Diamond Nanoparticles Downregulate Expression of CycD and CycE in Glioma Cells
Abstract
:1. Introduction
2. Results and Discussion
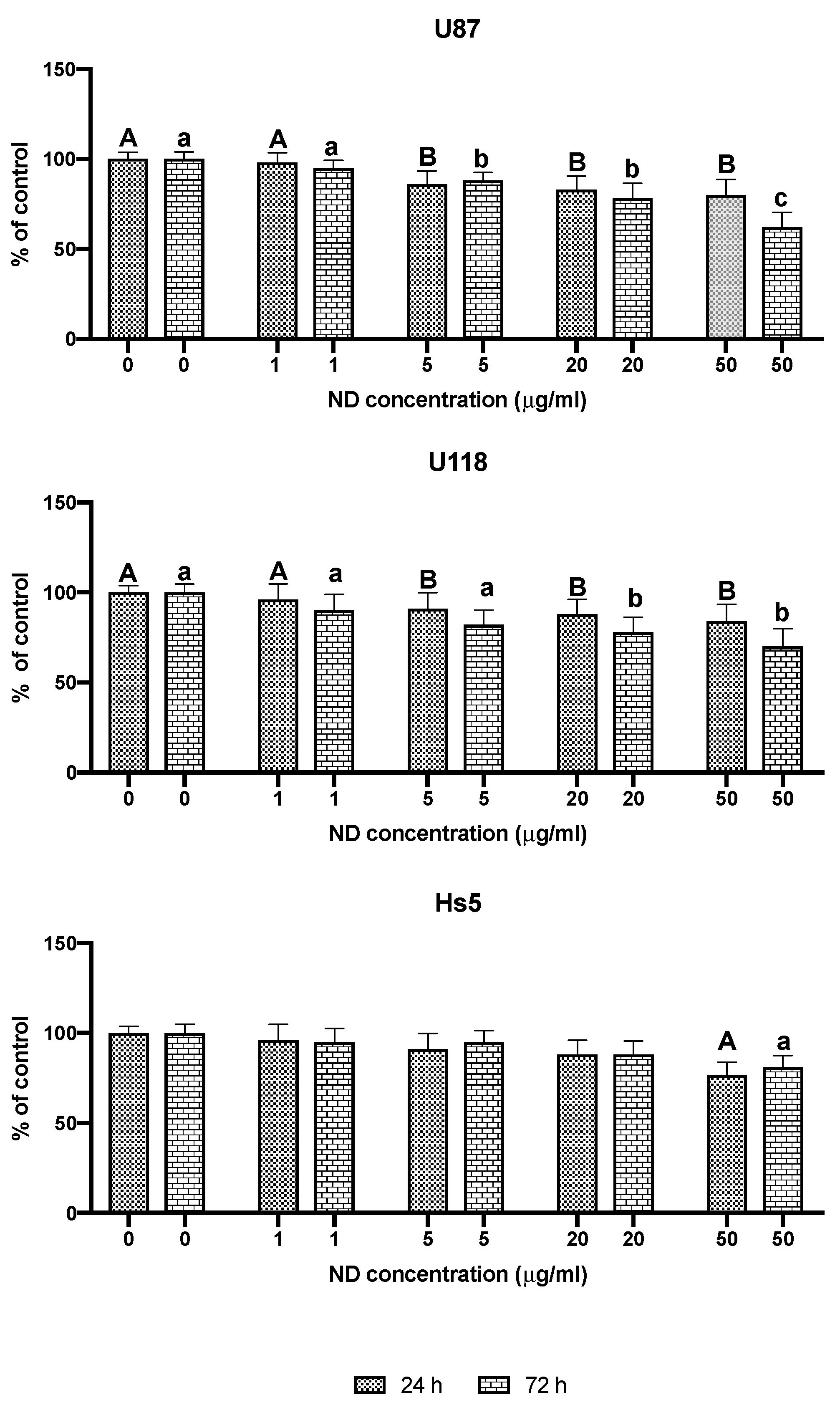
2.1. Characterization of NDs and Analysis of Cell Viability
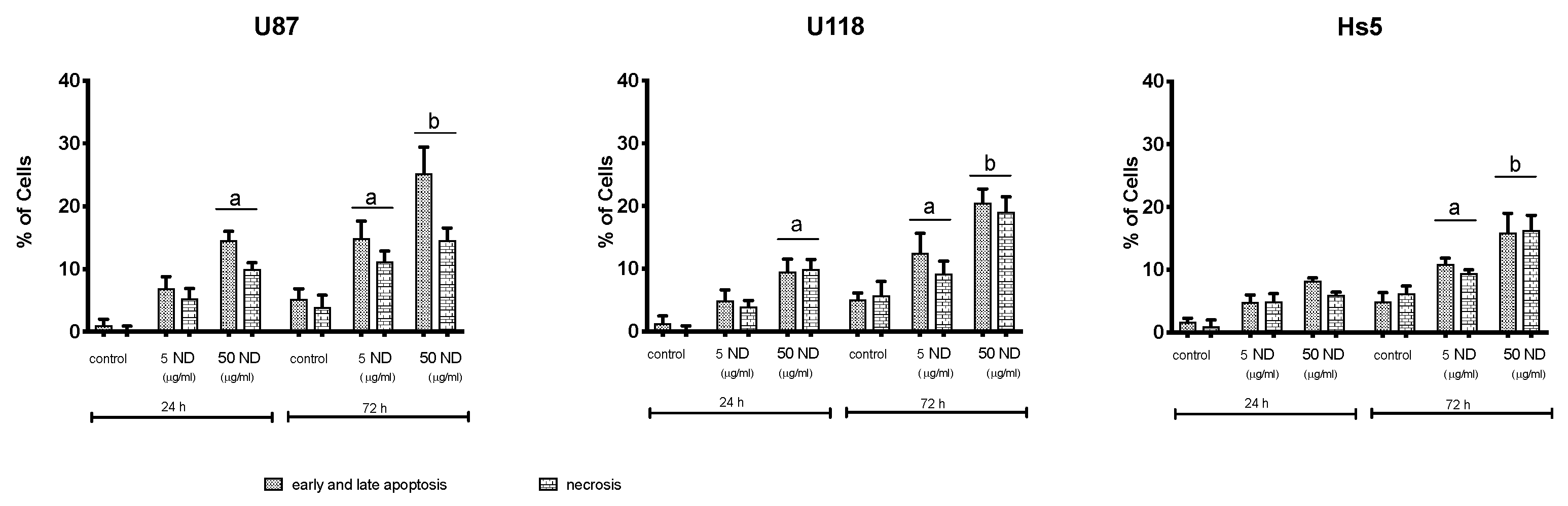
2.2. Analysis of Proliferation
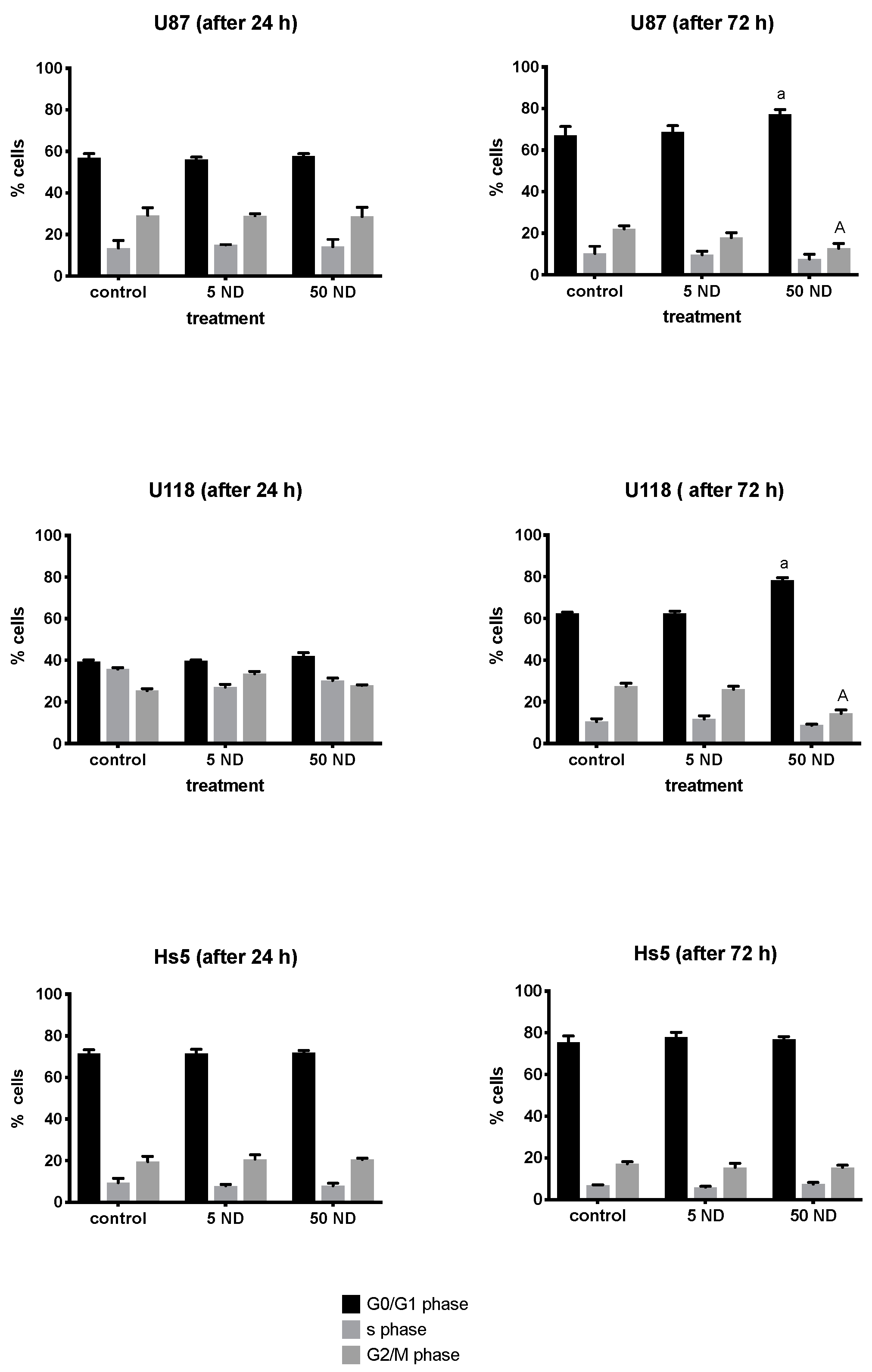
2.3. Analysis of Cell Cycle
3. Material and Methods
3.1. Nanomaterials
3.2. Visualization of Nanoparticles
3.3. Zeta Potential
3.4. Cells and Cell Cultures
3.5. Cell Morphology
3.6. Cell Viability Assays
3.7. Apoptosis and Necrosis Assay
3.8. Doubling Time Assay
3.9. Cell Proliferation Assay
3.10. c.DNA Synthesis and qPCR Analysis
3.11. Cell–Cycle Analysis
3.12. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Danilenko, V.V. On the history of the discovery of nanodiamond synthesis. Phys. Solid State 2004, 46, 595–599. [Google Scholar] [CrossRef]
- Holt, K.B. Diamond at the nanoscale: Applications of diamond nanoparticles from cellular biomarkers to quantum computing. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2007, 365, 2845–2861. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E. Glioblastoma: Overview of Disease and Treatment. Clin. J. Oncol. Nurs. 2016, 20, S2–S8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villà, S.; Balañà, C.; Comas, S. Radiation and concomitant chemotherapy for patients with glioblastoma multiforme. Chin. J. Cancer 2014, 33, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir-Kaynak, E.; Qutub, A.A.; Yesil-Celiktas, O. Advances in Glioblastoma Multiforme Treatment: New Models for Nanoparticle Therapy. Front. Physiol. 2018, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Bobo, R.H.; Laske, D.W.; Akbasak, A.; Morrison, P.F.; Dedrick, R.L.; Oldfield, E.H. Convection-enhanced delivery of macromolecules in the brain. Proc. Natl. Acad. Sci. USA 1994, 91, 2076–2080. