Assessment of Diversity in the Accessions of Setaria italica L. Based on Phytochemical and Morphological Traits and ISSR Markers
Abstract
:1. Introduction
2. Results and Discussion
2.1. Morphological Traits of S. italica Accessions
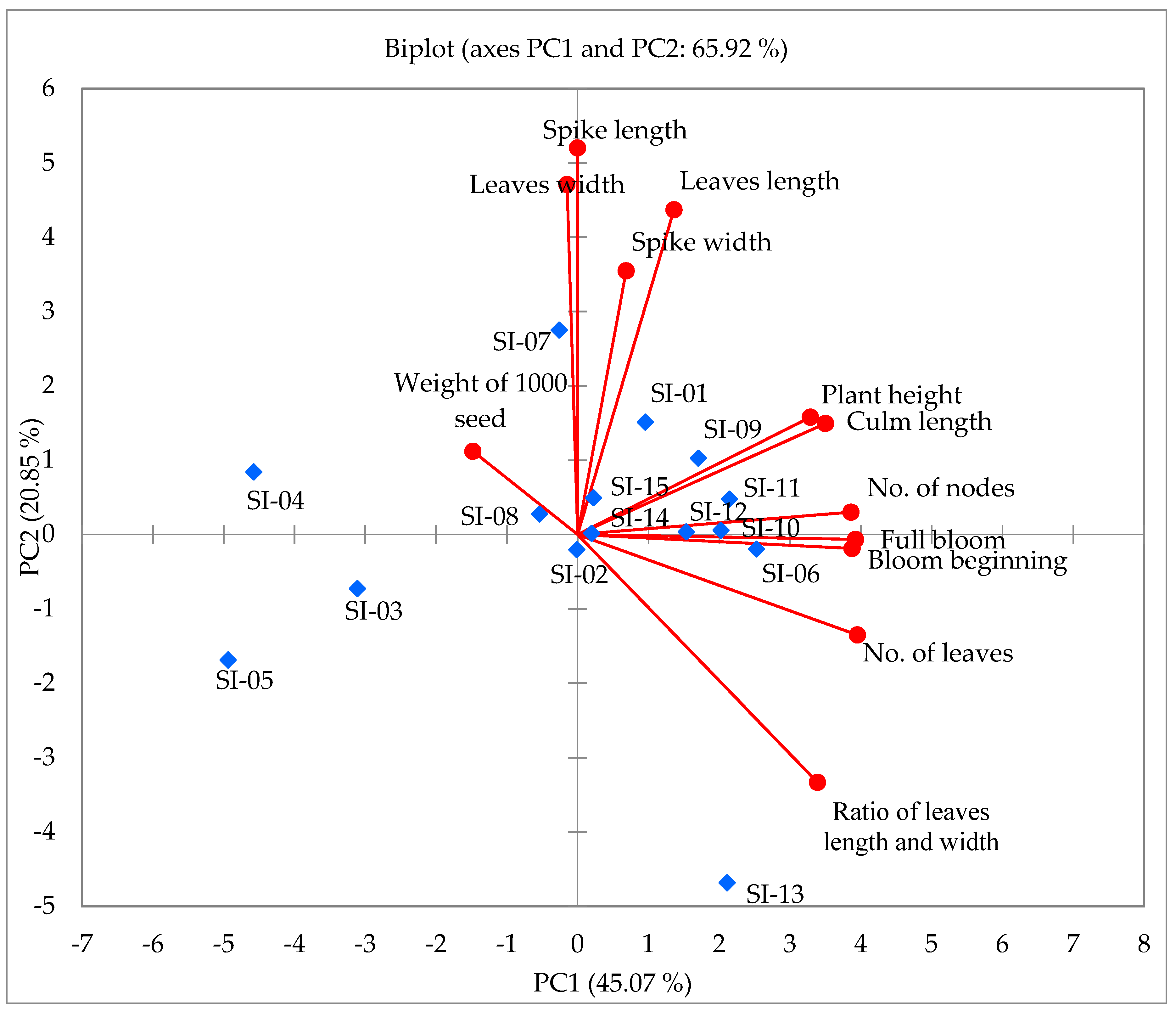
2.2. Principal Component Analysis
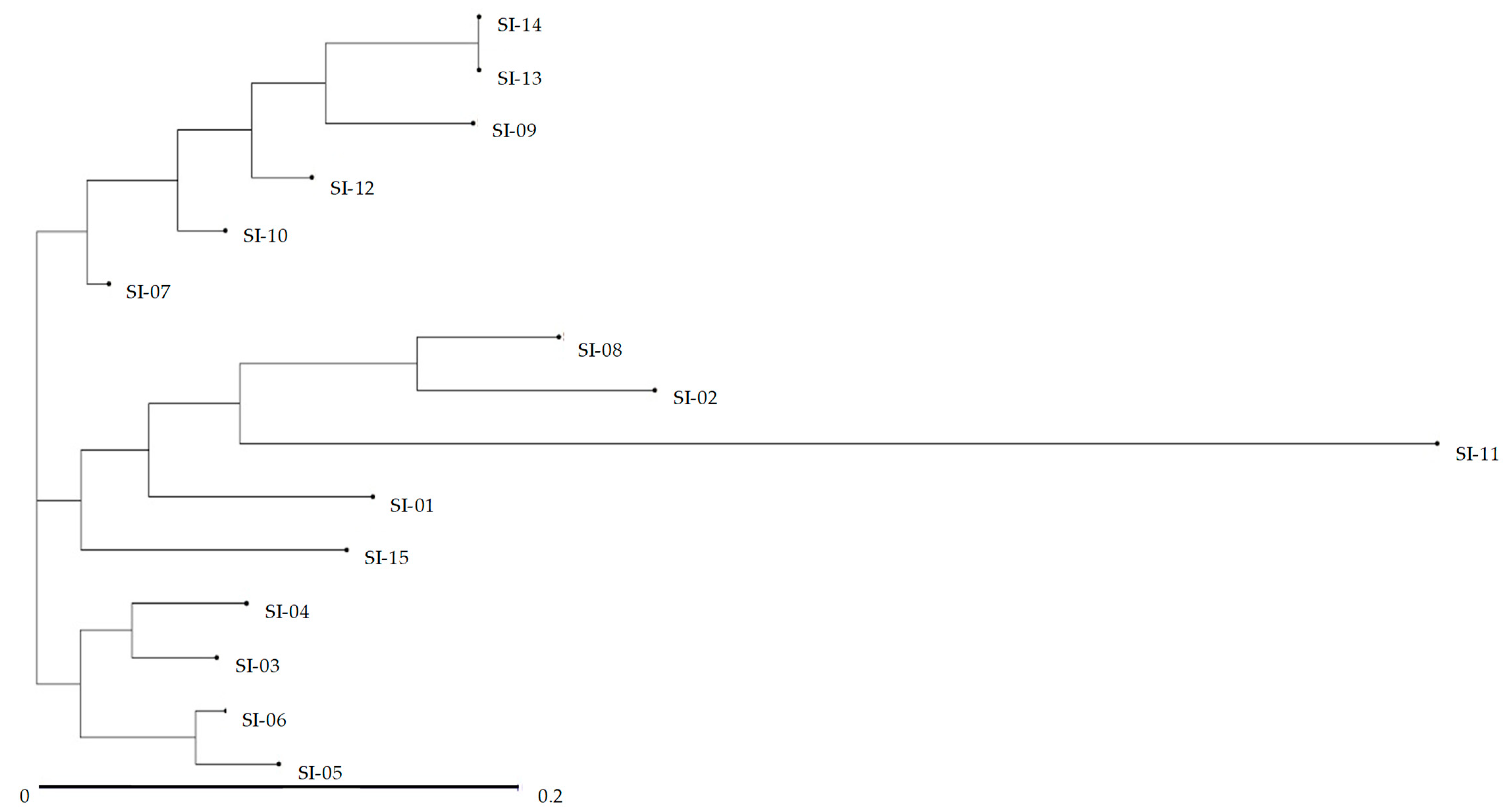
2.3. UPGMA Cluster Analysis
2.4. Identification and Quantification of Phenolic Compounds of S. italica by HPLC Analysis
2.5. In Vitro Antioxidant Capacity of S. italica Accessions
2.6. Screening of Antimicrobial Activity of S. italica Accessions Based on MIC
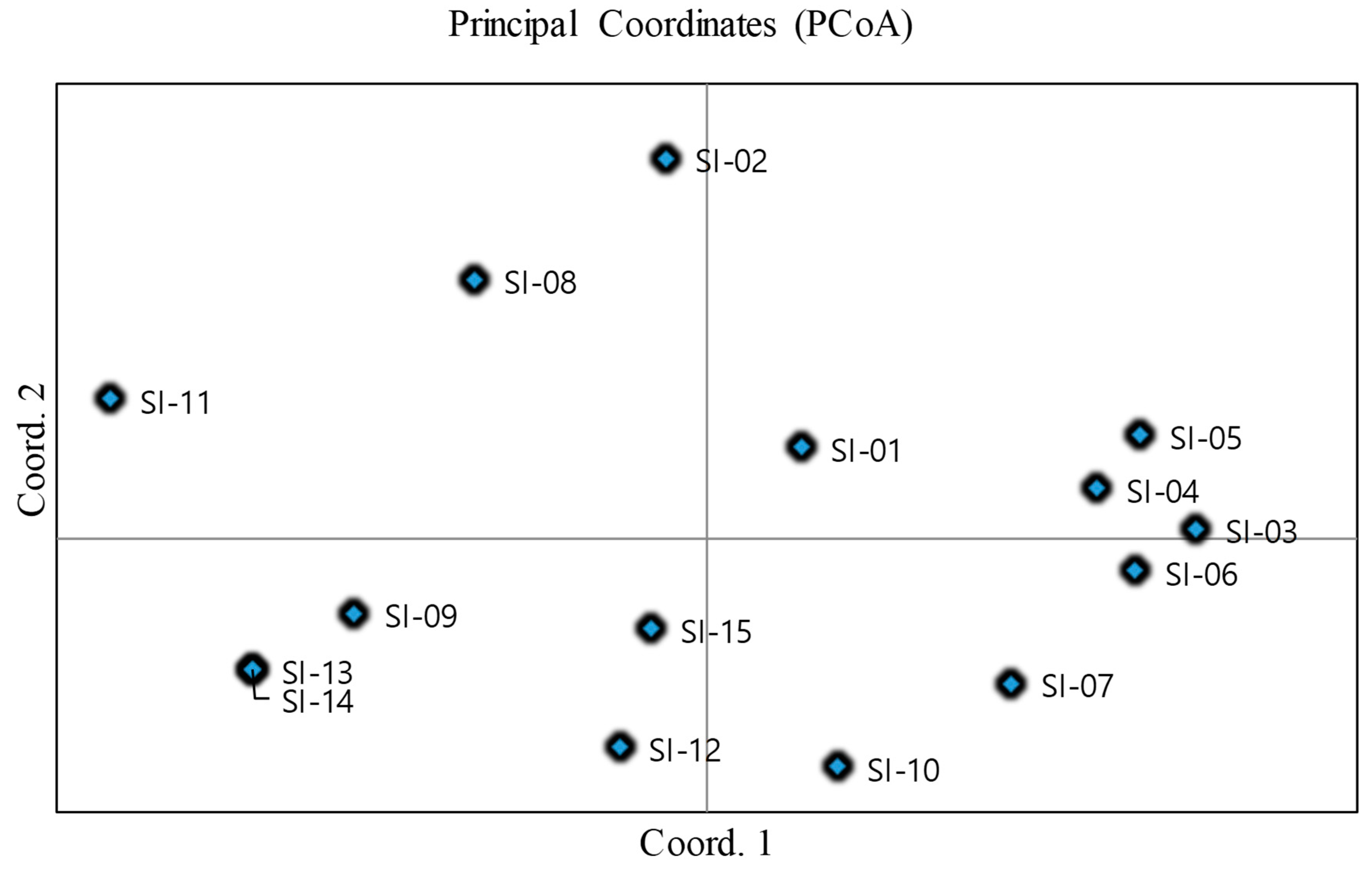
2.7. ISSR Marker Polymorphism
3. Material and Methods
3.1. Chemicals, Standard Compounds, and Solvents
3.2. Plant Materials
3.3. Identification and Quantification of Phenolic Compound Analysis by HPLC
3.4. Evaluation of Morphological Traits in S. italica Accessions
3.5. Screening of Antioxidant Activity of S. italica Accessions
3.6. Screening of Antimicrobial Activity of S. italica Accessions by Minimum Inhibitory Concentration (MIC)
3.6.1. Microbial Culture
3.6.2. DNA Extraction
3.6.3. PCR Amplification and Electrophoresis.
3.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sun, Y.L.; Zheng, S.L.; Lee, J.K.; Hong, S.K. Genetic diversity of foxtail millet (Setaria italica L.) from main Asian habitats based on the NRDNA its region. Pak. J. Bot. 2016, 48, 1637–1643. [Google Scholar]
- Paul, A.I.; Ibadapbiangshylla. Morphological and biochemical response to salinity stress on Setaria italica Seedlings. J. Appl. Adv. Res. 2017, 2, 235–248. [Google Scholar]
- Kumari, R.; Dikshit, N.; Sharma, D.; Bhat, K.V. Analysis of molecular genetic diversity in a representative collection of foxtail millet [Setaria italica (L.) P. Beauv.] from different agro-ecological regions of India. Physiol. Mol. Biol. Plants 2011, 17, 363–374. [Google Scholar] [CrossRef]
- Sakamoto, S. Origin and dispersal of common millet and foxtail millet. JARQ 1987, 21, 84–89. [Google Scholar]
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of Plants in the Old World: The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and the Mediterranean Basin; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Hirano, R.; Naito, K.; Fukunaga, K.; Watanabe, K.N.; Ohsawa, R.; Kawase, M. Genetic structure of landraces in foxtail millet (Setaria italica (L.) P. Beauv.) revealed with transposon display and interpretation to crop evolution of foxtail millet. Genome 2011, 54, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Doust, A.N.; Kellogg, E.A.; Devos, K.M.; Bennetzen, J.L. Foxtail millet: A sequence-driven grass model system. Plant Physiol. 2009, 149, 137–141. [Google Scholar] [CrossRef]
- Chandrasekara, A.; Naczk, M.; Shahidi, F. Effect of processing on the antioxidant activity of millet grains. Food Chem. 2012, 133, 1–9. [Google Scholar] [CrossRef]
