Identification and Functional Analysis of a Flavonol Synthase Gene from Grape Hyacinth
Abstract
1. Introduction
2. Results
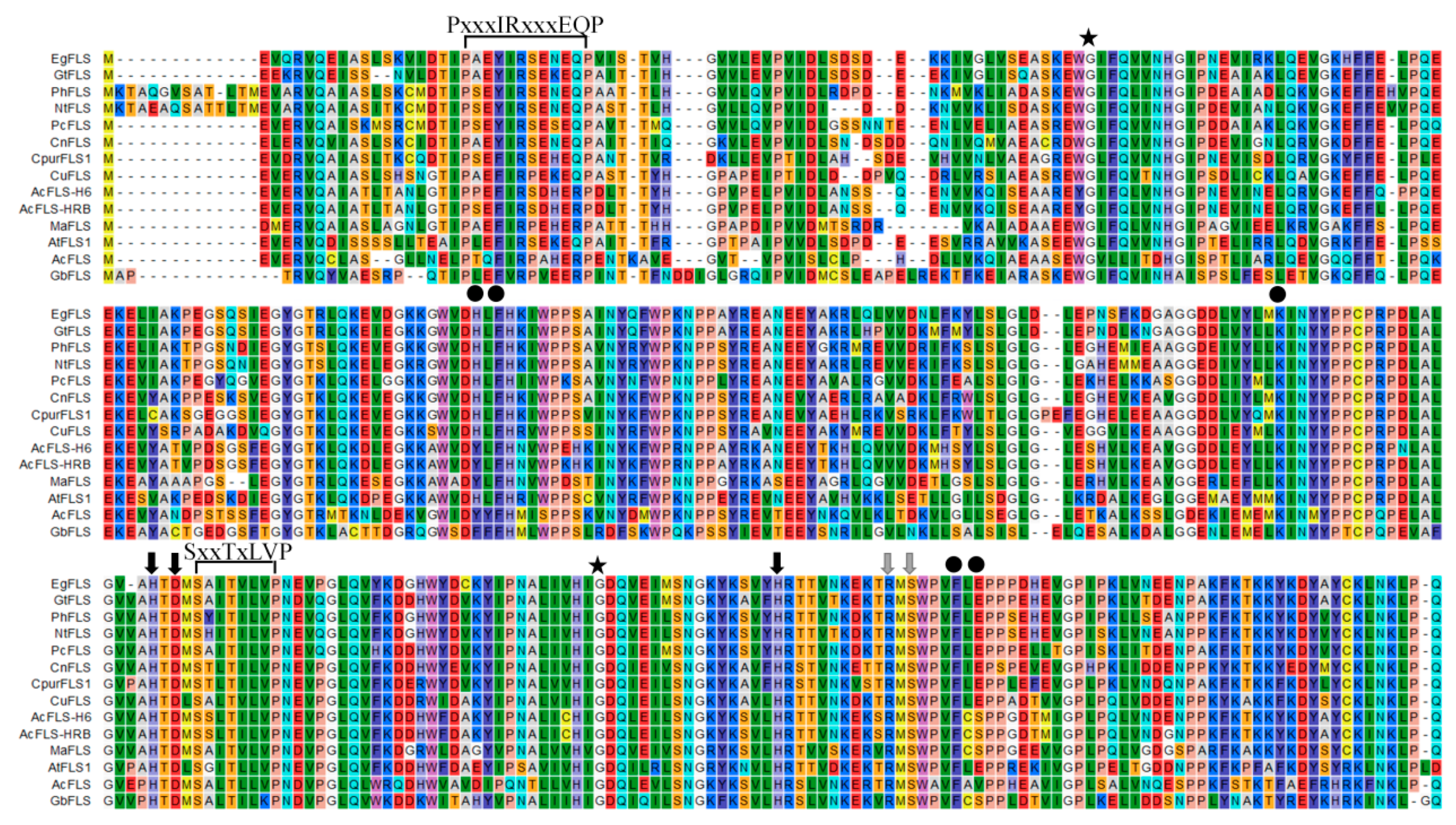
2.1. MaFLS Gene Cloning and Sequence Analysis
2.2. Correlation Analysis of MaFLS Expression Levels and Total Flavonol Content
2.3. In Vivo Localization of MaFLS
2.4. Heterologous Expression of MaFLS in Tobacco Alters Petal Color
2.5. Heterologous Expression of MaFLS in Tobacco Affects the Expression of Anthocyanin Pathway Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Cloning of Full-Length MaFLS cDNAs
4.3. Sequence Alignment and Phylogenetic Analysis
4.4. RNA Isolation and qRT-PCR Analysis
4.5. Subcellular Localization of MaFLS
4.6. Heterologous Expression Vector Construction and Stable Tobacco Transformation
4.7. HPLC Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| FLS | flavonol synthase. |
| DFR | dihydroflavonol 4-reductase |
| ANS | anthocyanidin synthase |
| qRT-PCR | quantitative real-time PCR |
| DHK | dihydrokaempferol |
| DHQ | dihydroquercetin |
| ORF | open reading frame |
| PCR | polymerase chain reaction |
| UFGT | glucosyltransferase |
| 2-ODD | 2-oxoglutarate-dependent dioxygenase |
References
- Nielsen, K.; Deroles, S.; Markham, K.; Bradley, M.; Podivinsky, E.; Manson, D. Antisense flavonol synthase alters copigmentation and flower color in lisianthus. Mol. Breed. 2002, 9, 217–229. [Google Scholar] [CrossRef]
- Davies, K.; Schwinn, K.; Deroles, S.; Manson, D.; Lewis, D.; Bloor, S.; Bradley, J. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 2003, 131, 259–268. [Google Scholar] [CrossRef]
- Brouillard, R.; Dangles, O. Flavonoids and flower colour. In The Flavonoids, Advances in Research since 1986; Harborne, J.B., Ed.; Chapman & Hall: London, UK, 1993; pp. 565–588. [Google Scholar]
- Zhou, X.; Fan, Z.; Chen, Y.; Zhu, Y.; Li, J.; Yin, H. Functional analyses of a flavonol synthase-like gene from Camellia nitidissima reveal its roles in flavonoid metabolism during floral pigmentation. J. Bio. Sci. 2013, 38, 593–604. [Google Scholar] [CrossRef]
- Britsch, L.; Heller, W.; Grisebach, H. Conversion of flavanone to flavone, dihydroflavonol and flavonol with an enzyme system from cell culture of parsley. Z. Naturforsch 1981, 36, 742–750. [Google Scholar] [CrossRef]
- Holton, T.; Brugliera, F.; Tanaka, Y. Cloning and expression of flavonol synthase from Petunia hybrida. Plant J. 1993, 4, 1003–1010. [Google Scholar] [CrossRef]
- Martens, S.; Preuß, A.; Matern, U. Multifunctional flavonoid dioxygenases: Flavonol and anthocyanin biosynthesisin Arabidopsis thaliana L. Phytochem. 2010, 71, 1040–1049. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, M.; Murrell, J.; Shirley, B. Characterization of flShirle synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis-further evidence for differential regulation of early and late genes. Plant Physiol. 1997, 113, 1437–1445. [Google Scholar] [CrossRef]