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Yamashita, Y.; Nishihara, M.; Sugiyama, S.; Sonoda, Y.; Kumabe, T.; Tominaga, T. Therapeutic efficacy of a polymeric micellar doxorubicin infused by convection-enhanced delivery against intracranial 9L brain tumor models. Neuro-Oncology 2009, 11, 151–157. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.M.; Zhang, Y.F.; Xie, Y.D.; Cai, Y.F.; Li, B.Y.; Li, W.; Yu, R.T. Hypoxia-responsive ionizable liposome delivery siRNA for glioma therapy. Int. J. Nanomed. 2017, 12, 1065–1083. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.; Dai, L. Nanodiamonds for nanomedicine. Nanomedicine 2009, 4, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Shkurupy, V.A.; Arkhipov, S.A.; Neshchadim, D.V.; Akhramenko, E.S.; Troitskii, A.V. In Vitro Effects of Nanosized Diamond Particles on Macrophages. Bull. Exp. Biol. Med. 2015, 158, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Kurantowicz, N.; Strojny, B.; Sawosz, E.; Jaworski, S.; Kutwin, M.; Grodzik, M.; Wierzbicki, M.; Lipińska, L.; Mitura, K.; Chwalibog, A. Biodistribution of a High Dose of Diamond, Graphite, and Graphene Oxide Nanoparticles After Multiple Intraperitoneal Injections in Rats. Nanoscale Res. Lett. 2015, 10, 398. [Google Scholar] [CrossRef] [PubMed]
- Strojny, B.; Kurantowicz, N.; Sawosz, E.; Grodzik, M.; Jaworski, S.; Kutwin, M.; Wierzbicki, M.; Hotowy, A.; Lipinska, L.; Chwalibog, A. Long Term Influence of Carbon Nanoparticles on Health and Liver Status in Rats. PLoS ONE 2015, 10, e0144821. [Google Scholar] [CrossRef] [PubMed]
