Tuscan Varieties of Sweet Cherry Are Rich Sources of Ursolic and Oleanolic Acid: Protein Modeling Coupled to Targeted Gene Expression and Metabolite Analyses
Abstract
1. Introduction
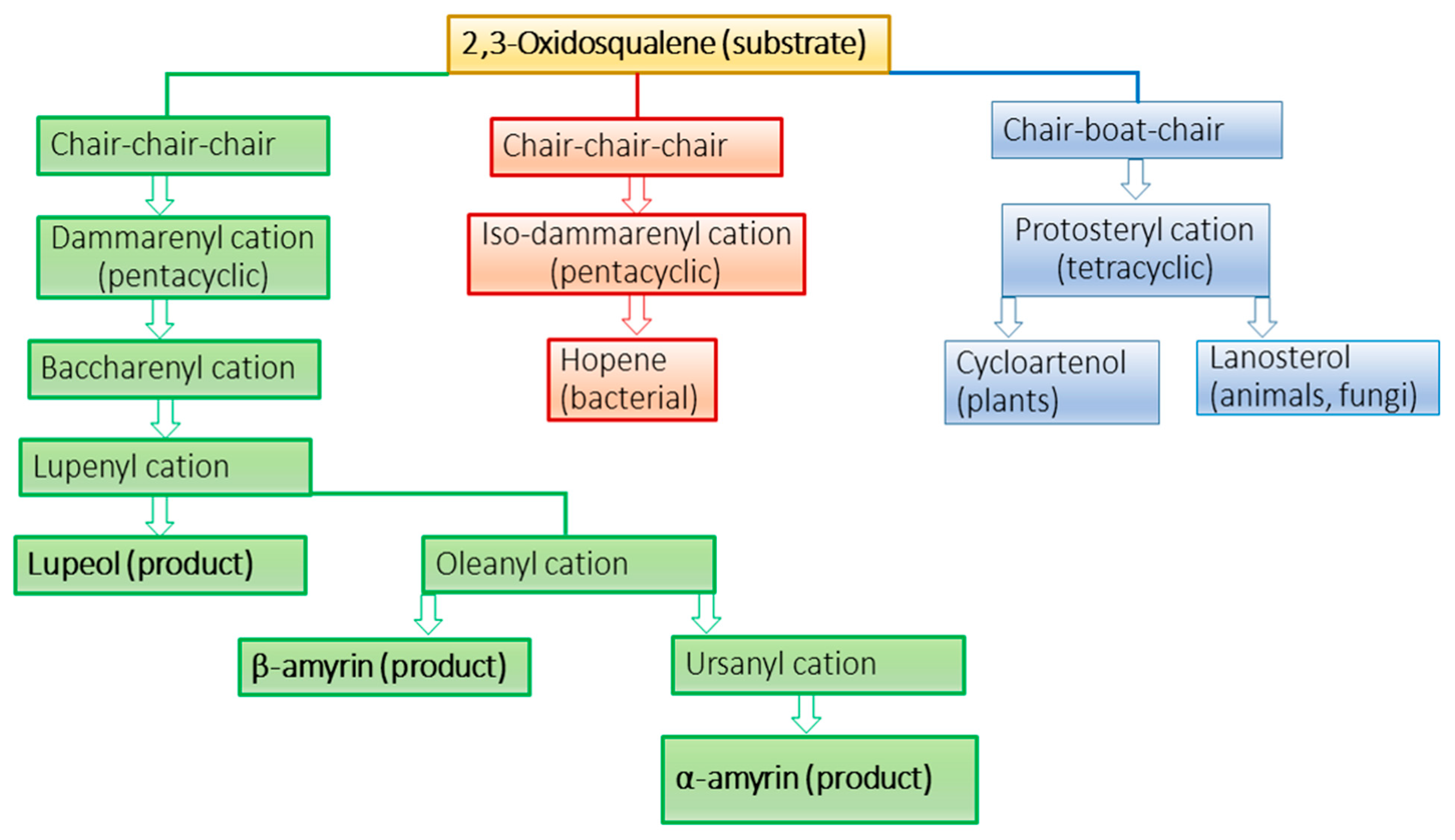
2. Results and Discussion
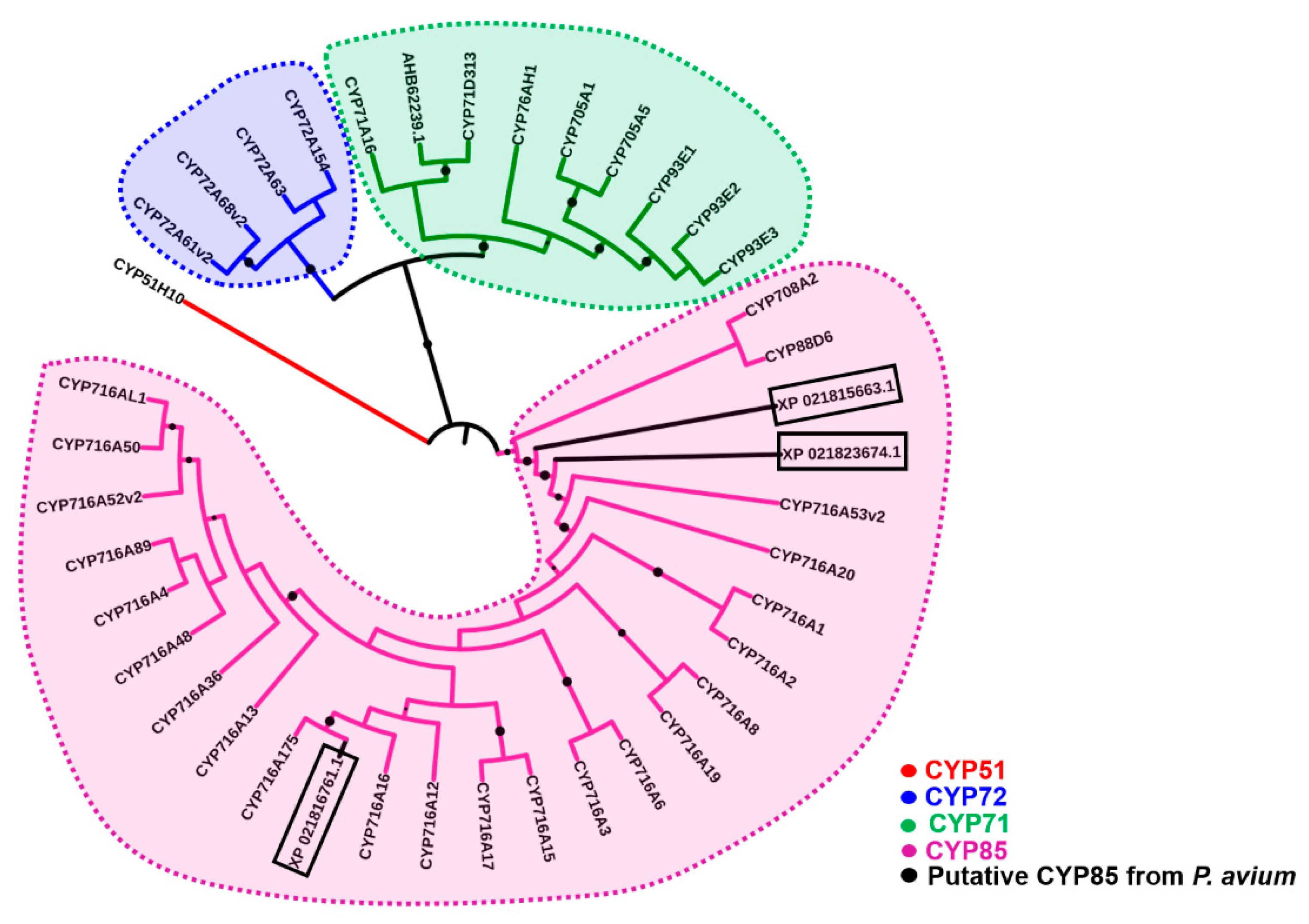
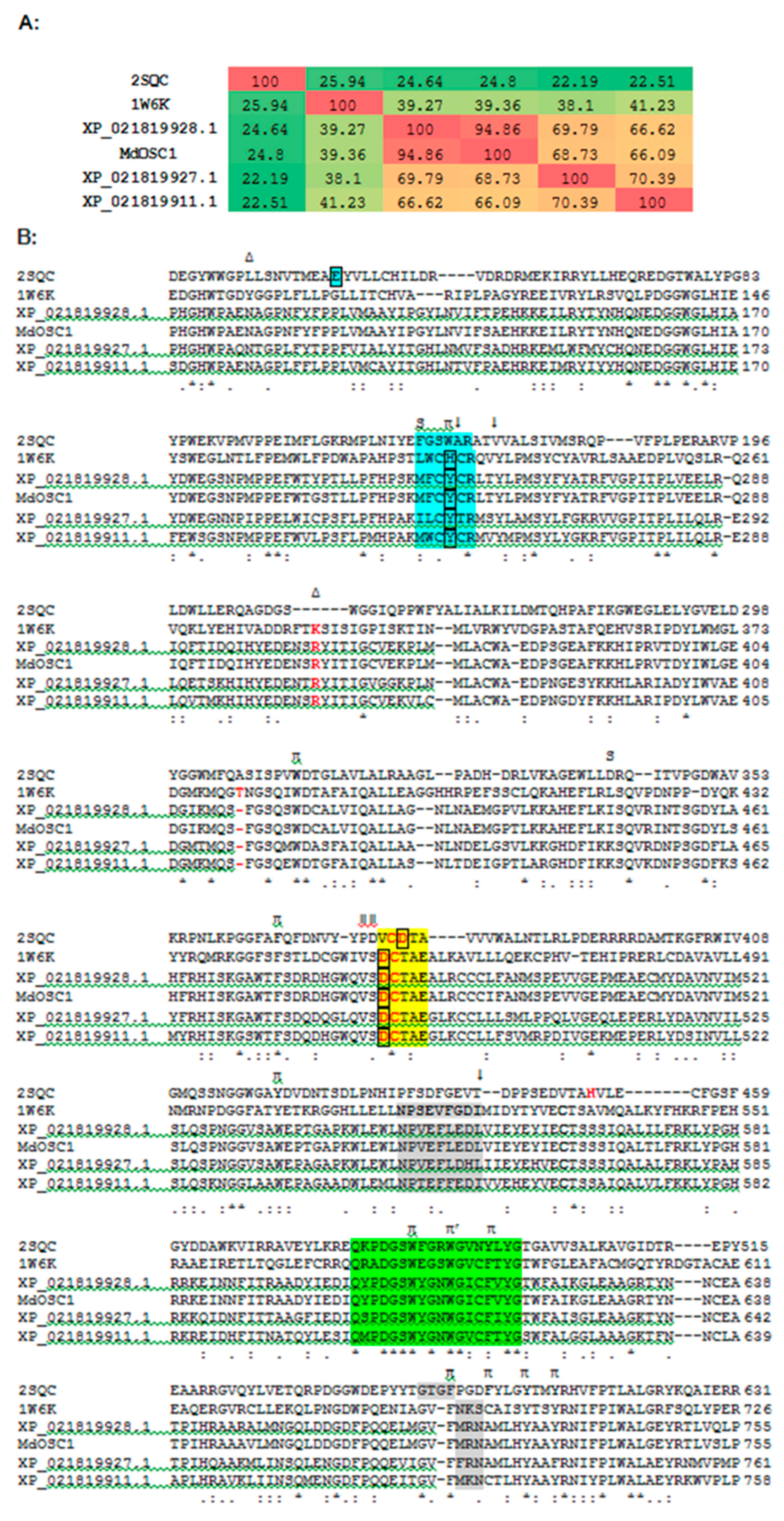
2.1. Bioinformatics
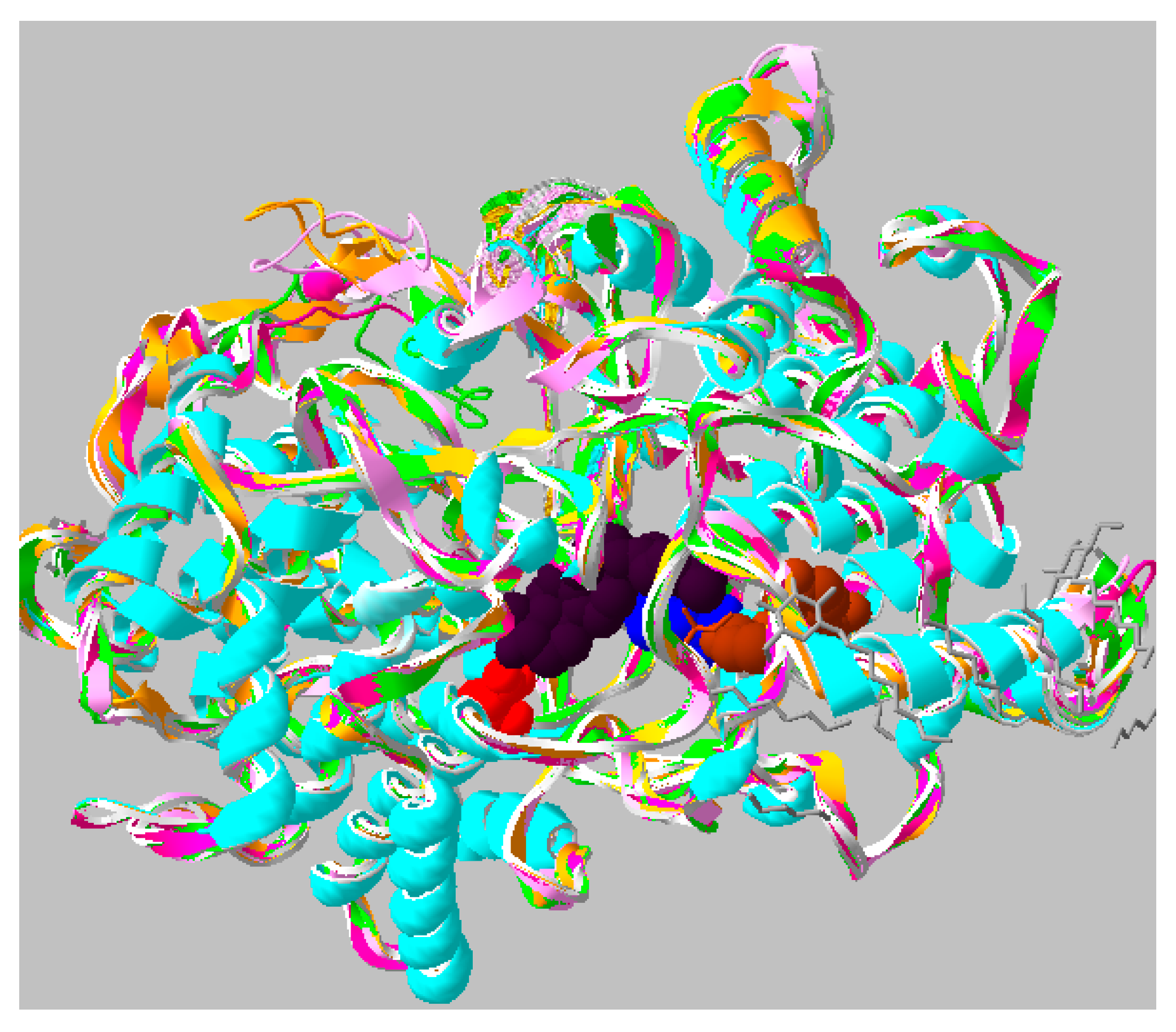
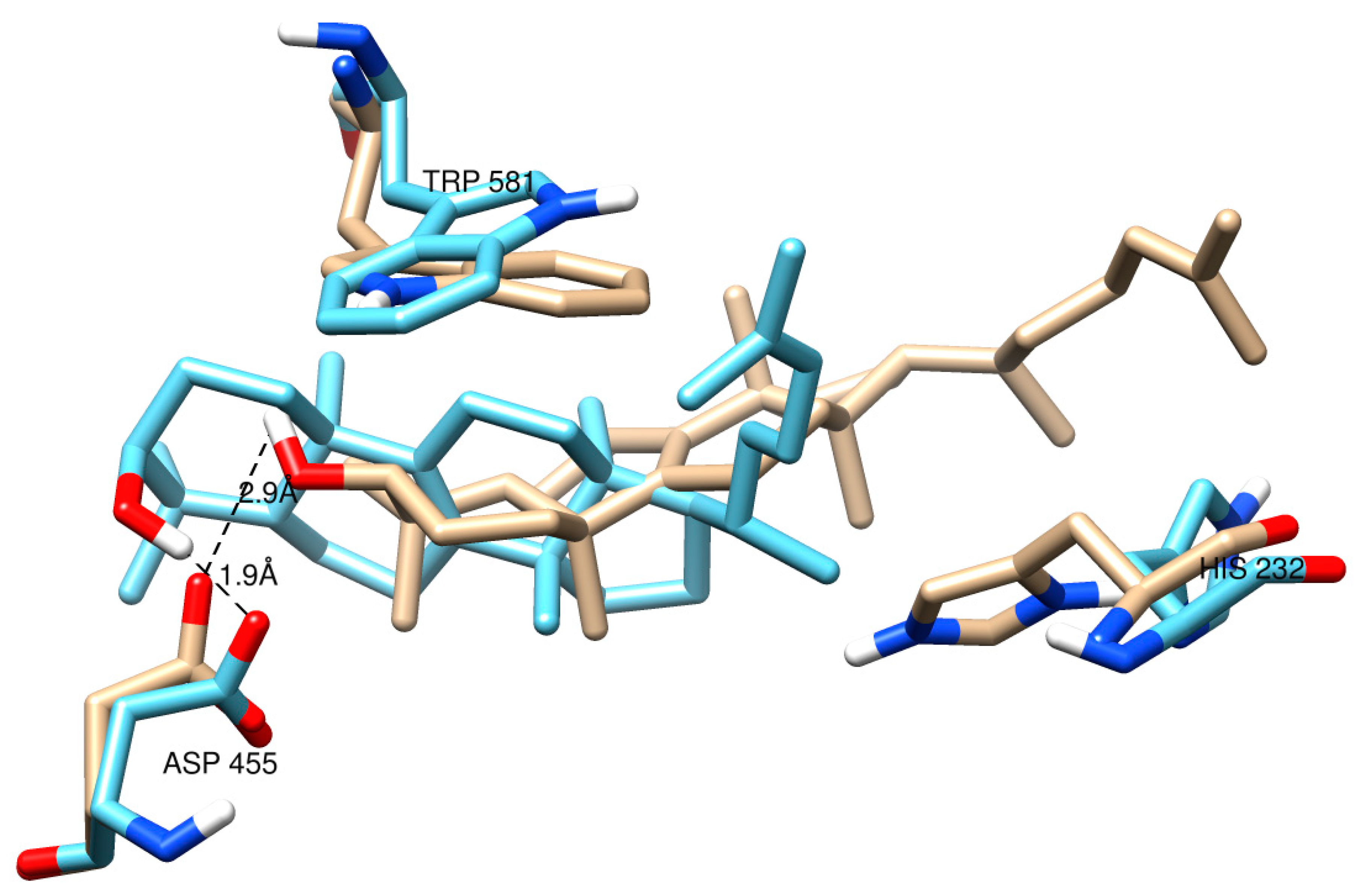
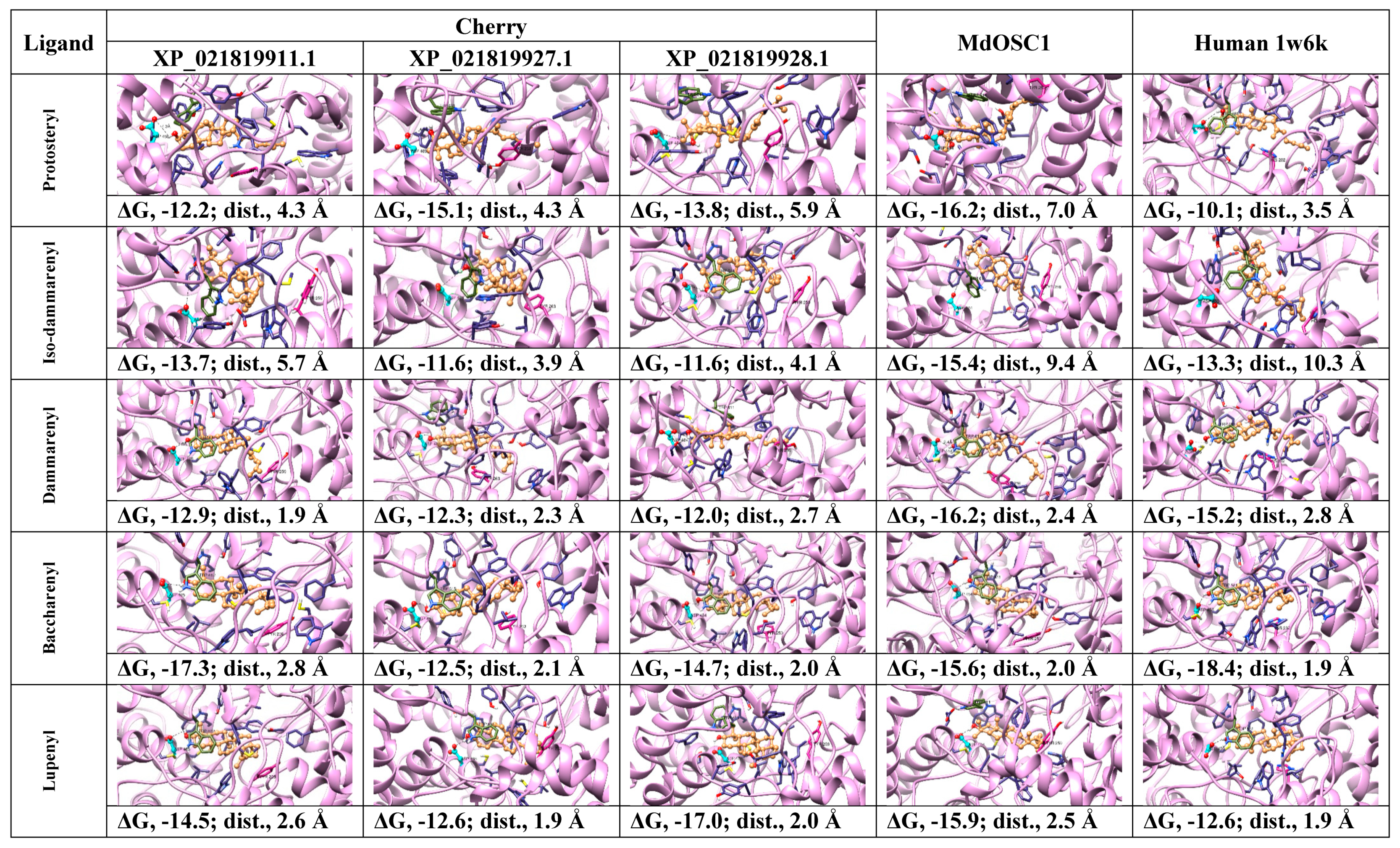
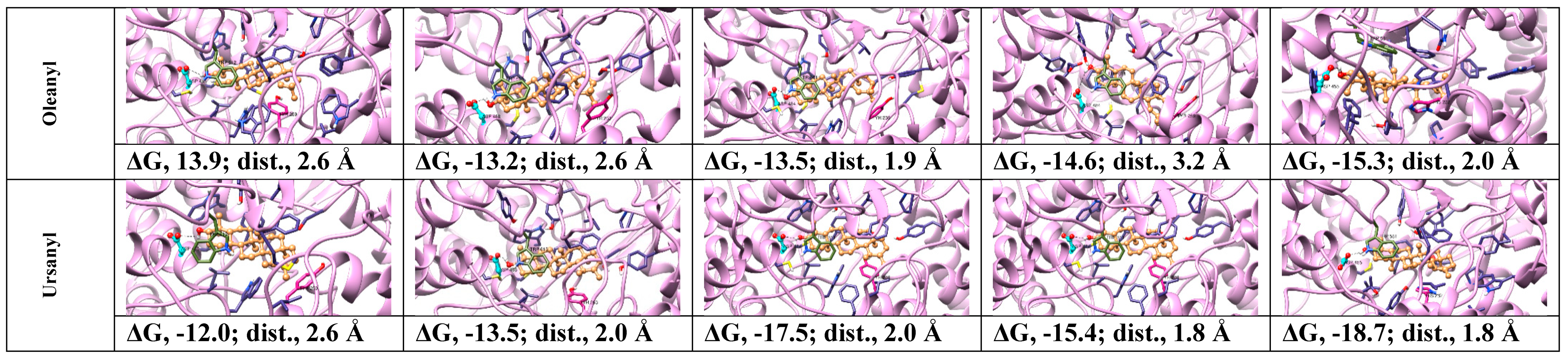
2.2. Protein Modeling
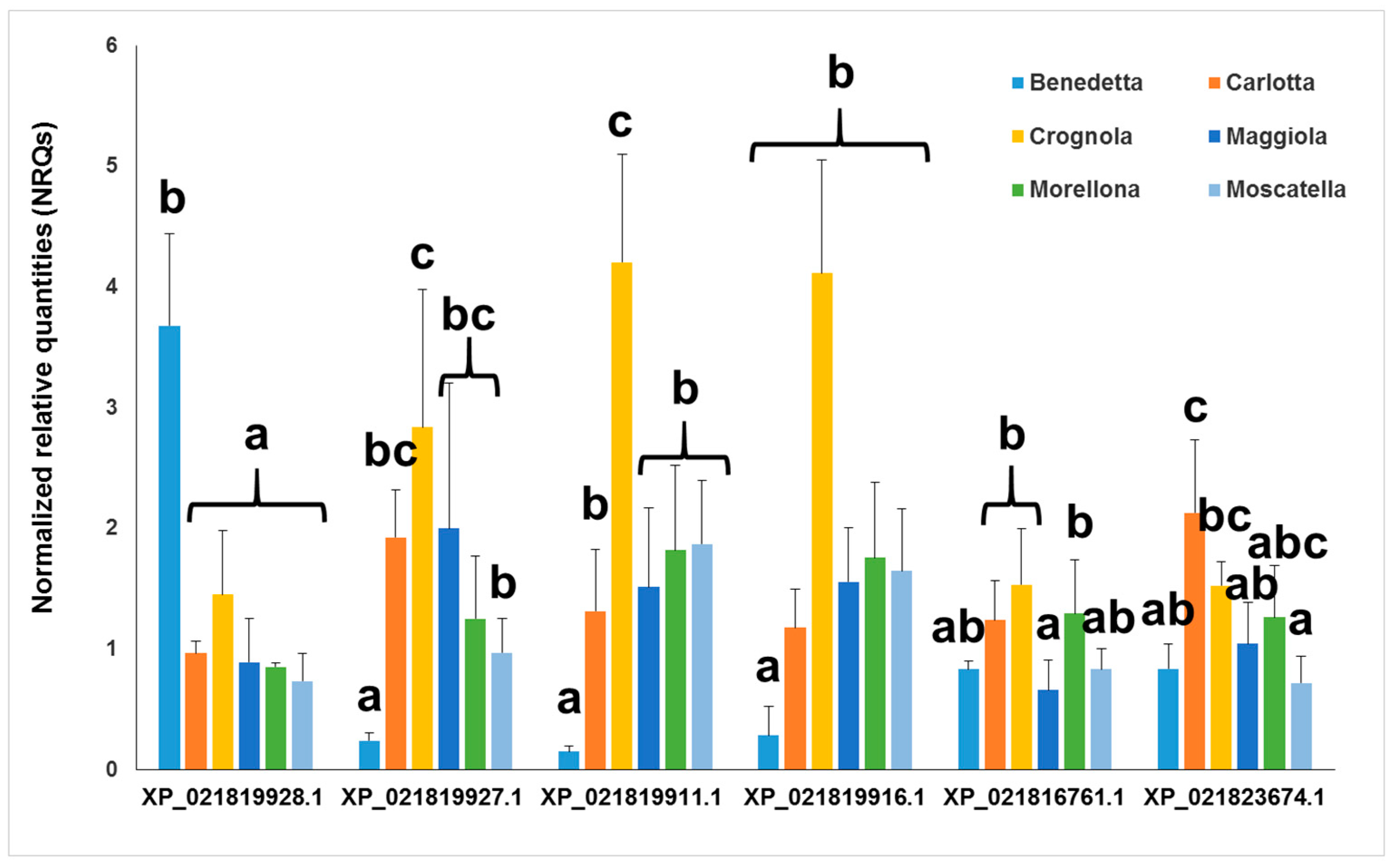
2.3. Gene Expression Analysis
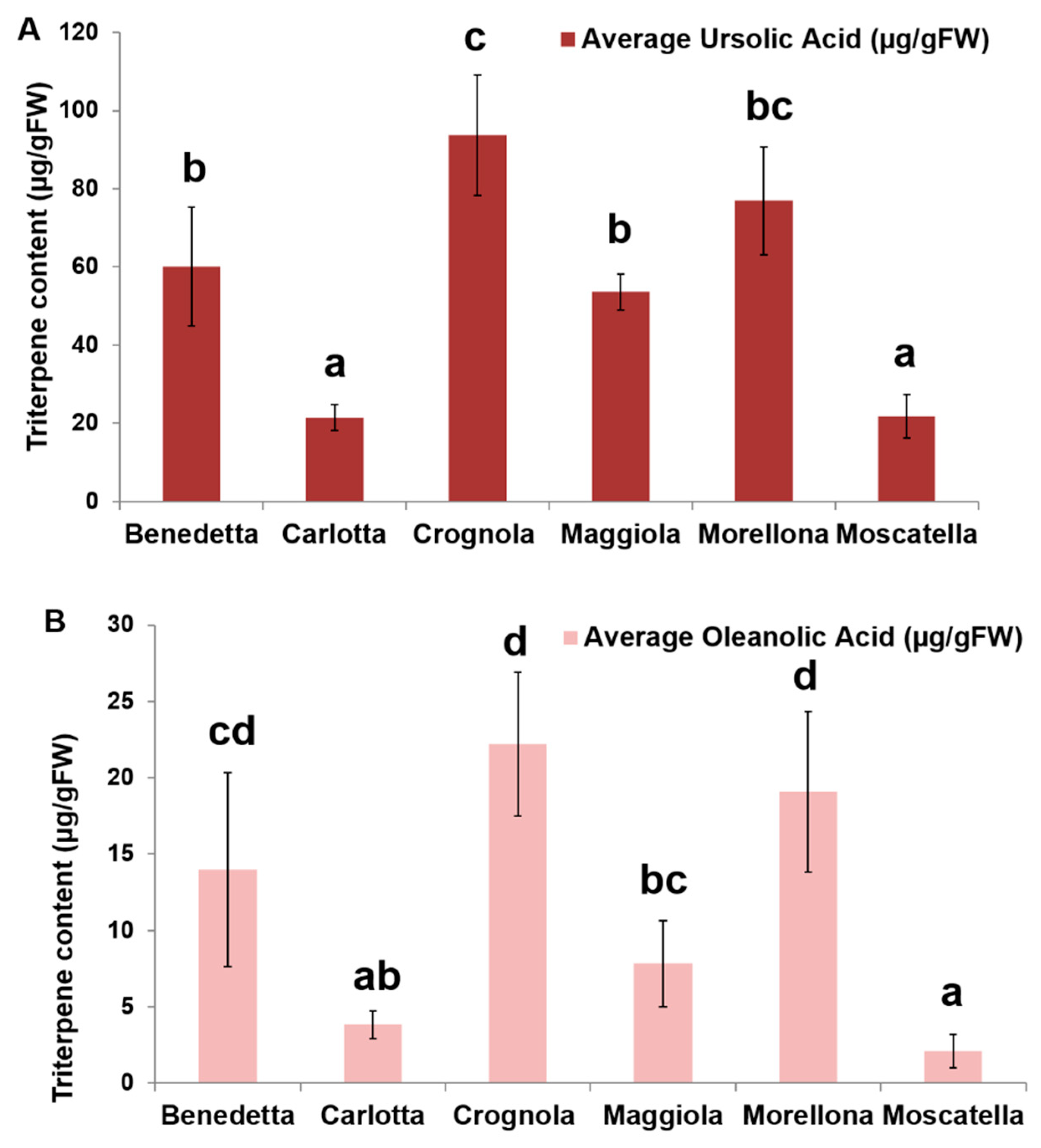
2.4. Pentacyclic Triterpene Content
3. Materials and Methods
3.1. Fruit Harvesting
3.2. RNA Extraction, Primer Design and Real-Time PCR Data Analysis
3.3. Bioinformatics
3.4. Extraction and Analysis of Pentacyclic Triterpenes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ballistreri, G.; Continella, A.; Gentile, A.; Amenta, M.; Fabroni, S.; Rapisarda, P. Fruit quality and bioactive compounds relevant to human health of sweet cherry (Prunus avium L.) cultivars grown in Italy. Food Chem. 2013, 140, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, A.C.; Rodrigues, M.; Santos, A.O.; Alves, G.; Silva, L.R. Antioxidant Status, Antidiabetic Properties and Effects on Caco-2 Cells of Colored and Non-Colored Enriched Extracts of Sweet Cherry Fruits. Nutrients 2018, 10, 1688. [Google Scholar] [CrossRef]
