated hybridization with a small number of cholesteryl RNA oligonucleotides directed enhanced in vivo transfection. Biomaterials 2019, 197, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Sato, K.; Asai, K.; Akutsu, T. Rtips: Fast and accurate tools for RNA 2D structure prediction using integer programming. Nucleic Acids Res. 2012, 40, W29-34. [Google Scholar] [CrossRef] [PubMed]
- Petros, R.A.; DeSimone, J.M. Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Discov. 2010, 9, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Anraku, Y.; Kishimura, A.; Kobayashi, A.; Oba, M.; Kataoka, K. Size-controlled long-circulating PICsome as a ruler to measure critical cut-off disposition size into normal and tumor tissues. Chem. Commun. 2011, 47, 6054–6056. [Google Scholar] [CrossRef] [PubMed]
- Cabral, H.; Matsumoto, Y.; Mizuno, K.; Chen, Q.; Murakami, M.; Kimura, M.; Terada, Y.; Kano, M.R.; Miyazono, K.; Uesaka, M.; et al. Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nat. Nanotechnol. 2011, 6, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Chollet, P.; Favrot, M.C.; Hurbin, A.; Coll, J.L. Side-effects of a systemic injection of linear polyethylenimine-DNA complexes. J. Gene Med. 2002, 4, 84–91. [Google Scholar] [CrossRef]
- Petsch, B.; Schnee, M.; Vogel, A.B.; Lange, E.; Hoffmann, B.; Voss, D.; Schlake, T.; Thess, A.; Kallen, K.J.; Stitz, L.; et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012, 30, 1210–1216. [Google Scholar] [CrossRef]
- Seelenfreund, E.; Robinson, W.A.; Amato, C.M.; Tan, A.-C.; Kim, J.; Robinson, S.E. Long Term Storage of Dry versus Frozen RNA for Next Generation Molecular Studies. PLoS ONE 2014, 9, e111827. [Google Scholar] [CrossRef]
- Rouskin, S.; Zubradt, M.; Washietl, S.; Kellis, M.; Weissman, J.S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 2014, 505, 701–705. [Google Scholar] [CrossRef]
- Jokerst, J.V.; Lobovkina, T.; Zare, R.N.; Gambhir, S.S. Nanoparticle PEGylation for imaging and therapy. Nanomedicine 2011, 6, 715–728. [Google Scholar] [CrossRef]
- Hatakeyama, H.; Akita, H.; Harashima, H. A multifunctional envelope type nano device (MEND) for gene delivery to tumours based on the EPR effect: a strategy for overcoming the PEG dilemma. Adv. Drug Deliv. Rev. 2011, 63, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, H.; Akita, H.; Kogure, K.; Oishi, M.; Nagasaki, Y.; Kihira, Y.; Ueno, M.; Kobayashi, H.; Kikuchi, H.; Harashima, H. Development of a novel systemic gene delivery system for cancer therapy with a tumor-specific cleavable PEG-lipid. Gene Ther. 2007, 14, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Takae, S.; Miyata, K.; Oba, M.; Ishii, T.; Nishiyama, N.; Itaka, K.; Yamasaki, Y.; Koyama, H.; Kataoka, K. PEG-detachable polyplex micelles based on disulfide-linked block catiomers as bioresponsive nonviral gene vectors. J. Am. Chem. Soc. 2008, 130, 6001–6009. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, H.; Akita, H.; Ito, E.; Hayashi, Y.; Oishi, M.; Nagasaki, Y.; Danev, R.; Nagayama, K.; Kaji, N.; Kikuchi, H.; et al. Systemic delivery of siRNA to tumors using a lipid nanoparticle containing a tumor-specific cleavable PEG-lipid. Biomaterials 2011, 32, 4306–4316. [Google Scholar] [CrossRef]