- Brož, A.; Bačáková, L.; Štenclová, P.; Kromka, A.; Potocký, Š.; Berbezier, I. Uptake and intracellular accumulation of diamond nanoparticles—A metabolic and cytotoxic study. Beilstein J. Nanotechnol. 2017, 8, 1649–1657. [Google Scholar] [CrossRef]
- Solarska, K.; Gajewska, A.; Bartosz, G.; Mitura, K. Induction of apoptosis in human endothelial cells by nanodiamond particles. J. Nanosci. Nanotechnol. 2012, 12, 5117–5121. [Google Scholar] [CrossRef]
- Niemiec, T.; Szmidt, M.; Sawosz, E.; Grodzik, M.; Mitura, K. The effect of diamond nanoparticles on redox and immune parameters in rats. J. Nanosci. Nanotechnol. 2011, 11, 9072–9077. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Nishikawa, M.; Liu, M.; Tei, T.; Kaul, S.C.; Wadhawa, R.; Zhang, M.; Takahashi, J.; Miyako, E. Self-assembled nanodiamond supraparticles for anticancer chemotherapy. Nanoscale 2018, 10, 8969–8978. [Google Scholar] [CrossRef]
- Wang, X.; Low, X.C.; Hou, W.; Abdullah, L.N.; Toh, T.B.; Mohd Abdul Rashid, M.; Ho, D.; Chow, E.K. Epirubicin-adsorbed nanodiamonds kill chemoresistant hepatic cancer stem cells. ACS Nano 2014, 8, 12151–12166. [Google Scholar] [CrossRef] [PubMed]
- Grodzik, M.; Sawosz, E.; Wierzbicki, M.; Orlowski, P.; Hotowy, A.; Niemiec, T.; Szmidt, M.; Mitura, K.; Chwalibog, A. Nanoparticles of carbon allotropes inhibit glioblastoma multiforme angiogenesis in ovo. Int. J. Nanomed. 2011, 6, 3041. [Google Scholar] [CrossRef]
- Grodzik, M. Changes in glioblastoma multiforme ultrastructure after diamond nanoparticles treatment. Experimental model in ovo. Ann. Warsaw Univ. Life Sci. SGGW Anim. Sci. 2013, 52, 29–35. [Google Scholar]
- Wierzbicki, M.; Jaworski, S.; Kutwin, M.; Grodzik, M.; Strojny, B.; Kurantowicz, N.; Zdunek, K.; Chodun, R.; Chwalibog, A.; Sawosz, E. Diamond, graphite, and graphene oxide nanoparticles decrease migration and invasiveness in glioblastoma cell lines by impairing extracellular adhesion. Int. J. Nanomed. 2017, 2, 7241–7254. [Google Scholar] [CrossRef]
