Synthesis of Chromen-4-One-Oxadiazole Substituted Analogs as Potent β-Glucuronidase Inhibitors
Abstract
:1. Introduction
2. Result and Discussions
2.1. Chemistry
2.2. β-Glucuronidase Activity
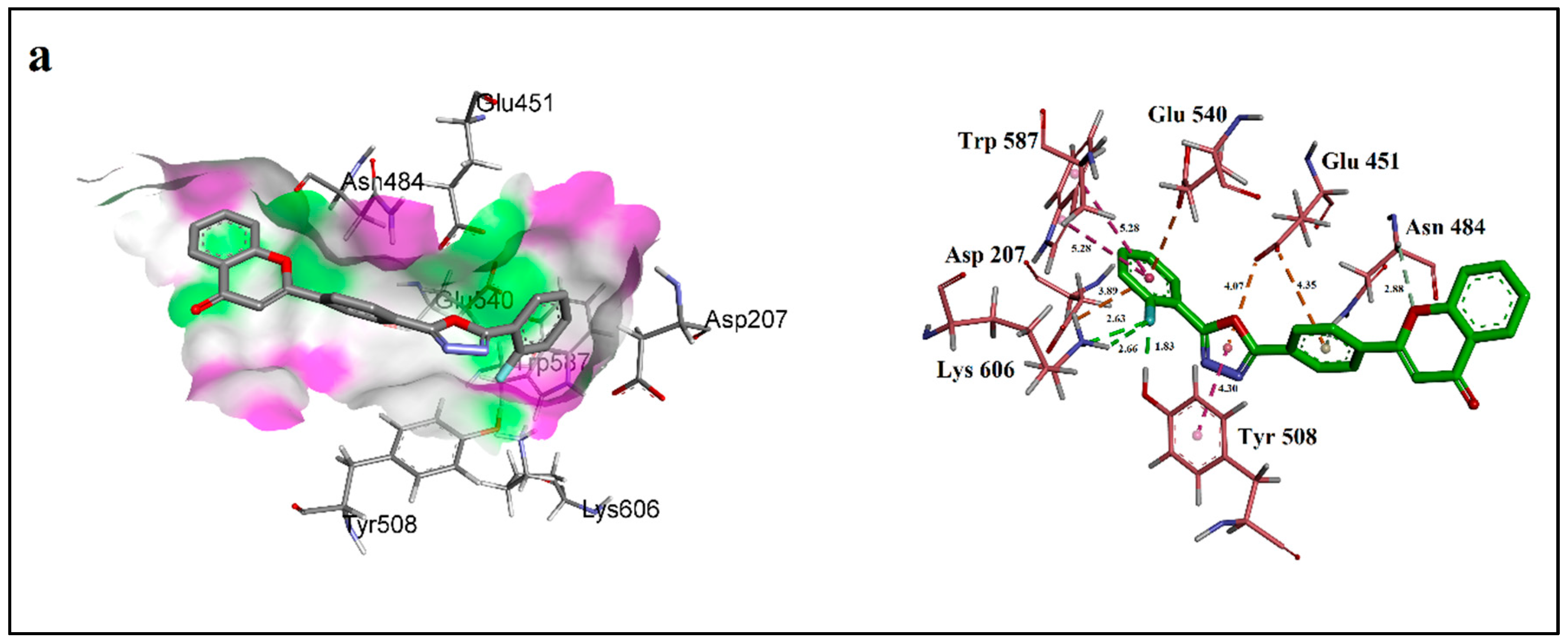
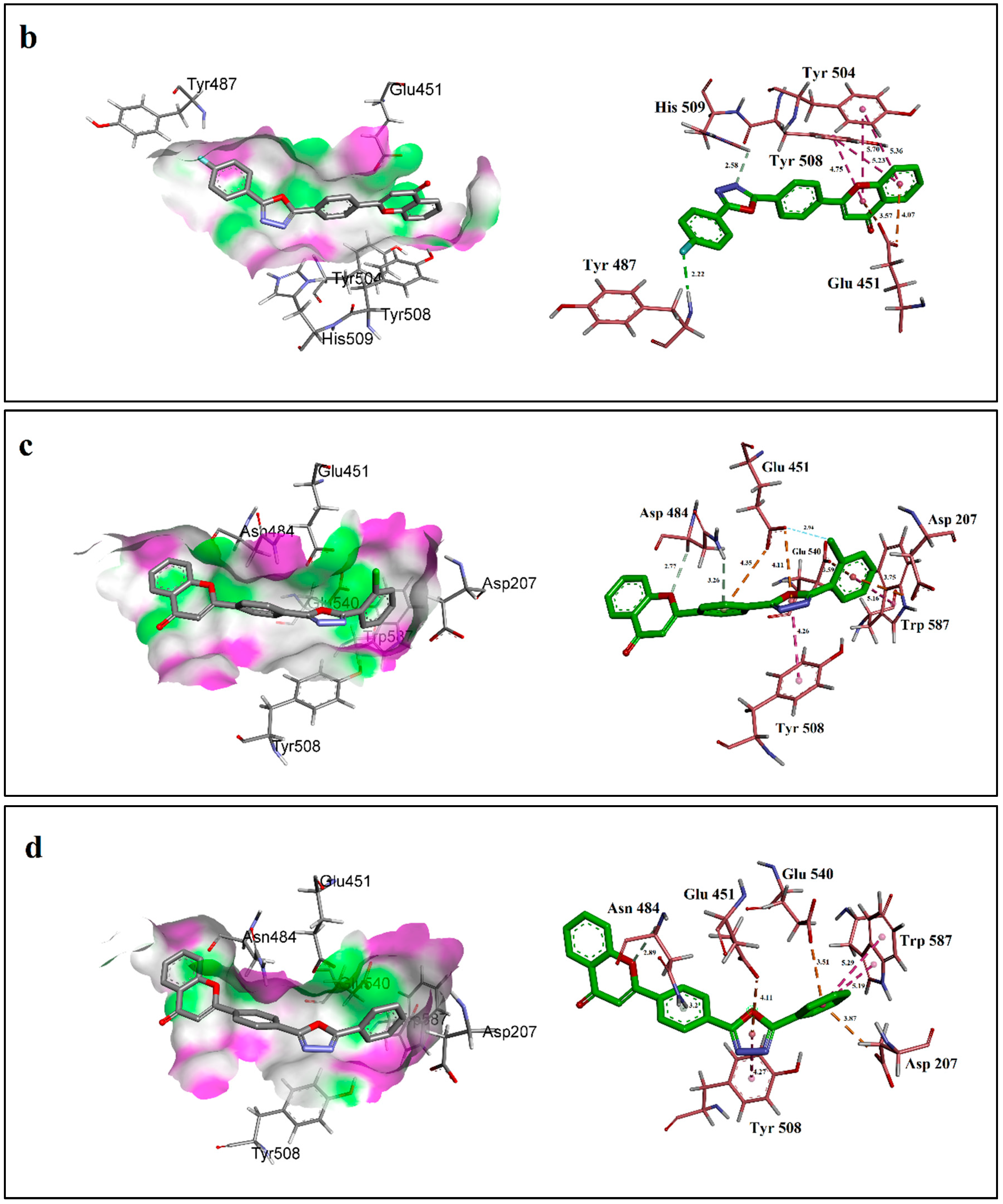
2.3. Molecular Docking Studies
3. Conclusions
4. Experimental
4.1. General Information
4.2. General Procedure for Synthesis of Flavone-Based Oxadiazoles
4.3. 2-(4-(5-(4-Chlorophenyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (1)
4.4. 2-(4-(5-(3-Chlorophenyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (2)
4.5. 2-(4-(5-(2-Chlorobenzyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (3)
4.6. 2-(4-(5-(4-Fluorophenyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (4)
4.7. 2-(4-(5-(3-Fluorophenyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (5)
4.8. 2-(4-(5-(2-Fluorophenyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (6)
4.9. 2-(4-(5-(p-Tolyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (7)
4.10. 2-(4-(5-(m-Tolyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (8)
4.11. 2-(4-(5-(o-Tolyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (9)
4.12. 2-(4-(5-(4-Nitrophenyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (10)
4.13. 2-(4-(5-(3-Nitrophenyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (11)
4.14. 2-(4-(5-(2-Nitrophenyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (12)
4.15. 2-(4-(5-(4-Methoxyphenyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (13)
4.16. 2-(4-(5-(3-Methoxyphenyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (14)
4.17. 2-(4-(5-(2-Methoxyphenyl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (15)
4.18. 2-(4-(5-(Pyridin-2-yl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (16)
4.19. 2-(4-(5-(Pyridin-3-yl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (17)
4.20. 2-(4-(5-(Pyridin-4-yl)-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (18)
4.21. 2-(4-(5-Phenyl-1,3,4-oxadiazol-2-yl)phenyl)-4H-chromen-4-one (19)
4.22. β-Glucuronidase Assay
4.23. Molecular Docking Studies Assay
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fishman, H.W. Beta-Glucuronidase; Academic Press: New York, NY, USA, 1983; p. 929. [Google Scholar]