- Correia, S.; Schouten, R.; Silva, A.P.; Gonçalves, B. Factors Affecting Quality and Health Promoting Compounds during Growth and Postharvest Life of Sweet Cherry (Prunus avium L.). Front. Plant Sci. 2017, 8, 2166. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-O.; Heo, H.J.; Kim, Y.J.; Yang, H.S.; Lee, C.Y. Sweet and Sour Cherry Phenolics and Their Protective Effects on Neuronal Cells. J. Agric. Food Chem. 2005, 53, 9921–9927. [Google Scholar] [CrossRef] [PubMed]
- Commisso, M.; Bianconi, M.; Di Carlo, F.; Poletti, S.; Bulgarini, A.; Munari, F.; Negri, S.; Stocchero, M.; Ceoldo, S.; Avesani, L.; et al. Multi-approach metabolomics analysis and artificial simplified phytocomplexes reveal cultivar-dependent synergy between polyphenols and ascorbic acid in fruits of the sweet cherry (Prunus avium L.). PLoS ONE 2017, 12, e0180889. [Google Scholar] [CrossRef]
- Peschel, S.; Franke, R.; Schreiber, L.; Knoche, M. Composition of the cuticle of developing sweet cherry fruit. Phytochemistry 2007, 68, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Szakiel, A.; Pączkowski, C.; Pensec, F.; Bertsch, C. Fruit cuticular waxes as a source of biologically active triterpenoids. Phytochem. Rev. 2012, 11, 263–284. [Google Scholar] [CrossRef]
- Buschhaus, C.; Jetter, R. Composition differences between epicuticular and intracuticular wax substructures: how do plants seal their epidermal surfaces? J. Exp. Bot. 2011, 62, 841–853. [Google Scholar] [CrossRef]
- Furtado, N.A.J.C.; Pirson, L.; Edelberg, H.; M Miranda, L.; Loira-Pastoriza, C.; Preat, V.; Larondelle, Y.; André, C.M. Pentacyclic Triterpene Bioavailability: An Overview of In Vitro and In Vivo Studies. Mol. Basel Switz. 2017, 22, 400. [Google Scholar] [CrossRef] [PubMed]