- Olmeda, D.; Castel, S.; Vilaró, S.; Cano, A. Beta-catenin regulation during the cell cycle: Implications in G2/M and apoptosis. Mol. Biol. Cell 2003, 14, 2844–2860. [Google Scholar] [CrossRef] [PubMed]
- Clogston, J.D.; Patri, A.K. Zeta potential measurement. Methods Mol. Biol. 2011, 697, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Yur’ev, G.S.; Dolmatov, V.Y. X-ray diffraction study of detonation nanodiamonds. J. Superhard Mater. 2010, 32, 311–328. [Google Scholar] [CrossRef]
- Kurantowicz, N.; Sawosz, E.; Halik, G.; Strojny, B.; Hotowy, A.; Grodzik, M.; Piast, R.; Pasanphan, W.; Chwalibog, A. Toxicity studies of six types of carbon nanoparticles in a chicken-embryo model. Int. J. Nanomed. 2017, 12, 2887–2898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chwalibog, A.; Sawosz, E.; Hotowy, A.; Szeliga, J.; Mitura, S.; Mitura, K.; Grodzik, M.; Orlowski, P.; Sokolowska, A. Visualization of interaction between inorganic nanoparticles and bacteria or fungi. Int. J. Nanomed. 2010, 5, 1085. [Google Scholar] [CrossRef] [PubMed]
- Hinzmann, M.; Jaworski, S.; Kutwin, M.; Jagiełło, J.; Koziński, R.; Wierzbicki, M.; Grodzik, M.; Lipińska, L.; Sawosz, E.; Chwalibog, A. Nanoparticles containing allotropes of carbon have genotoxic effects on glioblastoma multiforme cells. Int. J. Nanomed. 2014, 9, 2409–2417. [Google Scholar] [CrossRef] [Green Version]
- Keremidarska, M.; Ganeva, A.; Mitev, D.; Hikov, T.; Presker, R.; Pramatarova, L.; Krasteva, N. Comparative study of cytotoxicity of detonation nanodiamond particles with an osteosarcoma cell line and primary mesenchymal stem cells. Biotechnol. Biotechnol. Equip. 