- Khan, K.M.; Ambreen, N.; Taha, M.; Halim, S.A. Structure-based design, synthesis and biological evaluation of β-glucuronidase inhibitors. J. Comput. Aided Mol. Des. 2014, 28, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Kunert-Keil, C.; Ritter, C.A.; Kroemer, H.K.; Sperker, B. Deconjugating enzymes: Sulphatases and glucuronidases. In Enzyme Systems that Metabolise Drugs and Other Xenobiotics; Ioannides, C., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2002; Volume 2, p. 521. [Google Scholar]
- Gonick, H.C.; Kramer, H.J.; Schapiro, A.E. Urinary β-Glucuronidase Activity in Renal Disease. Arch. Intern. Med. 1973, 132, 63–69. [Google Scholar] [CrossRef]
- Perez, J.L.; Berrocal, C.I.; Berrocal, L. Evaluation of a commercial beta-glucuronidase test for the rapid and economical identification of Escherichia coli. J. Appl. Bacteriol. 1986, 61, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.H.; Zhang, H.D.; Xu, C.J.; Bian, Y.J.; Xu, X.J.; Xie, Q.M.; Zhang, R.H. Neuroprotective effects of flavonoids extracted from licorice on kainate-induced seizure in mice through their antioxidant properties. J. Zhejiang Univ. Sci. B 2013, 14, 1004–1012. [Google Scholar] [CrossRef] [PubMed]
- Pantscheva-Haschen, R.; Schulze, R.; Haerting, J.; Langkopf, B. An attempt to optimize the diagnosis of rejection after allogeneic kidney transplantation using urinary enzymes and multivariate statistics. Med. Lab. Diagn. 1988, 29, 179–187. [Google Scholar]
- Paigen, K.; Peterson, J.; Paigen, B. Role of Urinary β-Glucuronidase in Human Bladder Cancer. Cancer Res. 1984, 44, 3620–3623. [Google Scholar] [PubMed]
- Boyland, E.; Gasson, J.E.; Williams, D.C. Enzyme Activity in Relation to Cancer. Br. J. Cancer 1957, 11, 578–589. [Google Scholar] [CrossRef] [Green Version]
- Gloux, K.; Barteau, O.; Eloumami, H.; Beguet, F.; Leclerc, M.; Dore, J. A metagenomic β-glucuronidase uncovers a coreadaptive function of the human intestinal microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 4539–4546. [Google Scholar] [CrossRef]
- Zhu, Q.; Gao, R.; Wu, W.; Qin, H. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumor Biol. 2013, 34, 1285–1300. [Google Scholar] [CrossRef]