- Brendolise, C.; Yauk, Y.-K.; Eberhard, E.D.; Wang, M.; Chagne, D.; Andre, C.; Greenwood, D.R.; Beuning, L.L. An unusual plant triterpene synthase with predominant α-amyrin-producing activity identified by characterizing oxidosqualene cyclases from Malus × domestica. FEBS J. 2011, 278, 2485–2499. [Google Scholar] [CrossRef]
- Andre, C.M.; Legay, S.; Deleruelle, A.; Nieuwenhuizen, N.; Punter, M.; Brendolise, C.; Cooney, J.M.; Lateur, M.; Hausman, J.-F.; Larondelle, Y.; et al. Multifunctional oxidosqualene cyclases and cytochrome P450 involved in the biosynthesis of apple fruit triterpenic acids. New Phytol. 2016, 211, 1279–1294. [Google Scholar] [CrossRef]
- Belge, B.; Llovera, M.; Comabella, E.; Gatius, F.; Guillén, P.; Graell, J.; Lara, I. Characterization of Cuticle Composition after Cold Storage of “Celeste” and “Somerset” Sweet Cherry Fruit. J. Agric. Food Chem. 2014, 62, 8722–8729. [Google Scholar] [CrossRef] [PubMed]
- Quero-Garcia, J.; Iezzoni, A.; Pulawska, J.; Lang, G.A. Cherries: Botany, Production and Uses; CABI Editions: Oxford, UK, 2017; ISBN 978-1-78064-837-8. [Google Scholar]
- Ghiselli, L.; Rossi, E.; Whittaker, A.; Dinelli, G.; Baglio, A.P.; Andrenelli, L.; Benedettelli, S. Nutritional characteristics of ancient Tuscan varieties of Triticum aestivum L. Ital. J. Agron. 2016, 11, 237–245. [Google Scholar] [CrossRef]
- Agnoletti, M.; Emanueli, F. Biocultural Diversity in Europe; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-26315-1. [Google Scholar]
- Berni, R.; Cantini, C.; Romi, M.; Hausman, J.-F.; Guerriero, G.; Cai, G. Agrobiotechnology Goes Wild: Ancient Local Varieties as Sources of Bioactives. Int. J. Mol. Sci. 2018, 19, 2248. [Google Scholar] [CrossRef]