- Hegde, P.S.; Chandra, T.S. ESR spectroscopic study reveals higher free radical quenching potential in kodo millet (Paspalum scrobiculatum) compared to other millets. Food Chem. 2005, 92, 177–182. [Google Scholar] [CrossRef]
- Sireesha, Y.; Kasetti, R.B.; Nabi, S.A.; Swapna, S.; Apparao, C. Antihyperglycemic and hypolipidemic activities of Setaria italica seeds in STZ diabetic rats. Pathophysiology 2011, 18, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.Y.; Li, P.; Lin, Q.B. Research evolution on chemical component and physical character of foxtail millet. J. Chin. Cereals Oils Assoc. 2008, 22, 51–56. [Google Scholar]
- Brink, M.; Belay, G. Plant Resources of Tropical Africa 1. Cereals and Pulses; PROTA Foundation: Wageningen, The Netherlands, 2006. [Google Scholar]
- Kim, S.K.; Sohn, E.Y.; Lee, I.J. Starch properties of native foxtail millet, Setaria italica Beauv. J. Crop Sci. Biotech. 2009, 12, 59–62. [Google Scholar] [CrossRef]
- Gluchoff-fiasson, K.; Jay, M.; Viricel, M.R. Flavone 0- and c-glycosides from Setaria italica. Phytochemistry 1989, 28, 2471–2475. [Google Scholar] [CrossRef]
- Amadou, I.; Le, G.W.; Amza, T.; Sun, J.; Shi, Y.H. Purification and characterization of foxtail millet derived peptides with antioxidant and antimicrobial activities. Food Res. Int. 2013, 51, 422–428. [Google Scholar] [CrossRef]
- Monteiro, P.V.; Gopal, D.H.; Virupaksha, T.K.; Ramachandra, R. Chemical composition and in vitro protein digestibility of Italian millet (Setaria italica). Food Chem. 1988, 29, 19–26. [Google Scholar] [CrossRef]
- Zhang, L.Z.; Liu, R.H. Phenolic and carotenoid profiles and antiproliferative activity of foxtail millet. Food Chem. 2015, 174, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ma, Y.; Zhang, R.; Ma, H.; Liu, B. Preparation and characterization of foxtail millet bran oil using subcritical propane and supercritical carbon dioxide extraction. J. Food Sci. Technol. 2015, 52, 3099–3104. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Yang, G.; Ma, Y. Chemical characteristics and fatty acid profile of foxtail millet bran oil. J. Am. Oil Chem. Soc. 2010, 87, 63–67. [Google Scholar] [CrossRef]
- Thompson, L.V. Potential health benefits and problems associated with antinutrients in foods. Food Res. Int. 1993, 26, 131–149. [Google Scholar] [CrossRef]
- Suma, P.F.; Urooj, A. Antioxidant activity of extracts from foxtail millet (Setaria italica). J. Food Sci. Technol. 2012, 49, 500–504. [Google Scholar] [CrossRef]
- Pawar, V.S.; Pawar, V.D. Malting characteristics and biochemical changes of foxtail millet. J. Food Sci. Technol. 1997, 34, 416–418. [Google Scholar]
- Pang, M.; He, S.; Wang, L.; Cao, X.; Cao, L.; Jiang, S. Physicochemical properties, antioxidant activities and protective effect against acute ethanol-induced hepatic injury in mice of foxtail millet (Setaria italica) bran oil. Food Funct. 2014, 5, 1763–1770. [Google Scholar] [CrossRef]
- Sorghum and Millets in Human Nutrition; Food and Agriculture Organization of the United Nations: Rome, Italy, 2008.