- Uchida, S.; Itaka, K.; Chen, Q.; Osada, K.; Ishii, T.; Shibata, M.A.; Harada-Shiba, M.; Kataoka, K. PEGylated Polyplex With Optimized PEG Shielding Enhances Gene Introduction in Lungs by Minimizing Inflammatory Responses. Mol. Ther. 2012, 20, 1196–1203. [Google Scholar] [CrossRef]
- Ge, Z.; Chen, Q.; Osada, K.; Liu, X.; Tockary, T.A.; Uchida, S.; Dirisala, A.; Ishii, T.; Nomoto, T.; Toh, K.; et al. Targeted gene delivery by polyplex micelles with crowded PEG palisade and cRGD moiety for systemic treatment of pancreatic tumors. Biomaterials 2014, 35, 3416–3426. [Google Scholar] [CrossRef]
- Gjetting, T.; Arildsen, N.S.; Christensen, C.L.; Poulsen, T.T.; Roth, J.A.; Handlos, V.N.; Poulsen, H.S. In vitro and in vivo effects of polyethylene glycol (PEG)-modified lipid in DOTAP/cholesterol-mediated gene transfection. Int. J. Nanomed. 2010, 5, 371–383. [Google Scholar]
- Pitard, B.; Oudrhiri, N.; Lambert, O.; Vivien, E.; Masson, C.; Wetzer, B.; Hauchecorne, M.; Scherman, D.; Rigaud, J.L.; Vigneron, J.P.; et al. Sterically stabilized BGTC-based lipoplexes: structural features and gene transfection into the mouse airways in vivo. J. Gene Med. 2001, 3, 478–487. [Google Scholar] [CrossRef]
- Su, X.; Fricke, J.; Kavanagh, D.G.; Irvine, D.J. In Vitro and in Vivo mRNA Delivery Using Lipid-Enveloped pH-Responsive Polymer Nanoparticles. Mol. Pharmaceut. 2011, 8, 774–787. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Kinoh, H.; Ishii, T.; Matsui, A.; Tockary, T.A.; Takeda, K.M.; Uchida, H.; Osada, K.; Itaka, K.; Kataoka, K. Systemic delivery of messenger RNA for the treatment of pancreatic cancer using polyplex nanomicelles with a cholesterol moiety. Biomaterials 2016, 82, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, W.; He, Y.; Li, Y.; Yan, E.Z.; Zhang, K.; Irvine, D.J.; Hammond, P.T. Structurally Programmed Assembly of Translation Initiation Nanoplex for Superior mRNA Delivery. ACS Nano 2017, 11, 2531–2544. [Google Scholar] [CrossRef] [PubMed]
- Perche, F.; Uchida, S.; Akiba, H.; Lin, C.Y.; Ikegami, M.; Dirisala, A.; Nakashima, T.; Itaka, K.; Tsumoto, K.; Kataoka, K. Improved Brain Expression of Anti-Amyloid beta scFv by Complexation of mRNA Including a Secretion Sequence with PEG-based Block Catiomer. Curr. Alzheimer Res. 2017, 14, 295–302. [Google Scholar] [CrossRef]
- Cabral, H.; Miyata, K.; Osada, K.; Kataoka, K. Block Copolymer Micelles in Nanomedicine Applications. Chem. Rev. 2018, 118, 6844–6892. [Google Scholar] [CrossRef] [PubMed]