- BASIQ – Bottega Alimentare (grocery) for Sustainability, Identity and Quality | ecodynamicsgroup. Available online: http://www.valdimersegreen.com/en/basiq/ (accessed on 20 April 2019).
- Shirasawa, K.; Isuzugawa, K.; Ikenaga, M.; Saito, Y.; Yamamoto, T.; Hirakawa, H.; Isobe, S. The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2017, 24, 499–508. [Google Scholar] [CrossRef]
- Siedenburg, G.; Jendrossek, D. Squalene-Hopene Cyclases. Appl. Environ. Microbiol. 2011, 77, 3905–3915. [Google Scholar] [CrossRef] [PubMed]
- Salmon, M.; Thimmappa, R.B.; Minto, R.E.; Melton, R.E.; Hughes, R.K.; O’Maille, P.E.; Hemmings, A.M.; Osbourn, A. A conserved amino acid residue critical for product and substrate specificity in plant triterpene synthases. Proc. Natl. Acad. Sci. USA 2016, 113, E4407–E4414. [Google Scholar] [CrossRef] [PubMed]
- Ito, R.; Masukawa, Y.; Hoshino, T. Purification, kinetics, inhibitors and CD for recombinant β-amyrin synthase from Euphorbia tirucalli L and functional analysis of the DCTA motif, which is highly conserved among oxidosqualene cyclases. FEBS J. 2013, 280, 1267–1280. [Google Scholar] [CrossRef]
- Reinert, D.J.; Balliano, G.; Schulz, G.E. Conversion of squalene to the pentacarbocyclic hopene. Chem. Biol. 2004, 11, 121–126. [Google Scholar] [CrossRef][Green Version]
- Iturbe-Ormaetxe, I.; Haralampidis, K.; Papadopoulou, K.; Osbourn, A.E. Molecular cloning and characterization of triterpene synthases from Medicago truncatula and Lotus japonicus. Plant Mol. Biol. 2003, 51, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Thoma, R.; Schulz-Gasch, T.; D’Arcy, B.; Benz, J.; Aebi, J.; Dehmlow, H.; Hennig, M.; Stihle, M.; Ruf, A. Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature 2004, 432, 118–122. [Google Scholar] [CrossRef]