- Owens, D.; Alerding, A.; Crosby, K.; Bandara, A.; Westwood, J.; Winkel, B. Functional analysis of a predicted favonol synthase gene family in Arabidopsis. Plant Physiol. 2008, 147, 1046–1061. [Google Scholar] [CrossRef]
- Preuß, A.; Stracke, R.; Weisshaar, B.; Hillebrecht, A.; Matern, U.; Martens, S. Arabidopsis thaliana expresses a second functional flavonol synthase. FEBS Lett. 2009, 583, 1981–1986. [Google Scholar] [CrossRef] [PubMed]
- Moriguchi, T.; Kita, M.; Ogawa, K.; Tomono, Y.; Endo, T.; Omura, M. Flavonol synthase gene expression during citrus fruit development. Physiol. Plant 2002, 114, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.; Githiri, S.; Hatayama, K.; Dubouzet, E.; Shimada, N.; Aoki, T.; Ayabe, S.; Iwashina, T.; Toda, K.; Matsumura, H. A single-base deletion in soybean flavonol synthase gene is associated with magenta flower color. Plant Mol. Biol. 2007, 63, 125–135. [Google Scholar] [CrossRef]
- Kimura, S.; Nakatsuka, T.; Yamada, E.; Saito, M.; Nishihara, M. A flavonol synthase gene GtFLS defines anther-specific flavonol accumulation in gentian. Plant Biotechnol. 2010, 28, 211–221. [Google Scholar] [CrossRef]
- Xu, F.; Li, L.; Zhang, W.; Cheng, H.; Sun, N.; Cheng, S.; Wang, Y. Isolation, characterization, and function analysis of a flavonol synthase gene from Ginkgo biloba. Mol. Biol. Rep. 2012, 39, 2285–2296. [Google Scholar] [CrossRef]
- Ferreyra, F.; Rius, S.; Emiliani, J.; Pourcel, L.; Feller, A.; Morohashi, K.; Casati, P.; Grotewold, E. Cloning and characterization of a UV-B inducible maize fl E. Cl synthase. Plant J. 2010, 62, 77–91. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, H.; Jia, C.; Liu, Y.; Wang, F.; Wang, J. Cloning, characterization and functional analysis of a flavonol synthase from Vaccinium corymbosum. Trees 2016, 30, 1595–1605. [Google Scholar] [CrossRef]
- Park, S.; Kim, D.; Lee, J.; Ha, S.; Lim, S. Comparative analysis of two flavonol synthases from different-colored onions provides insight into flavonoid biosynthesis. J. Agric. Food Chem. 2017, 65, 5287–5298. [Google Scholar] [CrossRef]
- Akita, Y.; Kitamura, S.; Mikami, R.; Ishizaka, H. Identification of functional flavonol synthase genes from fragrant wild cyclamen (Cyclamen purpurascens). J. Plant Biochem. Biotechnol. 2018, 27, 147–155. [Google Scholar] [CrossRef]
- Liu, W.; Xiao, Z.; Fan, C.; Jiang, N.; Meng, X.; Xiang, X. Cloning and characterization of a flavonol synthase gene from Litchi chinensis and its variation among litchi cultivars with different fruit maturation periods. Front. Plant Sci. 2018, 9, 567. [Google Scholar] [CrossRef]