- Kim, D.H.; Jin, Y.H. Intestinal bacterial β-glucuronidase activity of patients with colon cancer. Arch. Pharm. Res. 2001, 24, 564–567. [Google Scholar] [CrossRef]
- Huang, W.; Liu, M.-Z.; Li, Y.; Tan, Y.; Yang, G.-F. Design, syntheses, and antitumor activity of novel chromone and aurone derivatives. Bioorg. Med. Chem. 2007, 15, 5191–5197. [Google Scholar] [CrossRef]
- Awadallah, F.M.; El-Waei, T.A.; Hanna, M.M.; Abbas, S.E.; Ceruso, M.; Oz, B.E.; Guler, O.O.; Supuran, C.T. Synthesis, carbonic anhydrase inhibition and cytotoxic activity of novel chromone-based sulfonamide derivatives. Eur. J. Med. Chem. 2015, 96, 425–435. [Google Scholar] [CrossRef] [Green Version]
- Dias, M.M.; Machado, N.F.; Marques, M.P. Dietary chromones as antioxidants—The structural variable. Food Funct. 2011, 2, 595. [Google Scholar] [CrossRef]
- Gomes, A.; Fernandes, E.; Silva, A.M.S.; Pinto, D.C.G.A.; Santos, C.M.M.; Cavaleiro, J.A.S.; Lima, J.L.F.C. Anti-inflammatory potential of 2-styrylchromones regarding their interference with arachidonic acid metabolic pathways. Biochem. Pharmacol. 2009, 78, 171–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, O.; Kumar, R.; Parkash, V. Synthesis and antifungal activity of some new 3-hydroxy-2-(1-phenyl-3-aryl-4-pyrazolyl) chromones. Eur. J. Med. Chem. 2008, 43, 435–440. [Google Scholar] [CrossRef]
- Farshori, N.N.; Banday, M.R.; Ahmad, A.; Khan, A.U.; Rauf, A. Synthesis, characterization, and in vitro antimicrobial activities of 5-alkenyl/hydroxyalkenyl-2-phenylamine-1,3,4-oxadiazoles and thiadiazoles. Bioorg. Med. Chem. Lett. 2010, 20, 1933–1938. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.N.; Shi, Y.X.; Zhang, L.; Ling, Y.; Li, B.J.; Nishida, Y.; Yang, X.L. Synthesis and Fungicidal Activity of Novel 2,5-Disubstituted-1,3,4-oxadiazole Derivatives. J. Agric. Food Chem. 2012, 60, 11649–11656. [Google Scholar] [CrossRef]
- Li, Y.H.; Zhu, H.J.; Chen, K.; Liu, R.; Khallaf, A.; Zhang, X.N.; Ni, J.P. Synthesis, insecticidal activity, and structure–activity relationship (SAR) of anthranilic diamides analogs containing oxadiazole rings. Org. Biomol. Chem. 2013, 11, 3979–3988. [Google Scholar] [CrossRef]