2014, 28, 733–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitura, K.; Jedrzejewska-Szczerska, M.; Ceynowa, P.; Dudek, M.; Cicha, M.; Kotela, I.; Mitura, S. Haemocompatibility Of Non-Functionalized and Plasmachemical Functionalized Detonation Nanodiamond Particles. Arch. Metall. Mater. 2015, 60, 2183–2189. [Google Scholar] [CrossRef]
- Adach, K.; Fijalkowski, M.; Gajek, G.; Skolimowski, J.; Kontek, R.; Blaszczyk, A. Studies on the cytotoxicity of diamond nanoparticles against human cancer cells and lymphocytes. Chem. Biol. Interact. 2016, 254, 156–166. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, J.; Li, W.; Zhang, Y.; Yang, X.; Chen, N.; Sun, Y.; Zhao, Y.; Fan, C.; Huang, Q. The biocompatibility of nanodiamonds and their application in drug delivery systems. Theranostics 2012, 2, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Schrand, A.M.; Huang, H.; Carlson, C.; Schlager, J.J.; Osawa, E.; Hussain, S.M.; Dai, L. Are Diamond Nanoparticles Cytotoxic? J. Phys. Chem. B 2007, 111, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Strojny, B.; Grodzik, M.; Sawosz, E.; Winnicka, A.; Kurantowicz, N.; Jaworski, S.; Kutwin, M.; Urbańska, K.; Hotowy, A.; Wierzbicki, M.; Chwalibog, A.; et al. Diamond Nanoparticles Modify Curcumin Activity: In Vitro Studies on Cancer and Normal Cells and In Ovo Studies on Chicken Embryo Model. PLoS ONE 2016, 11, e0164637. [Google Scholar] [CrossRef] [PubMed]
- Alenzi, F.Q.B. Links between apoptosis, proliferation and the cell cycle. Br. J. Biomed. Sci. 2004, 61, 99–102. [Google Scholar] [CrossRef]
- Thomas, V.; Halloran, B.A.; Ambalavanan, N.; Catledge, S.A.; Vohra, Y.K. In vitro studies on the effect of particle size on macrophage responses to nanodiamond wear debris. Acta Biomater. 2012, 8, 1939–1947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholzen, T.; Gerdes, J. The Ki-67 protein: From the known and the unknown. J. Cell Physiol. 2000, 182, 311–322. [Google Scholar] [CrossRef]