- Luo, P.; Ning, G.; Wang, Z.; Shen, Y.; Jin, H.; Li, P.; Huang, S.; Zhao, J.; Bao, M. Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants. Front. Plant. Sci. 2016, 6, 1257. [Google Scholar] [CrossRef]
- Yuan, Y.; Rebocho, A.; Sagawa, J.; Stanley, L.; Bradshaw, H. Competition between anthocyanin and flavonol biosynthesis produces spatial pattern variation of floral pigments between Mimulus species. PNAS 2016, 113, 2448–2453. [Google Scholar] [CrossRef]
- Lou, Q.; Wang, L.; Liu, H.; Liu, Y. Anthocyanin profiles in flowers of grape hyacinth. Molecules 2017, 22, 688. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Liu, H.; Lou, Q.; Liu, Y. Ectopic expression of the grape hyacinth (Muscari armeniacum) R2R3-MYB transcription factor gene, MaAN2, induces anthocyanin accumulation in tobacco. Front. Plant Sci. 2017, 8, 965. [Google Scholar] [CrossRef]
- Lou, Q.; Liu, Y.; Qi, Y.; Jiao, S.; Tian, F.; Jiang, L.; Wang, Y. Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. J. Exp. Bot. 2014, 65, 3157–3164. [Google Scholar] [CrossRef]
- Jiao, S.; Liu, Y.; Lou, Q.; Jiang, L. Cloning and expression analysis of dihydroflavonol 4-reductase gene (DFR) from grape hyacinth (Muscari armeniacum). J. Agric. Biotechnol. 2014, 22, 529–540. (in Chinese). [Google Scholar]
- Chua, C.; Biermann, D.; Goo, K.; Sim, T. Elucidation of active site residues of Arabidopsis thaliana flavonol synthase provides a molecular platform for engineering flavonols. Phytochemistry 2008, 69, 66–75. [Google Scholar] [CrossRef]
- Stracke, R.; De Vos, R.; Bartelniewoehner, L.; Ishihara, H.; Sagasser, M.; Martens, S.; Weisshaar, B. Metabolomic and genetic analyses of flavonol synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta 2009, 229, 427–445. [Google Scholar] [CrossRef]
- Stich, K.; Eidenberger, T.; Wurst, F. Flavonol synthase activity and the regulation of flavonol and anthocyanin biosynthesis during flower development in Dianthus caryophyllus L. (carnation). Z. Naturforsch. 1992, 47, 553–560. [Google Scholar] [CrossRef]
- Mori, S.; Asano, S.; Kobayashi, H.; Nakano, M. Analyses of anthocyanidins and anthocyanins in flowers of Muscari spp. Ngt. Dgk. Ngk. Knkyu. Hkk. 2002, 55, 13–18. [Google Scholar]
- Yoshida, K.; Aoki, H.; Kameda, K.; Kondo, T. Structure of muscarinin A, an acylated anthocyanin, from purplish blue spicate flower petals of Muscari arumeniacum. ITE Lett. Batter New Technol. Med. 2002, 1, 35–38. [Google Scholar]
- Qi, Y.; Lou, Q.; Li, H.; Yue, J.; Liu, Y.; Wang, Y. Anatomical and biochemical studies of bicolored flower development in Muscari latifolium. Protoplasma 2013, 250, 1273–1281. [Google Scholar] [CrossRef]
- Markham, K.; Ofman, D. Lisianthus flavonoid pigments and factors influencing their expression in flower colour. Phytochemistry 1993, 34, 679–685. [Google Scholar] [CrossRef]