- Akano, Y.; Shiga, F.; Asano, J.; Hori, W.; Anraku, T.; Uno, T. Synthesis and AMPA receptor antagonistic activity of a novel 7-imidazolyl-6-trifluoromethyl quinoxalinecarboxylic acid with a substituted phenyl group and improved its good physicochemical properties by introduced CF3 group. J. Bioorg. Med. Chem. 2004, 14, 5107–5111. [Google Scholar]
- Bansal, S.; Bala, M.; Suthar, S.S.; Choudhary, S.; Bhattacharya, S.; Bhardwaj, V.; Singla, S.; Joseph, A. Design and synthesis of novel 2-phenyl-5-(1,3-diphenyl-1H-pyrazx; fol-4-yl)-1,3,4-oxadiazoles as selective COX-2 inhibitors with potent anti-inflammatory activity. Eur. J. Med. Chem. 2014, 80, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Kucukguzel, S.G.; Oruc, E.E.; Rollas, S.; Sahin, F.; Ozbek, A. Synthesis, characterisation and biological activity of novel 4-thiazolidinones, 1,3,4-oxadiazoles and some related compounds. Eur. J. Med. Chem. 2002, 37, 197–206. [Google Scholar] [CrossRef]
- Bondock, S.; Adel, S.; Etman, H.A.; Badria, F.A. Synthesis and antitumor evaluation of some new 1,3,4-oxadiazole-based heterocycles. Eur. J. Med. Chem. 2012, 48, 192–199. [Google Scholar] [CrossRef]
- Husain, A.; Ajmal, M. Synthesis of novel 1,3,4-oxadiazole derivatives and their biological properties. Acta Pharm. 2009, 59, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Rahim, F.; Ullah, H.; Javid, M.T.; Wadood, A.; Taha, M.; Ashraf, M.; Shaukat, A.; Junaid, M.; Hussain, S.; Rehman, W.; et al. Synthesis, in vitro evaluation and molecular docking studies of thiazole derivatives as new inhibitors of α-glucosidase. Bioorg. Chem. 2015, 62, 15–21. [Google Scholar] [CrossRef]
- Ullah, H.; Rahim, F.; Taha, M.; Uddin, I.; Wadood, A.; Ali Shah, S.A.; Farooq, R.K.; Nawaz, M.; Wahab, Z.; Khan, K.M. Synthesis, molecular docking study and in vitro thymidine phosphorylase inhibitory potential of oxadiazole derivatives. Bioorg. Chem. 2018, 78, 58–67. [Google Scholar] [CrossRef]
- Javid, M.T.; Rahim, F.; Taha, M.; Rehman, H.U.; Nawaz, M.; Wadood, A.; Imran, S.; Uddin, I.; Mosaddik, A.; Khan, K.M. Synthesis, in vitro α-glucosidase inhibitory potential and molecular docking study of thiadiazole analogs. Bioorg. Chem. 2018, 78, 201–209. [Google Scholar] [CrossRef]