- Segura, M.J.R.; Jackson, B.E.; Matsuda, S.P.T. Mutagenesis approaches to deduce structure-function relationships in terpene synthases. Nat. Prod. Rep. 2003, 20, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Hermann, J.C.; Ghanem, E.; Li, Y.; Raushel, F.M.; Irwin, J.J.; Shoichet, B.K. Predicting substrates by docking high-energy intermediates to enzyme structures. J. Am. Chem. Soc. 2006, 128, 15882–15891. [Google Scholar] [CrossRef]
- Schulz-Gasch, T.; Stahl, M. Mechanistic insights into oxidosqualene cyclizations through homology modeling. J. Comput. Chem. 2003, 24, 741–753. [Google Scholar] [CrossRef]
- Tian, B.-X.; Wallrapp, F.H.; Holiday, G.L.; Chow, J.-Y.; Babbitt, P.C.; Poulter, C.D.; Jacobson, M.P. Predicting the functions and specificity of triterpenoid synthases: a mechanism-based multi-intermediate docking approach. PLoS Comput. Biol. 2014, 10, e1003874. [Google Scholar] [CrossRef]
- Berni, R.; Piasecki, E.; Legay, S.; Hausman, J.-F.; Siddiqui, K.S.; Cai, G.; Guerriero, G. Identification of the laccase-like multicopper oxidase gene family of sweet cherry (Prunus avium L.) and expression analysis in six ancient Tuscan varieties. Sci. Rep. 2019, 9, 3557. [Google Scholar] [CrossRef] [PubMed]
- Feenstra, K.A.; Starikov, E.B.; Urlacher, V.B.; Commandeur, J.N.M.; Vermeulen, N.P.E. Combining substrate dynamics, binding statistics, and energy barriers to rationalize regioselective hydroxylation of octane and lauric acid by CYP102A1 and mutants. Protein Sci. Publ. Protein Soc. 2007, 16, 420–431. [Google Scholar] [CrossRef]