- Dasgupta, T.; Qais, M.A.N. Pharmacological Potential Investigation of Setaria Italica (Poaceae). Lambert Acad. Pub. 2012.
- Rajeswari, J.R.; Guha, M.; Jayadeep, A.; Sathyendra, R.B.V. Effect of Alkaline Cooking on Proximate, Phenolics and Antioxidant activity of foxtail millet (Setaria italica). World Appl. Sci. J. 2015, 33, 146–152. [Google Scholar]
- Mathew, S.; Abraham, E.T. Studies on the antioxidant activities of cinnamon (Cinnamonum verum) bank extracts, through various in vitro models. Food Chem. 2006, 94, 520–528. [Google Scholar] [CrossRef]
- Sikora, E.; Cieslik, E.; Topolska, K. The source of natural antioxidants. Acta Scientiarum Polonorum Technologia Alimentaria 2008, 7, 5–17. [Google Scholar]
- Yanishlieva-Maslarova, N.V.; Heinonen, I.M. Sources of natural antioxidants: Vegetables, fruits, herbs, spices and teas. In Antioxidants in Food, Practical Applications; Pokorny, J., Yanishlieva, N., Gordon, M., Eds.; Woodhead Publishing: Cambridge, UK, 2001; pp. 210–266. [Google Scholar]
- Negi, P.S.; Jayaprakash, G.K.; Jena, B.S. Antioxidant and antimutagenic activi- ties of pomegranate peel extracts. Food Chem. 2003, 80, 393–397. [Google Scholar] [CrossRef]
- Zahin, M.; Aqil, F.; Ahmad, I. Broad spectrum antimutagenic activity of antioxidant active fraction of Punica granatum L. peel extract. Mutat. Res. 2010, 703, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Chandra, H.; Bishnoi, P.; Yadav, A.; Patni, B.; Mishra, A.P.; Nautiyal, A.R. Antimicrobial resistance and the alternative resources with special emphasis on plant-based antimicrobials—A review. Plants 2017, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Cowan, M.M. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 1999, 12, 564–582. [Google Scholar] [CrossRef] [PubMed]
- Dahanukar, S.; Kulkarni, R.; Rege, N. Pharmacology of medicinal plants and natural products. Indian J. Pharmacol. 2000, 32, S81–S118. [Google Scholar]
- Gabor, M. Anti-inflammatory and anti-allergic properties of flavonoids. Prog. Clin. Biol. Res. 1986, 213, 471–480. [Google Scholar] [PubMed]
- Gul, H.I.; Sahin, F.; Gul, M.; Ozturk, S.; Yerdelen, K.O. Evaluation of antimicrobial activities of several Mannich bases and their derivatives. Arch. Pharm. 2005, 338, 335–338. [Google Scholar] [CrossRef]
- Ramasamy, S.; Manoharan, A.C. Antibacterial effect of volatile components of selected medicinal plants against human pathogens. Asian J. Microbiol. Biotechnol. Environ. Sci. 2004, 6, 209–210. [Google Scholar]
- Esquinas Alcazar, J. Protecting crop genetic diversity for food security: Political, ethical and technical challenges. Nat. Rev. Genet. 2005, 6, 946–953. [Google Scholar] [CrossRef]
- Cao, K.; Wang, L.; Zhu, G.; Fang, W.; Chen, C.; Luo, J. Genetic diversity, linkage disequilibrium, and association mapping analyses of peach (Prunus persica) landraces in China. Tree Genet. Genomes. 2012, 8, 975–990. [Google Scholar] [CrossRef]
- Giraldo, E.; López-Corrales, M.; Hormaza, J.I. Selection of the most discriminating morphological qualitative variables for characterization of fig germplasm. J. Am. Soc. Hortic. Sci. 2010, 135, 240–249. [Google Scholar] [CrossRef]
- Khadivi-Khub, A.; Zamani, Z.; Fatahi, M.R. Multivariate analysis of Prunus subgen. Cerasus germplasm in Iran using morphological variables. Genet. Resour. Crop Evol. 2011, 59, 909–926. [Google Scholar] [CrossRef]
- Maulión, E.; Arroyob, L.E.; Daorden, M.E.; Valentini, G.H.; Cervigni, G.D.L. Performance profiling of Prunus persica (L.) Batsch collection and comprehensive association among fruit quality, agronomic and phenological traits. Sci. Hort. 2016, 198, 385–397. [Google Scholar] [CrossRef]
- Ognjanov, V.; Vujanic-Varga, D.; Gasic, K.; Nadj, B. Disease resistance in apple, pear and peach germplasm originating from the Balkan Peninsula. Acta Hort. 2000, 513, 63–68. [Google Scholar] [CrossRef]
- Paunovic, A.S.; Paunovic, A.S.; Milosevic, T.M.; Tisma, M.T.; Obradovic, A. Selection of native vineyard peach germplasm. Acta Hort. 1992, 315, 133–140. [Google Scholar] [CrossRef]
- Ognjanov, V.; Cerovic, S.; Bozovic, D.J.; Ninic-Todorovic, J.; Golosin, B. Selection of vineyard peach and myrobalan seedling (in Serbian). Pomology 2008, 42, 17–22. [Google Scholar]
- Rakonjac, V.; Nikolic, D.; Milutinovic, M.; Fotiric, M. Suitability of different vineyard peach genotypes for generative rootstocks production. Acta Hort. 2008, 771, 225–229. [Google Scholar] [CrossRef]
- Zec, G.; Colic, S.; Marinkovic, D.; Nikolic, D. Variability of vineyard peach tree characteristics. Genetika 2008, 40, 1–7. [Google Scholar] [CrossRef]
- Widelski, J.; Popova, M.; Graikou, K.; Glowniak, K.; Chinou, I. Coumarins from Angelica lucida L.—Antibacterial Activities. Molecules 2009, 14, 2729–2734. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Shrivastava, N.; Padh, H. Advances in molecular marker techniques and their replications in plant sciences. Plant Cell Rep. 2008, 27, 617–631. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Feuerstein, U.; Luesink, W.; Schulze, S.; Asp, T.; Studer, B.; Becker, H.C.; Dehmer, K.J. DArT, SNP, and SSR analyses of genetic diversity in Lolium perenne L. using bulk sampling. BMC Genet. 2018, 19, 10. [Google Scholar] [CrossRef] [PubMed]
- Feitosa Souza, C.P.; Ferreira, C.F.; de Souza, E.H.; Alfredo Neto, R.S.; Marconcini, J.M.; da Silva Ledo, C.A.; Duarte Souza, F.V. Genetic diversity and ISSR marker association with the quality of pineapple fiber for use in industry. Ind. Crops Prod. 2017, 104, 263–268. [Google Scholar] [CrossRef]