- Taha, M.; Ismail, N.H.; Imran, S.; Selvaraj, M.; Rashwan, H.; Farhanah, F.U.; Rahim, F.; Kesavanarayanan, K.S.; Ali, M. Synthesis of benzimidazole derivatives as potent β-glucuronidase inhibitors. Bioorg. Chem. 2015, 61, 36–44. [Google Scholar] [CrossRef]
- Taha, M.; Shah, S.A.A.; Afifi, M.; Imran, S.; Sultan, S.; Rahim, F.; Khan, K.M. Synthesis, α-glucosidase inhibition and molecular docking study of coumarin based derivatives. Bioorg. Chem. 2018, 77, 586–592. [Google Scholar] [CrossRef]
- Taha, M.; Shah, S.A.A.; Afifi, M.; Imran, S.; Sultan, S.; Rahim, F.; Ismail, N.H.; Khan, K.M. Synthesis, molecular docking study and thymidine phosphorylase inhibitory activity of 3-formylcoumarin derivatives. Bioorg. Chem. 2018, 78, 17–23. [Google Scholar] [CrossRef]
- Taha, M.; Ismail, N.H.; Imran, S.; Wadood, A.; Rahim, F.; Khan, K.M.; Riaz, M. Hybrid benzothiazole analogs as antiurease agent: Synthesis and molecular docking studies. Bioorg. Chem. 2016, 66, 80–87. [Google Scholar] [CrossRef]
- Hu, C.; Solomon, V.R.; Ulibarri, G.; Lee, H. The efficacy and selectivity of tumor cell killing by Akt inhibitors are substantially increased by chloroquine. Bioorg. Med. Chem. 2008, 16, 7888–7893. [Google Scholar] [CrossRef]
- Hu, C.; Solomon, V.R.; Cano, P.; Lee, H. A 4-aminoquinoline derivative that markedly sensitizes tumor cell killing by Akt inhibitors with a minimum cytotoxicity to non-cancer cells. Eur. J. Med. Chem. 2010, 45, 705–709. [Google Scholar] [CrossRef]
- Keith, C.T.; Borisy, A.A.; Stockwell, B.R. Multicomponent therapeutics for networked systems. Nat. Rev. Drug Discov. 2005, 4, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Mayur, Y.C.; Peters, G.J.; Prasad, V.V.S.R.; Lemos, C.; Sathish, N.K. Design of New Drug Molecules to be Used in Reversing Multidrug Resistance in Cancer Cells. Curr. Cancer Drug Targets 2009, 9, 298–306. [Google Scholar] [CrossRef]
- Solomon, V.R.; Hu, C.; Lee, H. Hybrid pharmacophore design and synthesis of isatin–benzothiazole analogs for their anti-breast cancer activity. Bioorg. Med. Chem. 2009, 17, 7585–7592. [Google Scholar] [CrossRef]