- Memmel, S.; Sukhorukov, V.L.; Höring, M.; Westerling, K.; Fiedler, V.; Katzer, A.; Krohne, G.; Flentje, M.; Djuzenova, C.S. Cell Surface Area and Membrane Folding in Glioblastoma Cell Lines Differing in PTEN and p53 Status. PLoS ONE 2014, 9, e87052. [Google Scholar] [CrossRef] [PubMed]
- Mastronardi, L.; Guiducci, A.; Puzzilli, F.; Ruggeri, A. Relationship between Ki-67 labeling index and survival in high-grade glioma patients treated after surgery with tamoxifen. J. Neurosurg. Sci. 1999, 43, 263–270. [Google Scholar]
- Cooper, G.M. The Eukaryotic Cell Cycle. In The Cell: A Molecular Approach; Sinauer Associates: Sunderland, MA, USA, 2000; ISBN 0878931066. [Google Scholar]
- Foster, D.A.; Yellen, P.; Xu, L.; Saqcena, M. Regulation of G1 Cell Cycle Progression: Distinguishing the Restriction Point from a Nutrient-Sensing Cell Growth Checkpoint(s). Genes Cancer 2010, 1, 1124–1131. [Google Scholar] [CrossRef]
- Grodzik, M.; Sawosz, E.; Wierzbicki, M.; Hotowy, A.M.; Prasek, M.; Jaworski, J.; Chwalibog, A. VEGF-dependent mechanism of anti-angiogenic action of diamond nanoparticles in glioblastoma multiforme tumor. TechConnect Briefs 2012, 3, 218–221. [Google Scholar]
- Downward, J. Cell cycle: Routine role for Ras. Curr. Biol. 1997, 7, R258–R260. [Google Scholar] [CrossRef] [Green Version]
- Carrasco-García, E.; Saceda, M.; Martínez-Lacaci, I. Role of receptor tyrosine kinases and their ligands in glioblastoma. Cells 2014, 3, 199–235. [Google Scholar] [CrossRef] [PubMed]
- Doubling Time. Available online: http://www.doubling-time.com/compute.php (accessed on 16 April 2019).