- Cui, C.; Li, Y.; Liu, Y.; Li, X.; Luo, S.; Zhang, Z.; Wu, R.; Liang, G.; Sun, J.; Peng, J.; et al. Determination of genetic diversity among Saccharina germplasm using ISSR and RAPD markers. C. R. Biol. 2017, 340, 76–86. [Google Scholar] [CrossRef]
- Amgai, R.B.; Pantha, S.; Chhetri, T.B.; Budhathoki, S.K.; Khatiwada, S.P.; Mudwari, A. Variation on agro-morphological traits in Nepalese foxtail millet (Setaria italica (L) P Beauv). J. Agron Nepal 2011, 2, 133–138. [Google Scholar] [CrossRef]
- Palakurthi, R.; Gurulakshmi, K.; Mallikarjuna, G.; Obul, R.P.C.; Banavath, J.N.; Sekhar, A.C. Comprehensive phenotypic and statistical based analysis of foxtail millet (Setaria italica L.) lines for parental line selection. Ann. Plant Sci. 2017, 6, 1736–1741. [Google Scholar]
- Chakraborty, S.; Das, P.K.; Guha, B.; Barman, B.; Sarmah, K.K. Coheritability, correlation and path analysis of yield components in boro rice. Oryza 2001, 38, 99–101. [Google Scholar]
- Sharma, A.K.; Sharma, R.N. Genetic variability and character association in early maturing rice. Oryza 2007, 44, 300–303. [Google Scholar]
- Duvick, D.N. Genetic progress in yield of United States maize (Zea mays L.). Maydica 1990, 50, 193–202. [Google Scholar]
- Mansfield, B.D.; Mumm, R.H. Survey of plant density tolerance in US maize germplasm. Crop Sci. 2014, 54, 157–173. [Google Scholar] [CrossRef]
- Li, Y.; Jia, J.; Wang, Y.; Wu, S. Intraspecific and interspecific variation in Setaria revealed by RAPD analysis. Genet. Resour. Crop Evol. 1998, 45, 279–285. [Google Scholar] [CrossRef]
- Rao, K.E.P.; De Wet, J.M.J.; Reddy, V.G.; Mengesha, M.H. Diversity in the small millets collection at ICRISAT. Adv. Small Millets 1994, 331–345. [Google Scholar]
- Labra, M.; Miele, M.; Ledda, B.; Grassi, F.; Mazzei, M.; Sala, F. Morphological characterization: Essential oil composition and DNA genotyping of Ocimum basilicum L. Cultivars Plant Sci. 2004, 167, 725–731. [Google Scholar] [CrossRef]
- Zhang, K.; Fan, G.; Zhang, X.; Zhao, F.; Wei, W.; Du, G.; Feng, X.; Wang, X.; Feng, W.F.; Song, G.; et al. Identification of QTLs for 14 agronomically important traits in Setaria italica based on SNPs generated from high-throughput sequencing. G3: Gene Genom. Genet. 2017, 7, 1587–1594. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis, 2nd ed.; Springer: New York, NY, USA, 2002; Volume XXIX. [Google Scholar]
- Kumari, D.; Madhujith, T.; Chandrasekara, A. Comparison of phenolic content and antioxidant activities of millet varieties grown in different locations in Sri Lanka. Food Sci. Nutr. 2017, 5, 474–485. [Google Scholar] [CrossRef]
- Grinberg, L.N.; Newmark, H.; Kitrossky, N.; Rahamim, E.; Chevion, M.; Rachmilewitz, E.A. Protective effects of tea polyphenols against oxidative damage to red blood cells. Biochem. Pharmacol. 1997, 54, 973–978. [Google Scholar] [CrossRef]
- Guo, Q.; Zhao, B.; Li, M.; Shen, S.; Xin, W. Studies on protective mechanisms of four components of green tea polyphenols against lipid peroxidation in synaptosomes. Biochim. Biophys. Acta 1996, 1304, 210–222. [Google Scholar] [CrossRef]
- Kashima, M. Effects of catechins on superoxide and hydroxyl radical. Chem. Pharm. Bull. (Tokyo) 1999, 47, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Nanjo, F.; Mori, M.; Goto, K.; Hara, Y. Radical scavenging activity of tea catechins and their related compounds. Biosci. Biotechnol. Biochem. 1999, 63, 1621–1623. [Google Scholar] [CrossRef]
- Sang, S.; Cheng, X.; Stark, R.E.; Rosen, R.T.; Yang, C.S.; Ho, C.T. Chemical studies on antioxidant mechanism of tea catechins: Analysis of radical reaction products of catechin and epicatechin with 2,2-diphenyl-1-picrylhydrazyl. Bioorg. Med. Chem. 2002, 10, 2233–2237. [Google Scholar] [CrossRef]
- Sawai, Y.; Sakata, K. NMR analytical approach to clarify the antioxidative molecular mechanism of catechins using 1,1-diphenyl-2-picrylhydrazyl. J. Agric. Food Chem. 1998, 46, 111–114. [Google Scholar] [CrossRef]
- Seeram, N.P.; Nair, M.G. Inhibition of lipid peroxidation and structure–activity-related studies of the dietary constituents anthocyanins, anthocyanidins, and catechins. J. Agric. Food Chem. 2002, 50, 5308–5312. [Google Scholar] [CrossRef]
- Zhao, B.; Guo, Q.; Xin, W. Free radical scavenging by green tea polyphenols. Methods Enzymolol. 2001, 335, 217–231. [Google Scholar]
- Anderson, R.F.; Fisher, L.J.; Hara, Y.; Harris, T.; Mak, W.B.; Melton, L.D.; Packer, J.E. Green tea catechins partially protect DNA from (.) OH radical-induced strand breaks and base damage through fast chemical repair of DNA radicals. Carcinogenesis 2001, 22, 1189–1193. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, K.; Suzuki, M.; Sasaki, K.; Miyase, T.; Sano, M. Formation of antioxidants from (−)-epigallocatechin gallate in mild alkaline fluids, such as authentic intestinal juice and mouse plasma. J. Nutr. Biochem. 1999, 10, 223–229. [Google Scholar] [CrossRef]
- Morris, M.C.; Evans, D.A.; Bienias, J.L.; Tangney, C.C.; Bennett, D.A.; Aggarwal, N.; Wilson, R.S.; Scherr, P.A. Dietary intake of antioxidant nutrients and the risk of incident Alzheimer disease in a biracial community study. J. Am. Med. Assoc. 2002, 287, 3230–3237. [Google Scholar] [CrossRef]