- Hubschwerlen, C.; Specklin, J.L.; Baeschlin, D.K.; Borer, Y.; Haefeli, S.; Sigwalt, C.; Schroeder, S.; Locher, H.H. Structure–activity relationship in the oxazolidinone–quinolone hybrid series: Influence of the central spacer on the antibacterial activity and the mode of action. Bioorg. Med. Chem. Lett. 2003, 13, 4229–4233. [Google Scholar] [CrossRef]
- Bhanot, S.K.; Singh, M.; Chatterjee, N.R. The Chemical and Biological Aspects of Fluoroquinolones Reality and Dreams. Curr. Pharm. Des. 2001, 7, 311–335. [Google Scholar] [CrossRef]
- Adamec, J.; Beckert, R.; Weiss, D.; Klimesova, V.; Waisser, K.; Mollmann, U.; Kaustova, J.; Buchta, V. Hybrid molecules of estrone: New compounds with potential antibacterial, antifungal, and antiproliferative activities. Bioorg. Med. Chem. 2007, 15, 2898–2906. [Google Scholar] [CrossRef] [PubMed]
- Kouznetsov, V.V.; Gomez, A.-B. Recent developments in the design and synthesis of hybrid molecules basedon aminoquinoline ring and their antiplasmodial evaluation. Eur. J. Med. Chem. 2009, 44, 3091–3113. [Google Scholar] [CrossRef] [PubMed]
- Imran, S.; Taha, M.; Ismail, N.H.; Kashif, S.M.; Rahim, F.; Jamil, W.; Hariono, M.; Yusuf, M.; Wahab, H. Synthesis of novel flavone hydrazones: In-vitro evaluation of α-glucosidase inhibition, QSAR analysis and docking studies. Eur. J. Med. Chem. 2015, 105, 156–170. [Google Scholar] [CrossRef]
- Taha, M.; Ismail, N.H.; Imran, S.; Selvaraj, M.; Rahim, A.; Ali, M.; Siddiqui, S.; Rahim, F.; Khan, K.M. Synthesis of novel benzohydrazone-oxadiazole hybrids as β-glucuronidase inhibitors and molecular modeling studies. Bioorg. Med. Chem. 2015, 23, 7394–7404. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Ullah, H.; Muqarrabun, L.M.R.A.; Khan, M.N.; Rahim, F.; Ahmat, N.; Ali, M.; Perveen, S. Synthesis of bis-indolylmethanes as new potential inhibitors of β-glucuronidase and their molecular docking studies. Eur. J. Med. Chem. 2018, 143, 1757–1767. [Google Scholar] [CrossRef] [PubMed]
- Jamil, W.; Perveen, S.; Shah, S.A.A.; Taha, M.; Ismail, N.H.; Perveen, S.; Ambreen, N.; Khan, K.M.; Choudhary, M.I. Phenoxyacetohydrazide Schiff Bases: β-Glucuronidase Inhibitors. Molecules 2014, 19, 8788–8802. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 4.5, San Diego: Dassault Systèmes. 2015. Available online: https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/ (accessed on 20 December 2018).