- Gronquist, M.; Bezzerides, A.; Attygalle, A.; Meinwald, J.; Eisner, M.; Eisner, T. Attractive and defensive functions of the ultraviolet pigments of a flower (Hypericum calycinum). Proc. Natl. Acad. Sci. USA 2001, 98, 13745–13750. [Google Scholar] [CrossRef]
- Mo, Y.; Nagel, C.; Taylor, L. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc. Natl. Acad. Sci. USA 1992, 89, 7213–7217. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, B.; Geisler, M.; Bigler, L.; Ring, C. Flavonols accumulate asymmetrically and affect auxin transport in Arabidopsis. Plant Physiol. 2011, 156, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Saslowsky, D.; Warek, U.; Winkel, B. Nuclear localization of flavonoid enzymes in Arabidopsis. J. Biol. Chem. 2005, 280, 23735–23740. [Google Scholar] [CrossRef]
- Yoo, S.; Cho, Y.; Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Horsch, R.; Fry, J.; Hoffmann, N.; Eichholtz, D.; Rogers, S.; Fraley, R. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1232. [Google Scholar]
- Gao, L.; Mazza, G. Characterization, quantitation, and distribution of anthocyanins and colorless phenolics in sweet cherries. J. Agric. Food Chem. 1995, 43, 343–346. [Google Scholar] [CrossRef]
- Hashimoto, F.; Tanaka, M.; Maeda, H.; Shimizu, K.; Sakata, Y. Characterization of cyanic flower color of Delphinium cultivars. J. Jpn. Soc. Hort. Sci. 2000, 69, 428–434. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, Q.; Ji, D.; Niu, L.; Zhang, Y. Determination of the phenolic content, profile, and antioxidant activity of seeds from nine tree peony (Paeonia section Moutan DC.) species native to China. Food Res. Int. 2017, 97, 141–148. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds analyzed in the study are unavailable from the authors due to their isolation on a small scale. |






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Su, B.; Zhang, H.; Gong, J.; Zhang, B.; Liu, Y.; Du, L. Identification and Functional Analysis of a Flavonol Synthase Gene from Grape Hyacinth. Molecules 2019, 24, 1579. https://doi.org/10.3390/molecules24081579
Liu H, Su B, Zhang H, Gong J, Zhang B, Liu Y, Du L. Identification and Functional Analysis of a Flavonol Synthase Gene from Grape Hyacinth. Molecules. 2019; 24(8):1579. https://doi.org/10.3390/molecules24081579
Chicago/Turabian StyleLiu, Hongli, Beibei Su, Han Zhang, Jiaxin Gong, Boxiao Zhang, Yali Liu, and Lingjuan Du. 2019. "Identification and Functional Analysis of a Flavonol Synthase Gene from Grape Hyacinth" Molecules 24, no. 8: 1579. https://doi.org/10.3390/molecules24081579
APA StyleLiu, H., Su, B., Zhang, H., Gong, J., Zhang, B., Liu, Y., & Du, L. (2019). Identification and Functional Analysis of a Flavonol Synthase Gene from Grape Hyacinth. Molecules, 24(8), 1579. https://doi.org/10.3390/molecules24081579