- Lee, L.S.; Kim, S.H.; Kim, Y.B.; Kim, Y.C. Quantitative analysis of major constituents in green tea with different plucking periods and their antioxidant activity. Molecules 2014, 19, 9173–9186. [Google Scholar] [CrossRef]
- Husain, S.R.; Cillard, J.; Cillard, P. Hydroxyl radical scavenging activity of flavonoids. Phytochemistry 1987, 26, 2489–2491. [Google Scholar] [CrossRef]
- Duthie, S.J.; Collins, A.R.; Duthie, G.G.; Dobson, V.L. Quercetin and myricetin protect against hydrogen peroxide-induced DNA damage (strand breaks and oxidised pyrimidines) in human lymphocytes. Mutat. Res. 1997, 393, 223–231. [Google Scholar] [CrossRef]
- Mokrani, A.; Madani, K. Effect of solvent, time and temperature on the extraction of phenolic compounds and antioxidant capacity of peach (Prunus persica L.) fruit. Sep. Purif. 2016, 162, 68–76. [Google Scholar] [CrossRef]
- Deepti, K.; Umadevi, P.; Vijayalakshmi, G.; Vinod, B.; Polarao, B. Antimicrobial Activity and Phytochemical Analysis of Morinda tinctoria Roxb. Leaf Extracts. Asian Pac. J. Trop Biomed. 2012, S1440–S1442. [Google Scholar] [CrossRef]
- Kabir, F.; Katayama, S.; Tanji, N.; Nakamura, S. Antimicrobial effects of chlorogenic acid and related compounds. J. Korean Soc. Appl. Biol. Chem. 2014, 57, 359–365. [Google Scholar] [CrossRef]
- Lou, Z.; Wang, H.; Zhu, S.; Ma, C.; Wang, Z. Antibacterial activity and mechanism of action of chlorogenic acid. J. Food Sci. 2011, 76, 398–403. [Google Scholar] [CrossRef]
- Ikigai, H.; Nakae, T.; Hara, Y.; Shimamura, T. Bactericidal catechins damage the lipid bilayer. Biochim. Biophys. Acta 1993, 1147, 132–136. [Google Scholar] [CrossRef]
- Hermenean, A.; Ardelean, A.; Stan, M.; Hadaruga, N.; Mihali, C.V.; Costache, M.; Dinischiotu, A. Formulation in mice intoxicated with carbon tetrachloride: A comparative study. J. Med. Food. 2014, 17, 670–677. [Google Scholar] [CrossRef]
- Pinho, E.; Grootveld, M.; Soares, G.; Henriques, M. Cyclodextrins as encapsulation agents for plant bioactive compounds. Carbohyd. Polym. 2014, 101, 121–135. [Google Scholar] [CrossRef]
- Sirk, T.W.; Brown, E.F.; Sum, A.K.; Friedman, M. Molecular dynamics study on the biophysical interactions of seven green tea catechins with lipid bilayers of cell membranes. J. Agric. Food Chem. 2008, 56, 7750–7758. [Google Scholar] [CrossRef]
- Sirk, T.W.; Brown, E.F.; Friedman, M.; Sum, A.K. Molecular binding of catechins to biomembranes: Relationship to biological activity. J. Agric. Food Chem. 2009, 57, 6720–6728. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, S.; Sarethy, I.P.; Dang, S.; Gabrani, R. Green tea extract: Possible mechanism and antibacterial activity on skin pathogens. Food Chem. 2012, 135, 672–675. [Google Scholar] [CrossRef]
- Shah, S.; Stapleton, P.D.; Taylor, P.W. The polyphenol (−)- epicatechin gallate disrupts the secretion of virulence-related proteins by Staphylococcus aureus. Lett. Appl. Microbiol. 2008, 46, 181–185. [Google Scholar] [CrossRef]
- Okamoto, M.; Leumg, K.P.; Ansai, T.; Sugimoton, A.; Maeda, N. Inhibitory effects of green tea catechins on protein tyrosine phosphatase in Prevotella intermedia. Oral Microbiol. Immunol. 2003, 18, 192–195. [Google Scholar] [CrossRef]
- Okamoto, M.; Sugimoto, A.; Leung, K.P.; Nakayama, K.; Kamaguchi, A.; Maeda, N. Inhibitory effect of green tea catechins on cysteine eproteinases in Porphyromonas gingivalis. Oral Microbiol. Immunol. 2004, 19, 118–120. [Google Scholar] [CrossRef] [PubMed]
- Gradišar, H.; Pristovšek, P.; Plaper, A.; Jerala, R. Green tea inhibit bacterial DNA 304 gyrase by interaction with its ATP binding site. J. Med. Chem. 2007, 50, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Puupponen-Pimiä, R.; Nohynek, L.; Meier, C.; Kähkönen, M.; Heinonen, M.; Hopia, A.; Oksman-Caldentey, K.M. Antimicrobial properties of phenolic compounds from berries. J. Appl. Microbiol. 2001, 90, 494–507. [Google Scholar] [CrossRef]
- Barzegar, A. Antioxidant activity of polyphenolic myricetin in vitro cell free and cell-based systems. Mol. Biol. Res. Commun. 2016, 5, 87–95. [Google Scholar] [PubMed]
- Griep, M.A.; Blood, S.; Larso, M.A.; Koepsell, S.A.; Hinrichs, S.H. Myricetin inhibits Escherichia coli DnaB helicase but not primase. Bioorg. Med. Chem. 2007, 15, 7203–7208. [Google Scholar] [CrossRef]
- Dvorakova, Z.; Cepkova, P.H.; Janovska, D.; Viehmannova, I.; Svobodova, E.; Cusimamani, E.F.; Milella, L. Comparative analysis of genetic diversity of 8 millet genera revealed by ISSR markers. Emir. J. Food Agric. 2015, 27, 617–628. [Google Scholar]
- Ajitkumar, I.P.; Panneerselvam, R. Analysis of intra specific variation in Setaria italica (L.) P. Beauv landraces using RAPD and ISSR markers. Int. J. Res. Biochem. Biophy. 2013, 3, 15–20. [Google Scholar]
- Ardie, S.W.; Khumaida, N.; Fauziah, N.; Yudiansyah, Y. Biodiversity Assessment of Foxtail Millet (Setaria italica L.) Genotypes Based on RAPD Marker. J. Trop. Crop Sci. 2017, 4, 21–25. [Google Scholar]
- Sikdar, S.; Tiwari, S.; Tripathi, N.; Thakur, V.V. Genetic diversity in Indian foxtail millet [Setaria italica (L.) P. Beauv] revealed by random amplified polymorphic DNA markers. Int. J. Chem. Studies 2018, 6, 1458–1465. [Google Scholar]
- Pathak, A.K.; Singh, S.P.; Tuli, R. Amplified fragment length polymorphism finger printing to identify genetic relatedness among lychee cultivars and markers associated with small-seeded cultivars. J. Am. Soc. Hortic. Sci. 2014, 139, 657–668. [Google Scholar] [CrossRef]