- Almeida, A.; Dong, L.; Khakimov, B.; Bassard, J.-E.; Moses, T.; Lota, F.; Goossens, A.; Appendino, G.; Bak, S. A Single Oxidosqualene Cyclase Produces the Seco-Triterpenoid α-Onocerin. Plant Physiol. 2018, 176, 1469–1484. [Google Scholar] [CrossRef]
- DeLuca, S.; Khar, K.; Meiler, J. Fully Flexible Docking of Medium Sized Ligand Libraries with RosettaLigand. PLoS ONE 2015, 10, e0132508. [Google Scholar] [CrossRef]
- Berni, R.; Romi, M.; Cantini, C.; Hausman, J.-F.; Guerriero, G.; Cai, G. Functional Molecules in Locally-Adapted Crops: The Case Study of Tomatoes, Onions, and Sweet Cherry Fruits from Tuscany in Italy. Front. Plant Sci. 2019, 9, 1983. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B.; de Bakker, P.I.W.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Lyskov, S.; Chou, F.-C.; Conchúir, S.Ó.; Der, B.S.; Drew, K.; Kuroda, D.; Xu, J.; Weitzner, B.D.; Renfrew, P.D.; Sripakdeevong, P.; et al. Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE). PLoS ONE 2013, 8, e63906. [Google Scholar] [CrossRef] [PubMed]
- Kiss, R.; Sandor, M.; Szalai, F.A. http://Mcule.com: a public web service for drug discovery. J. Cheminform. 2012, 4, P17. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]

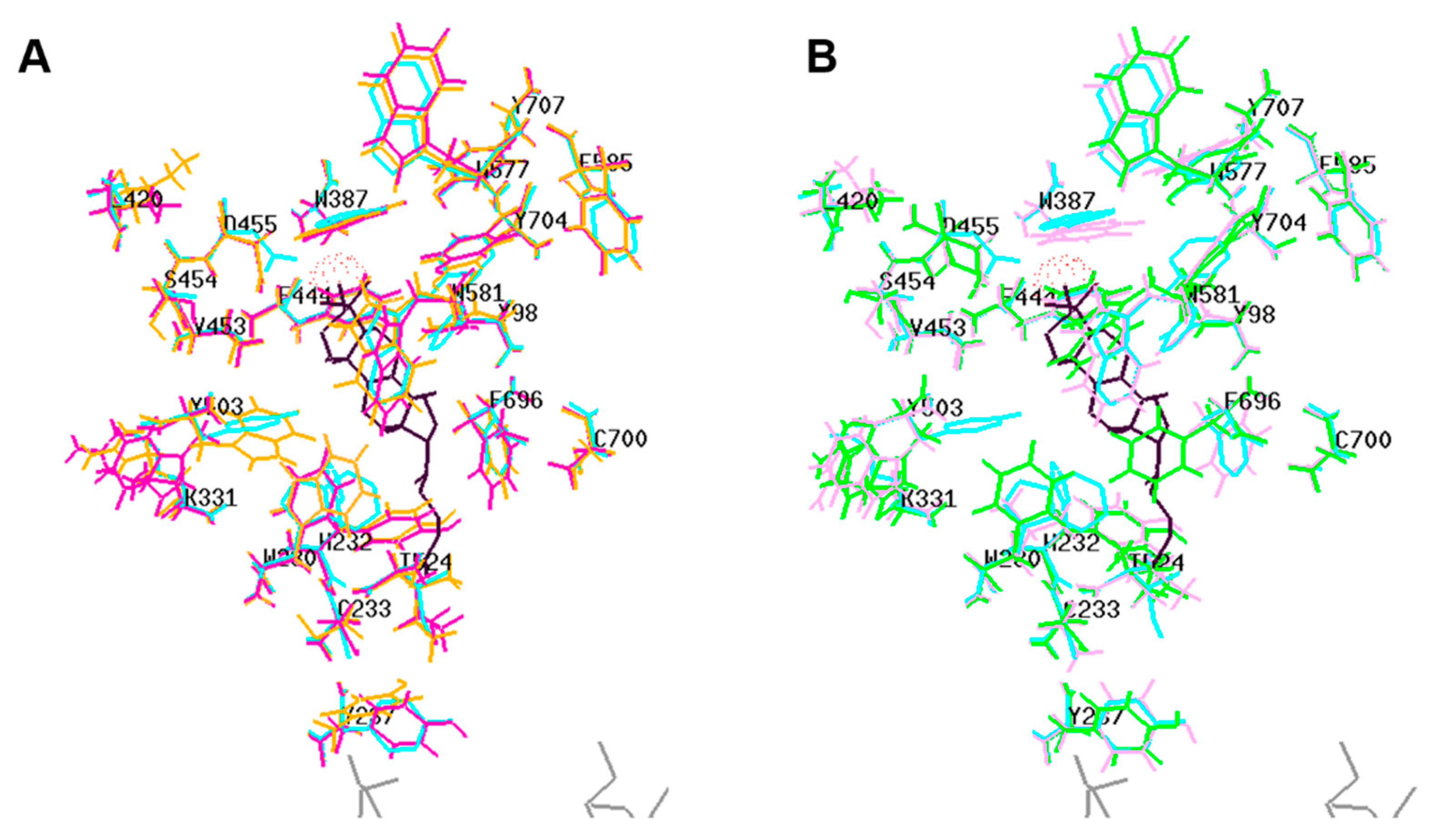
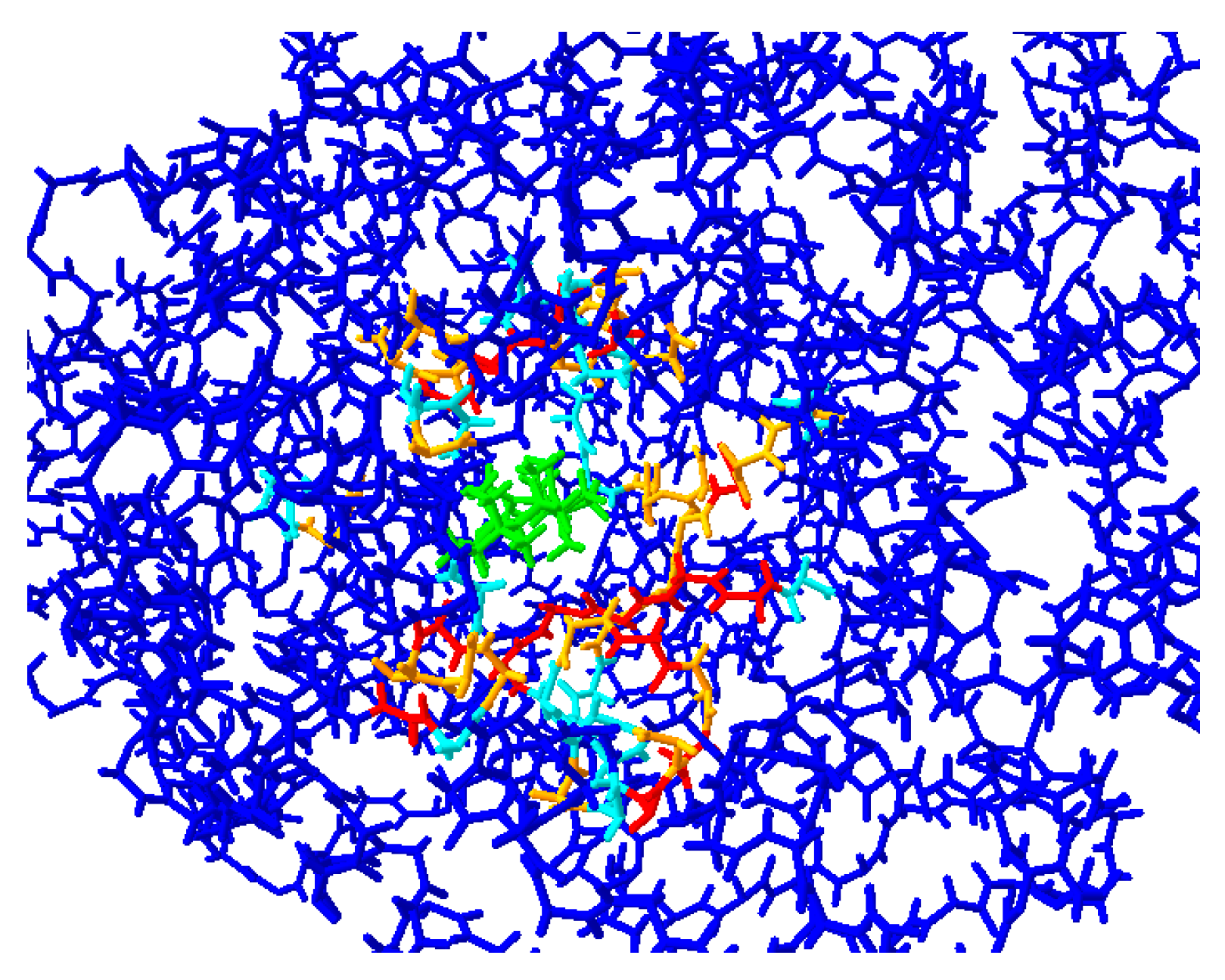

| OSC | Residue | RMSD (Å) | |
|---|---|---|---|
| Dammarenyl Cation | β-Amyrin Product | ||
| XP_021819911.1 | Y 259 | 0.368 | 0.254 |
| G 414 | 0.109 | 0.350 | |
| D 485 (protonating Asp) | 0.089 | 0.105 | |
| D 554 | 0.242 | 0.690 | |
| W 612 (binding center) | 0.395 | 0.288 | |