- Nomoto, T.; Matsumoto, Y.; Miyata, K.; Oba, M.; Fukushima, S.; Nishiyama, N.; Yamasoba, T.; Kataoka, K. In situ quantitative monitoring of polyplexes and polyplex micelles in the blood circulation using intravital real-time confocal laser scanning microscopy. J. Control. Release 2011, 151, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Itaka, K.; Uchida, H.; Hayakawa, K.; Ogata, T.; Ishii, T.; Fukushima, S.; Osada, K.; Kataoka, K. In vivo messenger RNA introduction into the central nervous system using polyplex nanomicelle. PLoS ONE 2013, 8, e56220. [Google Scholar] [CrossRef] [PubMed]
- Matsui, A.; Uchida, S.; Hayashi, A.; Kataoka, K.; Itaka, K. Prolonged engraftment of transplanted hepatocytes in the liver by transient pro-survival factor supplementation using ex vivo mRNA transfection. J. Control. Release 2018, 285, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, R. Nanoparticle-mediated gene delivery to the lung. Methods Mol. Biol. 2008, 433, 301–331. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |





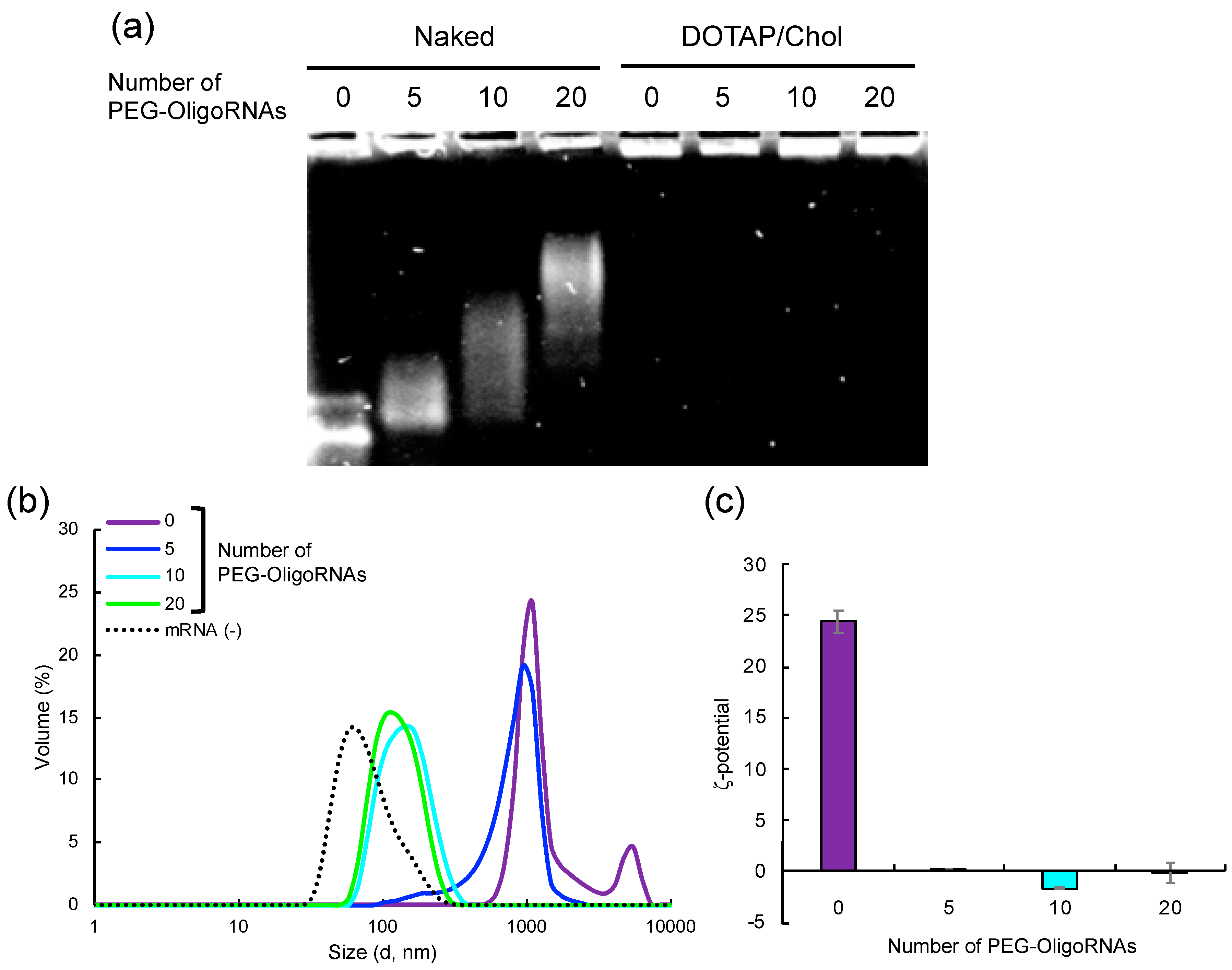

| Number of PEG-OligoRNAs | 5 | 10 | 20 |
| Hybridization efficiency (%) | 87 | 95 | 94 |
| Lipo-Min | Lipo-Max | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| mRNA (−) | Number of PEG-OligoRNAs | mRNA (−) | Number of PEG-OligoRNAs | |||||||
| 0 | 5 | 10 | 20 | 0 | 5 | 10 | 20 | |||
| Size (nm) a | 43 | 4071 | 234 | 90 | 78 | 25 | 254 | 132 | 68 | 57 |
| PDI b | 0.42 | 0.26 | 0.19 | 0.13 | 0.21 | 0.45 | 0.20 | 0.19 | 0.12 | 0.18 |
| mRNA (−) | Number of PEG-OligoRNAs | ||||
|---|---|---|---|---|---|
| 0 | 5 | 10 | 20 | ||
| Size (nm) a | 84 | 1698 | 853 | 152 | 132 |
| PDI b | 0.15 | 0.31 | 0.47 | 0.10 | 0.08 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurimoto, S.; Yoshinaga, N.; Igarashi, K.; Matsumoto, Y.; Cabral, H.; Uchida, S. PEG-OligoRNA Hybridization of mRNA for Developing Sterically Stable Lipid Nanoparticles toward In Vivo Administration. Molecules 2019, 24, 1303. https://doi.org/10.3390/molecules24071303
Kurimoto S, Yoshinaga N, Igarashi K, Matsumoto Y, Cabral H, Uchida S. PEG-OligoRNA Hybridization of mRNA for Developing Sterically Stable Lipid Nanoparticles toward In Vivo Administration. Molecules. 2019; 24(7):1303. https://doi.org/10.3390/molecules24071303
Chicago/Turabian StyleKurimoto, Shota, Naoto Yoshinaga, Kazunori Igarashi, Yu Matsumoto, Horacio Cabral, and Satoshi Uchida. 2019. "PEG-OligoRNA Hybridization of mRNA for Developing Sterically Stable Lipid Nanoparticles toward In Vivo Administration" Molecules 24, no. 7: 1303. https://doi.org/10.3390/molecules24071303
APA StyleKurimoto, S., Yoshinaga, N., Igarashi, K., Matsumoto, Y., Cabral, H., & Uchida, S. (2019). PEG-OligoRNA Hybridization of mRNA for Developing Sterically Stable Lipid Nanoparticles toward In Vivo Administration. Molecules, 24(7), 1303. https://doi.org/10.3390/molecules24071303