- Thiruvengadam, M.; Rekha, K.; Chung, I.M. Induction of hairy roots by Agrobacterium rhizogenes-mediated transformation of spine gourd (Momordica dioica Roxb. ex. willd) for the assessment of phenolic compounds and biological activities. Scientia Horticulturae 2016, 198, 132–141. [Google Scholar] [CrossRef]
- Ghimire, B.K.; Yu, C.Y.; Chung, I.M. Assessment of the phenolic profile, antimicrobial activity and oxidative stability of transgenic Perilla frutescens L. overexpressing tocopherol methyltransferase (g-tmt) gene. Plant Physiol. Biochem. 2017, 118, 77–87. [Google Scholar] [CrossRef]
- Kobayashi, M.; Kakizono, T.; Nagai, S. Enhanced carotenoid biosynthesis by oxidative stress in acetate-induced cyst cells of a green unicellular alga, Haematococcus pluvialis. Appl. Environ. Microbiol. 1993, 59, 867–873. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
Sample Availability: Samples of the compounds are available from the authors. |

| Accession Number | Landrace Name | Origin | Province |
|---|---|---|---|
| SI-01 | Cheongsilmichajo | Chuncheon-si, sudong-ri | Kangwon-Do, Korea |
| SI-02 | Hwangchajo (Jinju) | Hongcheon-gun, gulji-ri | Kangwon-Do, Korea |
| SI-03 | Mongdangjo | Gapyeong-gun, mokdong-ri | Gyeonggi-Do, Korea |
| SI-04 | Mejo | Wonju-si, hojeo-myeon | Kangwon-Do, Korea |
| SI-05 | Kanghwajo | Yeongwol-gun, macha 9-ri | Kangwon-Do, Korea |
| SI-06 | Neutjo | Yangyang-gun, josan-ri | Kangwon-Do, Korea |
| SI-07 | Kwangjugeurujo | Cheorwon-gun, yukdan 3-ri | Kangwon-Do, Korea |
| SI-08 | Kojangjo | Inje-gun, bupyeong-ri | Kangwon-Do, Korea |
| SI-09 | Eoreunchajo | Taebaek-si, samsu-dong | Kangwon-Do, Korea |
| SI-10 | Saljo | Sokcho-si, joyang-dong | Kangwon-Do, Korea |
| SI-11 | Hwangchajo | Gosung-gun, oho-ri | Kangwon-Do, Korea |
| SI-12 | Sanjeongjo | Pyeongchang-gun, haanmi-ri | Kangwon-Do, Korea |
| SI-13 | Eunchajo | Pocheon-si, udong 2-ri | Gyeonggi-Do, Korea |
| SI-14 | Boksimichajo | Jeongseon-gun, hoedong 5-ri | Kangwon-Do, Korea |
| SI-15 | Bokseulhwangchajo | Gangneung-si, sindang-ri | Kangwon-Do, Korea |
| Accessions | Plant Height (cm) ** | Culm Length (cm) | Number of Leaves | Leaves Length (cm) | Leaves Width (cm) | Ratio of Leaves Length and Width | Leaves Color | Leaf Orientation | Tiller Number |
|---|---|---|---|---|---|---|---|---|---|
| SI-01 | 143.17 ± 2.02 e | 130.17 ± 1.04 k | 11.00 ± 2.00 ef | 49.37 ± 0.55 h | 2.97 ± 0.16 fd | 3.95 ± 0.05 d | Green | 2 | 1 |
| SI-02 | 160.93 ± 3.00 h | 139.66 ± 2.08 l | 9.37 ± 0.71 cd | 38.50 ± 1.50 c | 2.70 ± 0.30 cdef | 3.92 ± 0.11 d | Green | 3 | |
| SI-03 | 115.83 ± 2.57 c | 98.17 ± 1.04 d | 8.33 ± 0.58 bc | 40.30 ± 1.13 d | 2.33 ± 0.15 abc | 3.59 ± 0.09 c | Light green | 2 | 1 |
| SI-04 | 99.33 ± 2.52 b | 80.00 ± 2.00 b | 7.17 ± 0.77 b | 37.30 ± 1.13 bc | 2.67 ± 0.15 bcdef | 2.47 ± 0.05 a | Green | 2 | 2 |
| SI-05 | 84.43 ± 1.50 a | 67.10 ± 2.01 a | 5.67 ± 0.29 a | 36.47 ± 1.50 b | 2.49 ± 0.80 abcde | 2.30 ± 0.10 a | Green | 1 | 2 |
| SI-06 | 167.67 ± 1.53 i | 150.10 ± 3.00 m | 12.17 ± 0.76 efg | 46.10 ± 1.02 fg | 2.43 ± 0.60 abcde | 4.76 ± 0.15 f | Light green | 1 | 1 |
| SI-07 | 144.10 ± 2.00 e | 121.43 ± 2.50 i | 9.17 ± 1.04 c | 48.60 ± 1.50 h | 3.87 ± 0.06 h | 2.51 ± 0.08 a | Green | 2 | 1 |
| SI-08 | 126.60 ± 2.25 d | 103.77 ± 1.66 e | 10.67 ± 0.28 de | 37.17 ± 0.76 bc | 2.73 ± 0.25 def | 3.33 ± 0.20 b | Green | 2 | 2 |
| SI-09 | 173.67 ± 1.52 j | 160.00 ± 2.00 n | 12.23 ± 0.92 efg | 46.27 ± 0.75 g | 3.20 ± 0.26 g | 4.07 ± 0.10 de | Light green | 1 | 2 |
| SI-10 | 152.50 ± 2.88 g | 130.50 ± 1.58 k | 12.37 ± 0.75 fg | 42.65 ± 0.93 e | 2.30 ± 0.24 ab | 5.36 ± 0.13 g | Light green | 2 | 1 |
| SI-11 | 148.00 ± 2.82 f | 128.00 ± 1.41 j | 13.20 ± 0.98 g | 43.50 ± 1.41 e | 2.45 ± 0.49 abcde | 5.30 ± 0.28 g | Green | 2 | 1 |
| SI-12 | 201.33 ± 1.52 k | 116.16 ± 2.03 h | 12.13 ± 0.70 efg | 34.30 ± 1.08 a | 2.40 ± 0.20 abcd | 5.13 ± 0.23 g | Green | 2 | 2 |
| SI-13 | 127.66 ± 2.51 d | 115.50 ± 1.50 g | 14.76 ± 0.25 h | 33.23 ± 0.87 a | 2.13 ± 0.15 a | 6.83 ± 0.20 h | Light green | 1 | 1 |
| SI-14 | 117.13 ± 1.00 c | 96.50 ± 0.50 c | 12.10 ± 0.36 efg | 44.53 ± 0.55 efg | 2.80 ± 0.10 ef | 4.26 ± 0.15 e | Green | 2 | 1 |
| SI-15 | 128.20 ± 0.72 d | 107.43 ± 0.60 f | 12.06 ± 0.30 efg | 44.30 ± 0.30 ef | 2.92 ± 0.06 fg | 4.27 ± 0.15 e | Green | 2 | 1 |
| Accessions | Number of Nodes ** | Bloom Beginning (Days) | Full Bloom (Days) | Spike Length (cm) | Spike Width (cm) | Weight of 1000 Seed (g) |
|---|---|---|---|---|---|---|
| SI-01 | 12.00 ± 2.00 cd | 121.00 ± 1.00 c | 128.00 ± 2.00 c | 22.47 ± 0.50 f | 2.90 ± 0.02 de | 1.63 ± 0.08 b |
| SI-02 | 10.00 ± 2.00 bcd | 114.00 ± 2.00 b | 121.33 ± 2.52 b | 20.43 ± 0.81 de | 2.40 ± 0.10 b | 1.64 ± 0.07 b |
| SI-03 | 7.67 ± 1.53 abc | 94.00 ± 2.00 a | 99.67 ± 1.53 a | 20.37 ± 0.55 cde | 2.30 ± 0.10 b | 2.17 ± 0.01 ef |
| SI-04 | 5.33 ± 1.53 a | 93.67 ± 3.51 a | 99.67 ± 1.53 a | 22.00 ± 1.00 f | 3.57 ± 0.12 hi | 2.91 ± 0.06 i |
| SI-05 | 6.00 ± 1.00 ab | 94.33 ± 1.53 a | 99.67 ± 1.53 a | 17.33 ± 0.58 b | 1.93 ± 0.06 a | 1.77 ± 0.01 bc |
| SI-06 | 14.67 ± 7.37 d | 127.67 ± 1.53 d | 135.00 ± 2.00 d | 19.33 ± 0.76 cd | 2.55 ± 0.05 bc | 1.83 ± 0.06 c |
| SI-07 | 14.00 ± 2.00 d | 114.33 ± 2.51 b | 121.00 ± 1.00 b | 22.00 ± 1.00 f | 2.58 ± 0.10 bc | 1.98 ± 0.02 d |
| SI-08 | 7.67 ± 1.53 abc | 128.00 ± 1.00 d | 134.67 ± 1.53 d | 21.30 ± 0.60 ef | 3.27 ± 0.15 fg | 2.50 ± 0.10 g |