| G 613 | 0.567 | 0.361 | |
| W 621 | 0.384 | 0.001 | |
| K 730 | 0.151 | 0.251 | |
| XP_021819927.1 | Y 263 | 0.266 | 0.105 |
| V 373 | 0.321 | 0.001 | |
| G 374 | 0.318 | 0.001 | |
| D 488 (protonating Asp) | 0.123 | 0.238 | |
| A 536 | 0.216 | 0.409 | |
| W 537 | 0.041 | 0.483 | |
| W 615 (binding center) | 0.161 | 0.245 | |
| F 732 | 0.124 | 0.336 | |
| XP_021819928.1 | Y 259 | 0.187 | 0.132 |
| C 369 | 1.245 | 0.793 | |
| F 473 | 0.354 | 0.237 | |
| D 484 (protonating Asp) | 0.578 | 0.144 | |
| A 532 | 0.301 | 0.246 | |
| W 611 (binding center) | 0.102 | 0.160 | |
| M 726 | 0.302 | 0.305 | |
| H 732 | 0.554 | 0.044 | |
| MdOSC1 | Y 259 | 0.198 | 0.161 |
| C 369 | 0.332 | 0.503 | |
| E 371 | 0.388 | 0.432 | |
| K 372 | 0.876 | 0.786 | |
| P 373 | 0.572 | 0.020 | |
| F 412 | 0.250 | 0.417 | |
| D 484 (protonating Asp) | 0.151 | 0.163 | |
| W 611 (binding center) | 0.115 | 0.214 | |
| Name | Sequence (5′→3′) | Amplicon Size | Amplification Efficiency |
|---|---|---|---|
| PaveTIF4E Fwd | GGCAAAGCCTCGATACAATG | 71 | 1.90 |
| PaveTIF4E Rev | TTGGTTATGGAGAGCGAAGAC | ||
| PavGAPDH Fwd | TATCAAAGCCACAGCCACTG | 118 | 1.88 |
| PavGAPDH Rev | TGCTATTCGGAGAACCAACC | ||
| XP_021819928.1 Fwd | AAGGCAGACATGGGAGTTTG | 135 | 1.94 |
| XP_021819928.1 Rev | ATCTGAAAACGCCAGAGGAG | ||
| XP_021819916.1 Fwd | ATGAGGGTTCAGCTTGATGC | 104 | 2.09 |
| XP_021819916.1 Rev | CCACCGTGATTGTGTGAATG | ||
| XP_021819927.1 Fwd | CGAAGAGTGTTGTCTGCTTAACG | 109 | 2.04 |
| XP_021819927.1 Rev | AAAGAGAATAGTCACCCCCTTG | ||
| XP_021819911.1 Fwd | TGTATTCCCAGCAGAGCATC | 74 | 1.90 |
| XP_021819911.1 Rev | CCCAACCACCATCTTCATTC | ||
| XP_021816761.1 Fwd | TGAGAAAGATGCTCCCCAAC | 107 | 1.90 |