- Aithal, M.G.; Rajeswari, N. Validation of housekeeping genes for gene expression analysis in glioblastoma using quantitative real-time polymerase chain reaction. Brain Tumor Res. Treat. 2015, 3, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, M.A.; Huang, Q.; Li, F.; Liu, X.; Li, C.-Y. Cell death-stimulated cell proliferation: A tissue regeneration mechanism usurped by tumors during radiotherapy. Semin. Radiat. Oncol. 2013, 23, 288–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| U87 | U118 | Hs5 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | 5 μg/mL ND | 50 μg/mL ND | Control | 5 μg/mL ND | 50 μg/mL ND | Control | 5 μg/mL ND | 50 μg/mL ND | |
| Incubation time, h | |||||||||
| 0 | 3.0 × 104 | 3.0 × 104 | 3.0 × 104 | 3.00 × 104 | 3.00 × 104 | 3.00 × 104 | 3.00 × 104 | 3.0 × 104 | 3.0 × 104 |
| 24 | 5.2 × 104 | 5.0 × 104 | 3.5 × 104 | 6.92 × 104 | 6.89 × 104 | 6.60 × 104 | 3.80 × 104 | 3.80 × 104 | 3.80 × 104 |
| 72 | 105 | 9.2 × 104 | 6.7 × 104 | 3.90 × 105 | 4.05 × 105 | 3.00 × 105 | 6.05 × 104 | 6.00 × 104 | 6.00 × 104 |
| Doubling time, h | 32 | 34 | 39 | 19 | 19 | 23 | 71 | 72 | 72 |
| Gene | Sequence | Source |
|---|---|---|
| TBP | F: GAGCTGTGATGTGAAGTTTCC R: TCTGGGTTTGATCATTCTGTAG | 44 |
| CycA2 | F: TTATTGCTGGAGCTGCCTTT R: CTCTGGTGGGTTGAGGAGAG | 45 |
| CycD2 | F: TACTTCAAGTGCGTGCAGAAGGAC R: TCCCACACTTCCAGTTGCGATCAT | sp |
| CycB1 | F: GGCTTCCTCTTCACCAGGCA R: CGCGATCGCCCTGGAAAC | sp |
| CycE2 | F: AATCAGGCAAAGGTGAAGGA R: CCCCAAGAAGCCCAGATAAT | sp |
| Rb1 | F: CGGGAGTCGGGAGAGGACGG R: CGAGAGGCAGGTCCTCCGGG | sp |
| E2F1 | F: ACCTTCGTAGCATTGCAGACC R: TTCTTGCTCCAGGCTGAGTAG | 46 |
| PCNA | F: CCATCCTCAAGAAGGTGTTGG R: GTGTCCCATATCCGCAATTTTAT | 47 |
| Ki-67 | F: CCACACTGTGTCGTCGTTTG R: CCGTGCGCTTATCCATTCA | sp |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grodzik, M.; Szczepaniak, J.; Strojny-Cieslak, B.; Hotowy, A.; Wierzbicki, M.; Jaworski, S.; Kutwin, M.; Soltan, E.; Mandat, T.; Lewicka, A.; et al. Diamond Nanoparticles Downregulate Expression of CycD and CycE in Glioma Cells. Molecules 2019, 24, 1549. https://doi.org/10.3390/molecules24081549
Grodzik M, Szczepaniak J, Strojny-Cieslak B, Hotowy A, Wierzbicki M, Jaworski S, Kutwin M, Soltan E, Mandat T, Lewicka A, et al. Diamond Nanoparticles Downregulate Expression of CycD and CycE in Glioma Cells. Molecules. 2019; 24(8):1549. https://doi.org/10.3390/molecules24081549
Chicago/Turabian StyleGrodzik, Marta, Jaroslaw Szczepaniak, Barbara Strojny-Cieslak, Anna Hotowy, Mateusz Wierzbicki, Slawomir Jaworski, Marta Kutwin, Emilia Soltan, Tomasz Mandat, Aneta Lewicka, and et al. 2019. "Diamond Nanoparticles Downregulate Expression of CycD and CycE in Glioma Cells" Molecules 24, no. 8: 1549. https://doi.org/10.3390/molecules24081549
APA StyleGrodzik, M., Szczepaniak, J., Strojny-Cieslak, B., Hotowy, A., Wierzbicki, M., Jaworski, S., Kutwin, M., Soltan, E., Mandat, T., Lewicka, A., & Chwalibog, A. (2019). Diamond Nanoparticles Downregulate Expression of CycD and CycE in Glioma Cells. Molecules, 24(8), 1549. https://doi.org/10.3390/molecules24081549