| SI-09 | 13.33 ± 1.52 d | 114.00 ± 2.00 b | 122.33 ± 1.52 b | 19.30 ± 0.20 cd | 2.80 ± 0.30 cd | 1.44 ± 0.05 a |
| SI-10 | 13.25 ± 1.25 d | 128.00 ± 1.63 d | 135.00 ± 0.82 d | 21.62 ± 0.47 f | 3.47 ± 0.25 gh | 2.13 ± 0.12 def |
| SI-11 | 13.25 ± 1.76 d | 128.00 ± 2.82 d | 135.00 ± 4.24 d | 21.95 ± 0.49 f | 3.80 ± 0.14 i | 2.05 ± 0.07 de |
| SI-12 | 13.33 ± 1.04 d | 121.67 ± 1.15 c | 127.33 ± 3.05 c | 24.06 ± 0.30 g | 3.16 ± 0.20 ef | 2.24 ± 0.21 f |
| SI-13 | 14.23 ± 1.56 d | 127.66 ± 1.52 d | 134.00 ± 1.73 d | 12.80 ± 0.20 a | 1.70 ± 0.20 a | 1.71 ± 0.07 bc |
| SI-14 | 14.06 ± 0.60 d | 121.30 ± 0.60 c | 128.16 ± 0.76 c | 19.26 ± 0.64 c | 2.81 ± 0.07 cd | 2.75 ± 0.05 h |
| SI-15 | 11.96 ± 0.45 cd | 122.33 ± 3.21 c | 128.00 ± 0.90 c | 22.10 ± 0.52 f | 2.36 ± 0.15 b | 2.62 ± 0.06 gh |
| Analytes | Plant Height | Culm Length | Number of Leaves | Leaves Length | Leaves Width | Ratio of Leaf L/W | Number of Nodes | Bloom Beginning | Full Bloom | Spike Length | Spike Width | Weight of 1000 Seed | Tiller Number |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plant height | 1 | 0.800 ** | 0.557 * | 0.187 | 0.072 | 0.471 | 0.653 ** | 0.551 * | 0.566 * | 0.348 | 0.478 | −0.379 | −0.019 |
| Culm length | 1 | 0.585 * | 0.517 * | 0.198 | 0.442 | 0.677 ** | 0.573 * | 0.607 * | 0.108 | 0.096 | −0.611 * | −0.298 | |
| Number of leaves | 1 | 0.135 | −0.195 | 0.893 ** | 0.823 ** | 0.873 ** | 0.874 ** | −0.156 | 0.168 | −0.129 | −0.421 | ||
| Leaves length | 1 | 0.636 * | −0.162 | 0.447 | 0.205 | 0.232 | 0.333 | −0.062 | −0.173 | −0.502 | |||
| Leaves width | 1 | −0.548 * | 0.147 | −0.067 | −0.046 | 0.332 | −0.070 | −0.050 | 0.010 | ||||
| Ratio of leaf L/W | 1 | 0.675 ** | 0.707 ** | 0.700 ** | −0.318 | 0.094 | −0.213 | −0.418 | |||||
| Number of nodes | 1 | 0.754 ** | 0.763 ** | −0.067 | 0.077 | −0.294 | −0.551 * | ||||||
| Bloom beginning | 1 | 0.998 ** | 0.020 | 0.228 | −0.100 | −0.381 | |||||||
| Full bloom | 1 | 0.017 | 0.224 | −0.125 | −0.378 | ||||||||
| Spike length | 1 | 0.732 ** | 0.365 | 0.105 | |||||||||
| Spike width | 1 | 0.366 | 0.329 | ||||||||||
| Weight of 1000 seed | 1 | 0.085 | |||||||||||
| Tiller number | 1 |
| Accessions | p-HY * | CH | CAT | CA | SY | SA | o-C | |
| --------------------------------------------------------------------------------------(µg/mL) **------------------------------------------------- | ||||||||
| SI-01 | 0.96 ± 0.06 c | 0 a | 31.41 ± 0.52 d | 0.41 ± 0.03 a | 8.63 ± 0.40 d | 9.92 ± 0.39 h | 0 a | |
| SI-02 | 0.41 ± 0.02 6b | 0 a | 0 a | 0 a | 0 a | 0.36 ± 0.04 ab | 0 a | |
| SI-03 | 0 a | 1.22 ± 2.00 bc | 0 a | 0 a | 0.16 ± 0.02 a | 0.84 ± 0.06 abc | 0 a | |
| SI-04 | 0 a | 0.35 ± 0.05 b | 21.27 ± 0.05 c | 1.50 ± 0.05 a | 7.32 ± 0.20 b | 5.26 ± 0.05 e | 0 a | |
| SI-05 | 1.40 ± 0.56 d | 0 a | 55.34 ± 0.58 e | 0.56 ± 0.05 a | 17.97 ± 0.06 f | 23.06 ± 1.01 k | 0 a | |
| SI-06 | 0 a | 0.35 ± 0.05 b | 34.53 ± 0.50 d | 0.43 ± 0.02 a | 7.79 ± 0.17 c | 9.37 ± 0.55 h | 0 a | |
| SI-07 | 0 a | 0 a | 0 a | 0 a | 0 a | 1.36 ± 0.15 bc | 0 a | |
| SI-08 | 0 a | 0 a | 11.62 ± 0.54 b | 6.83 ± 1.42 b | 0 a | 25.04 ± 1.00 l | 0 a | |
| SI-09 | 0 a | 9.28 ± 0.03 e | 67.93 ± 0.11 f | 12.25 ± 0.03 c | 0 a | 21.62 ± 1.19 j | 0 a | |
| SI-10 | 0 a | 0.59 ± 1.19 ab | 0 a | 0 a | 0 a | 1.69 ± 0.96 c | 0.18 ± 0.18 b | |
| SI-11 | 0 a | 2.45 ± 0.35 d | 0 a | 0 a | 0 a | 3.15 ± 0.05 d | 0.45 ± 0.04 c | |
| SI-12 | 0 a | 0.69 ± 0.04 ab | 20.25 ± 9.00 c | 0.29 ± 0.01 a | 7.36 ± 0.06 b | 7.15 ± 0.06 f | 0 a | |
| SI-13 | 0.44 ± 0.05 b | 0.28 ± 0.03 a | 35.21 ± 0.26 d | 0.55 ± 0.05 a | 10.48 ± 0.50 e | 8.19 ± 0.08 g | 0 a | |
| SI-14 | 0 a | 1.39 ± 0.13 c | 0 a | 0 a | 0 a | 0 a | 0 a | |
| SI-15 | 0 a | 0 a | 0 a | 0 a | 0 a | 0 a | 0 a | |
| Accessions | FE | NA | HN | MY | t-C | QU | NE | Total Phenolic Compound |
| --------------------------------------------------------------------------------------(µg/mL) *----------------------------------------------------------- | ||||||||
| SI-01 | 0 a | 0 a | 0 a | 0a | 0 a | 0 a | 0 a | 51.23 ± 1.08 a |
| SI-02 | 8.28 ± 0.20 bc | 0 a | 0 a | 1.79 ± 0.26 b | 0.05 ± 0.01 c | 0 a | 0 a | 10.63 ± 0.55 a |
| SI-03 | 3.27 ± 0.15 ab | 0.61 ± 0.10 cd | 0.76 ± 0.05 d | 2.30 ± 0.20 b | 0 a | 10.99 ± 0.08 d | 0 a | 20.09 ± 1.01 a |
| SI-04 | 0 a | 0.28 ± 0.03 ab | 0 a | 7.39 ± 0.17 h | 0 a | 0 a | 0 a | 44.37 ± 0.55 a |
| SI-05 | 0 a | 0.40 ± 0.05 bc | 0.37 ± 0.03 c | 3.06 ± 0.05 c | 0 a | 0 a | 0 a | 101.32 ± 1.14 ab |
| SI-06 | 2.10 ± 2.77 a | 0 a | 0.66 ± 0.05 d | 3.82 ± 0.11 d | 0 a | 0 a | 0 a | 57.14 ± 1.03 a |
| SI-07 | 1.98 ± 0.06 a | 0 a | 0 a | 2.21 ± 0.25 b | 0 a | 0.66 ± 0.05 c | 0 a | 6.21 ± 0.80 a |
| SI-08 | 0 a | 0 a | 0.91 ± 0.03 e | 5.61 ± 0.60 g | 0 a | 0 a | 0 a | 43.25 ± 1.09 a |
| SI-09 | 0 a | 9.32 ± 0.08 g | 3.77 ± 0.20 f | 23.96 ± 0.45 j | 0 a | 0 a | 0 a | 152.83 ± 1.06 b |
| SI-10 | 4.47 ± 8.94 ab | 0.25 ± 0.49 ab | 0.05 ± 0.10 a | 2.41 ± 0.89 bc | 0.02 ± 0.03 ab | 0.42 ± 0.28 b | 3.64 ± 2.42 b | 13.74 ± 10.06 a |
| SI-11 | 17.50 ± 0.71 d | 1.06 ± 0.08 e | 0.19 ± 0.01 b | 3.75 ± 0.35 d | 0.04 ± 0.01 bc | 0 a | 0 a | 28.50 ± 0.71 a |
| SI-12 | 0 a | 0 a | 0 a | 4.61 ± 0.34 ef | 0 a | 0 a | 0 a | 39.88 ± 0.83 a |
| SI-13 | 0 a | 0 a | 0 a | 8.70 ± 0.26 h | 0 a | 0 a | 0 a | 62.88 ± 2.59 ab |
| SI-14 | 11.43 ± 0.51 c | 0.75 ± 0.10 d | 0 a | 5.22 ± 0.25 fg | 0.72 ± 0.03 e | 0 a | 0 a | 20.12 ± 2.80 a |
| SI-15 | 20.52 ± 0.50 d | 1.69 ± 0.28 f | 0.25 ± 0.05 bc | 11.19 ± 0.27 i | 0.36 ± 0.03 d | 0 a | 0.52 ± 0.02 a | 34.48 ± 0.50 a |