| XP_021816761.1 Rev | CCTGTTTTCCCAACCATCAG | ||
| XP_021823674.1 Fwd | GAGGTTCAAATGGGAAGTGC | 96 | 1.90 |
| XP_021823674.1 Rev | TGACGATGAAGACGAACTGG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berni, R.; Hoque, M.Z.; Legay, S.; Cai, G.; Siddiqui, K.S.; Hausman, J.-F.; Andre, C.M.; Guerriero, G. Tuscan Varieties of Sweet Cherry Are Rich Sources of Ursolic and Oleanolic Acid: Protein Modeling Coupled to Targeted Gene Expression and Metabolite Analyses. Molecules 2019, 24, 1590. https://doi.org/10.3390/molecules24081590
Berni R, Hoque MZ, Legay S, Cai G, Siddiqui KS, Hausman J-F, Andre CM, Guerriero G. Tuscan Varieties of Sweet Cherry Are Rich Sources of Ursolic and Oleanolic Acid: Protein Modeling Coupled to Targeted Gene Expression and Metabolite Analyses. Molecules. 2019; 24(8):1590. https://doi.org/10.3390/molecules24081590
Chicago/Turabian StyleBerni, Roberto, Mubasher Zahir Hoque, Sylvain Legay, Giampiero Cai, Khawar Sohail Siddiqui, Jean-Francois Hausman, Christelle M. Andre, and Gea Guerriero. 2019. "Tuscan Varieties of Sweet Cherry Are Rich Sources of Ursolic and Oleanolic Acid: Protein Modeling Coupled to Targeted Gene Expression and Metabolite Analyses" Molecules 24, no. 8: 1590. https://doi.org/10.3390/molecules24081590
APA StyleBerni, R., Hoque, M. Z., Legay, S., Cai, G., Siddiqui, K. S., Hausman, J.-F., Andre, C. M., & Guerriero, G. (2019). Tuscan Varieties of Sweet Cherry Are Rich Sources of Ursolic and Oleanolic Acid: Protein Modeling Coupled to Targeted Gene Expression and Metabolite Analyses. Molecules, 24(8), 1590. https://doi.org/10.3390/molecules24081590