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Stierand, K.; Maass, P.C.; Rarey, M. Molecular complexes at a glance: Automated generation of two-dimensional complex diagrams. Bioinformatics 2006, 22, 1710–1716. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |



| S. No | R | IC50 ± SEM a | S. No | R | IC50 ±SEM a |
|---|---|---|---|---|---|
| 1 | 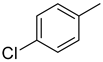 | 2.6 ± 0.1 | 11 | 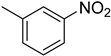 | 26.1 ± 0.5 |
| 2 | 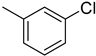 | 5.1 ± 0.2 | 12 | 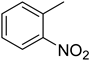 | 17.2 ± 0.4 |
| 3 | 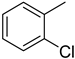 | 2.1 ± 0.1 | 13 | 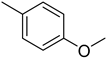 | 32.20 ± 0.6 |
| 4 |  | 1.1 ± 0.05 | 14 | 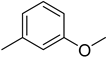 | 39.5 ± 0.7 |
| 5 | 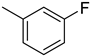 | 3.8 ± 0.2 | 15 | 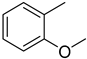 | 20.1 ± 0.5 |
| 6 | 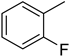 | 0.8 ± 0.1 | 16 | 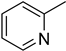 | 14.9 ± 0.4 |
| 7 | 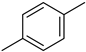 | 9.4 ± 0.3 | 17 | 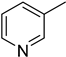 | 34.7 ± 0.6 |
| 8 | 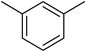 | 13.0 ± 0.3 | 18 | 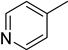 | 24.5 ± 0.5 |
| 9 | 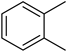 | 8.6 ± 0.3 | 19 |  | 42.3 ± 0.8 |
| 10 | 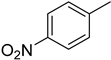 | 21.3 ± 0.4 | d-saccharic acid 1,4 lactone c | 48.1 ± 1.2 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taha, M.; Rahim, F.; Ali, M.; Khan, M.N.; Alqahtani, M.A.; Bamarouf, Y.A.; Gollapalli, M.; Farooq, R.K.; Shah, S.A.A.; Ahmed, Q.U.; et al. Synthesis of Chromen-4-One-Oxadiazole Substituted Analogs as Potent β-Glucuronidase Inhibitors. Molecules 2019, 24, 1528. https://doi.org/10.3390/molecules24081528
Taha M, Rahim F, Ali M, Khan MN, Alqahtani MA, Bamarouf YA, Gollapalli M, Farooq RK, Shah SAA, Ahmed QU, et al. Synthesis of Chromen-4-One-Oxadiazole Substituted Analogs as Potent β-Glucuronidase Inhibitors. Molecules. 2019; 24(8):1528. https://doi.org/10.3390/molecules24081528
Chicago/Turabian StyleTaha, Muhammad, Fazal Rahim, Muhammad Ali, Muhammad Naseem Khan, Mohammed A. Alqahtani, Yasser A. Bamarouf, Mohammed Gollapalli, Rai Khalid Farooq, Syed Adnan Ali Shah, Qamar Uddin Ahmed, and et al. 2019. "Synthesis of Chromen-4-One-Oxadiazole Substituted Analogs as Potent β-Glucuronidase Inhibitors" Molecules 24, no. 8: 1528. https://doi.org/10.3390/molecules24081528
APA StyleTaha, M., Rahim, F., Ali, M., Khan, M. N., Alqahtani, M. A., Bamarouf, Y. A., Gollapalli, M., Farooq, R. K., Shah, S. A. A., Ahmed, Q. U., & Zakaria, Z. A. (2019). Synthesis of Chromen-4-One-Oxadiazole Substituted Analogs as Potent β-Glucuronidase Inhibitors. Molecules, 24(8), 1528. https://doi.org/10.3390/molecules24081528