| Accessions | Root | Stem | Leaf | Seed |
|---|---|---|---|---|
| RC50 (µg mL−1) * | ||||
| SI-01 | 93.27 ± 0.25 g | 138.00 ± 1.00 f | 32.53 ± 1.20 a | 159.67 ± 1.50 b |
| SI-02 | 140.33 ± 0.58 j | 222.43 ± 2.14 k | 60.20 ± 1.05 e | 310.00 ± 1.00 i |
| SI-03 | 78.53 ± 0.50 d | 99.43 ± 0.51 a | 32.33 ± 1.53 a | 252.00 ± 2.00 d |
| SI-04 | 100.00 ± 1.00 h | 198.67 ± 1.16 i | 39.10 ± 1.02 b | 270.00 ± 2.00 f |
| SI-05 | 71.10 ± 1.02 c | 118.00 ± 1.00 c | 60.00 ± 1.00 e | 280.00 ± 2.00 g |
| SI-06 | 58.20 ± 1.06 ab | 198.33 ± 1.53 i | 59.10 ± 1.02 e | 314.67 ± 4.16 j |
| SI-07 | 118.53 ± 1.50 i | 142.20 ± 2.03 g | 37.10 ± 1.02 b | 210.33 ± 1.53 c |
| SI-08 | 56.20 ± 1.06 a | 120.77 ± 1.57 d | 72.17 ± 1.26 f | 309.83 ± 1.23 i |
| SI-09 | 88.00 ± 1.00 f | 107.67 ± 1.53 b | 47.90 ± 1.16 c | 135.17 ± 0.76 a |
| SI-10 | 176.21 ± 1.06 k | 132.67 ± 1.53 e | 87.87 ± 1.63 g | 581.33 ± 4.16 k |
| SI-11 | 58.17 ± 1.26 ab | 118.33 ± 1.53 cd | 48.77 ± 1.57 c | 270.00 ± 2.00 f |
| SI-12 | 79.83 ± 1.76 d | 119.67 ± 1.53 cd | 51.20 ± 1.06 d | 280.67 ± 3.05 g |
| SI-13 | 60.07 ± 1.40 b | 239.10 ± 1.02 l | 48.00 ± 1.00 c | 259.33 ± 5.03 e |
| SI-14 | 83.00 ± 3.00 e | 177.87 ± 0.23 h | 50.83 ± 0.76 d | 286.33 ± 2.52 h |
| SI-15 | 100.33 ± 1.53 h | 298.53 ± 1.50 m | 48.00 ± 2.00 c | 270.00 ± 2.00 f |
| Analytes | Phenolic Compound | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-HY 1 | CH | CAT | CA | SY | SA | o-C | FE | NA | HN | MY | t-C | QU | NE | |
| DPPH | 0.166 | 0.647 ** | 0.557 * | 0.576 * | 0.011 | 0.332 | −0.225 | −0.243 | 0.681 ** | 0.631 * | 0.568 * | −0.137 | −0.005 | −0.468 |
| Accessions | MIC (μg/mL) * | ||||
|---|---|---|---|---|---|
| S. aureus | B. subtilis | S. typhimurium | K. pneumonia | E. coli | |
| SI-01 | 1000 | 1000 | >1000 | >1000 | 500 |
| SI-02 | 1000 | 1000 | >1000 | >1000 | 500 |
| SI-03 | 1000 | >1000 | >1000 | >1000 | 1000 |
| SI-04 | 1000 | >1000 | >1000 | >1000 | 500 |
| SI-05 | 1000 | 1000 | >1000 | >1000 | 500 |
| SI-06 | 1000 | 1000 | >1000 | >1000 | 500 |
| SI-07 | 1000 | 1000 | >1000 | 1000 | 500 |
| SI-08 | 1000 | 1000 | 500 | >1000 | 500 |
| SI-09 | 1000 | 1000 | >1000 | >1000 | 500 |
| SI-10 | 1000 | >1000 | >1000 | >1000 | 500 |
| SI-11 | 1000 | 1000 | >1000 | >1000 | 500 |
| SI-12 | 1000 | 1000 | >1000 | >1000 | 500 |
| SI-13 | 1000 | 1000 | >1000 | >1000 | 500 |
| SI-14 | 1000 | 1000 | 500 | 500 | 500 |
| SI-15 | 1000 | 1000 | 1000 | 500 | 250 |
| ISSR Primer | Base Sequence (5′-3′) | Total Number of Bands | Number of Polymorphic Bands | Percentage of Polymorphism (%) |
|---|---|---|---|---|
| ISSR1 | (CTG)7G | 5 | 5 | 100 |
| ISSR2 | (GAG)6C | 2 | 2 | 100 |
| ISSR3 | (GAC)6T | 3 | 3 | 100 |
| ISSR4 | (GACA)5 | 6 | 4 | 66.67 |
| ISSR5 | (GTC)6A | 4 | 3 | 75.00 |
| ISSR6 | (GTG)6C | 3 | 3 | 100 |
| ISSR7 | (CTC)7 | 7 | 7 | 100 |
| ISSR8 | (GTG)6A | 6 | 5 | 83.33 |
| ISSR9 | A(CACA)3CACTG | 6 | 4 | 100 |
| ISSR10 | (GAC)6 | 5 | 5 | 100.00 |
| ISSR11 | (GACA)4 | 8 | 3 | 76.67 |
| ISSR12 | (CTC)7A | 6 | 5 | 70.00 |
| ISSR13 | (GACA)4A | 7 | 4 | 40.00 |
| ISSR14 | (TC)8AG | 6 | 7 | 75.70 |
| Total | 74 | 60 |
| Primer | P * | q | Na | Ne | I | He | uHe |
|---|---|---|---|---|---|---|---|
| ISSR1 | 0.106 | 0.894 | 2.000 | 1.233 | 0.337 | 0.189 | 0.195 |
| ISSR 2 | 0.484 | 0.516 | 2.000 | 1.998 | 0.693 | 0.499 | 0.517 |
| ISSR 3 | 0.553 | 0.447 | 2.000 | 1.978 | 0.688 | 0.494 | 0.511 |
| ISSR 4 | 1.000 | 0.000 | 1.000 | 1.000 | 0.000 | 0.000 | 0.000 |
| ISSR 5 | 1.000 | 0.000 | 1.000 | 1.000 | 0.000 | 0.000 | 0.000 |
| ISSR 6 | 0.368 | 0.632 | 2.000 | 1.869 | 0.658 | 0.465 | 0.481 |
| ISSR 7 | 0.184 | 0.816 | 2.000 | 1.428 | 0.477 | 0.300 | 0.310 |
| ISSR 8 | 0.368 | 0.632 | 2.000 | 1.869 | 0.658 | 0.465 | 0.481 |
| ISSR 9 | 0.742 | 0.258 | 2.000 | 1.621 | 0.571 | 0.383 | 0.396 |
| ISSR 10 | 0.742 | 0.258 | 2.000 | 1.621 | 0.571 | 0.383 | 0.396 |
| ISSR 11 | 0.106 | 0.894 | 2.000 | 1.233 | 0.337 | 0.189 | 0.195 |
| ISSR 12 | 0.368 | 0.632 | 2.000 | 1.869 | 0.658 | 0.465 | 0.481 |
| ISSR 13 | 0.368 | 0.632 | 2.000 | 1.869 | 0.658 | 0.465 | 0.481 |
| ISSR 14 | 0.270 | 0.730 | 2.000 | 1.650 | 0.583 | 0.394 | 0.408 |
| Mean | 0.475 | 0.524 | 1.857 | 1.588 | 0.492 | 0.335 | 0.347 |
| SE | 0.286 | 0.286 | 0.097 | 0.094 | 0.064 | 0.047 | 0.048 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghimire, B.K.; Yu, C.Y.; Kim, S.-H.; Chung, I.-M. Assessment of Diversity in the Accessions of Setaria italica L. Based on Phytochemical and Morphological Traits and ISSR Markers. Molecules 2019, 24, 1486. https://doi.org/10.3390/molecules24081486
Ghimire BK, Yu CY, Kim S-H, Chung I-M. Assessment of Diversity in the Accessions of Setaria italica L. Based on Phytochemical and Morphological Traits and ISSR Markers. Molecules. 2019; 24(8):1486. https://doi.org/10.3390/molecules24081486
Chicago/Turabian StyleGhimire, Bimal Kumar, Chang Yeon Yu, Seung-Hyun Kim, and Ill-Min Chung. 2019. "Assessment of Diversity in the Accessions of Setaria italica L. Based on Phytochemical and Morphological Traits and ISSR Markers" Molecules 24, no. 8: 1486. https://doi.org/10.3390/molecules24081486
APA StyleGhimire, B. K., Yu, C. Y., Kim, S.-H., & Chung, I.-M. (2019). Assessment of Diversity in the Accessions of Setaria italica L. Based on Phytochemical and Morphological Traits and ISSR Markers. Molecules, 24(8), 1486. https://doi.org/10.3390/molecules24081486









