Half-Wave Potentials and In Vitro Cytotoxic Evaluation of 3-Acylated 2,5-Bis(phenylamino)-1,4-benzoquinones on Cancer Cells
Abstract
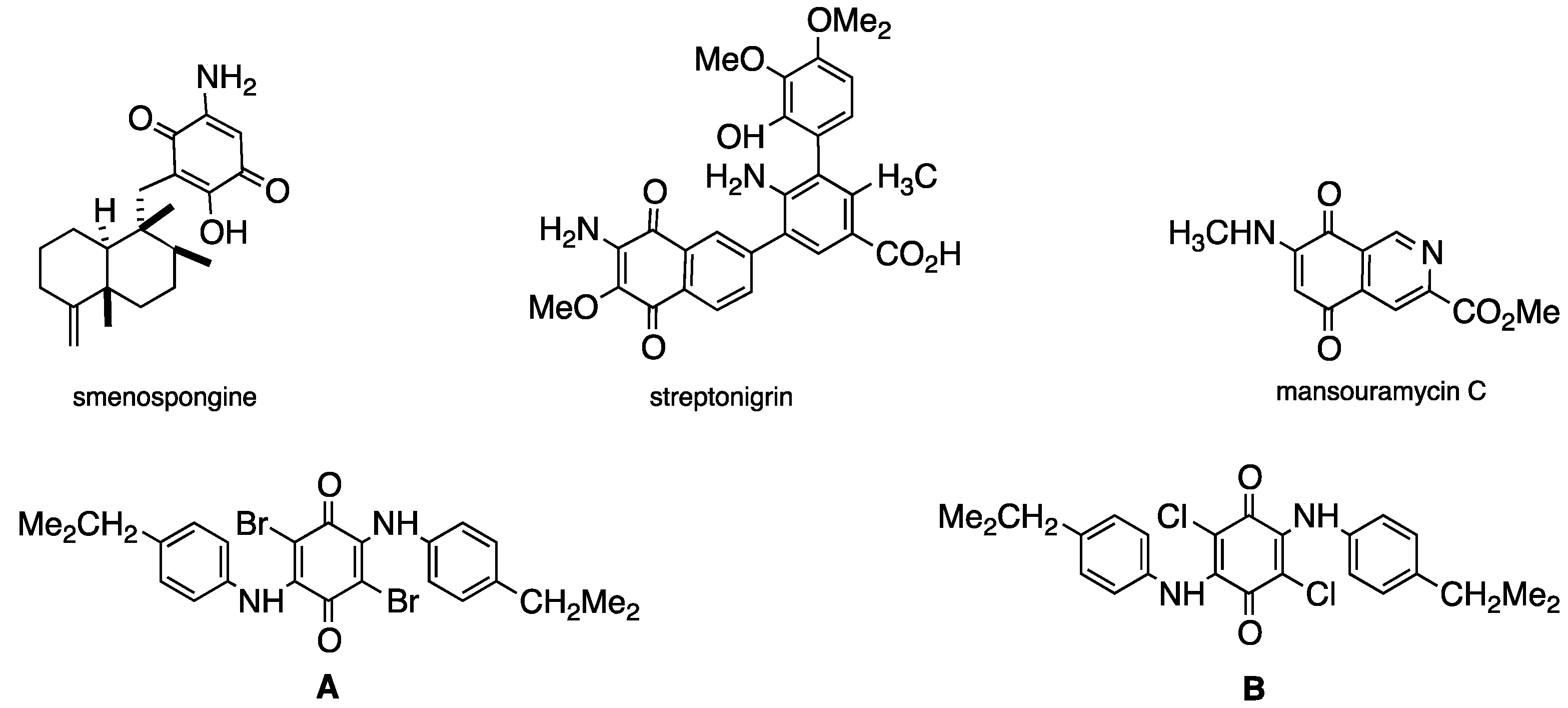
:1. Introduction
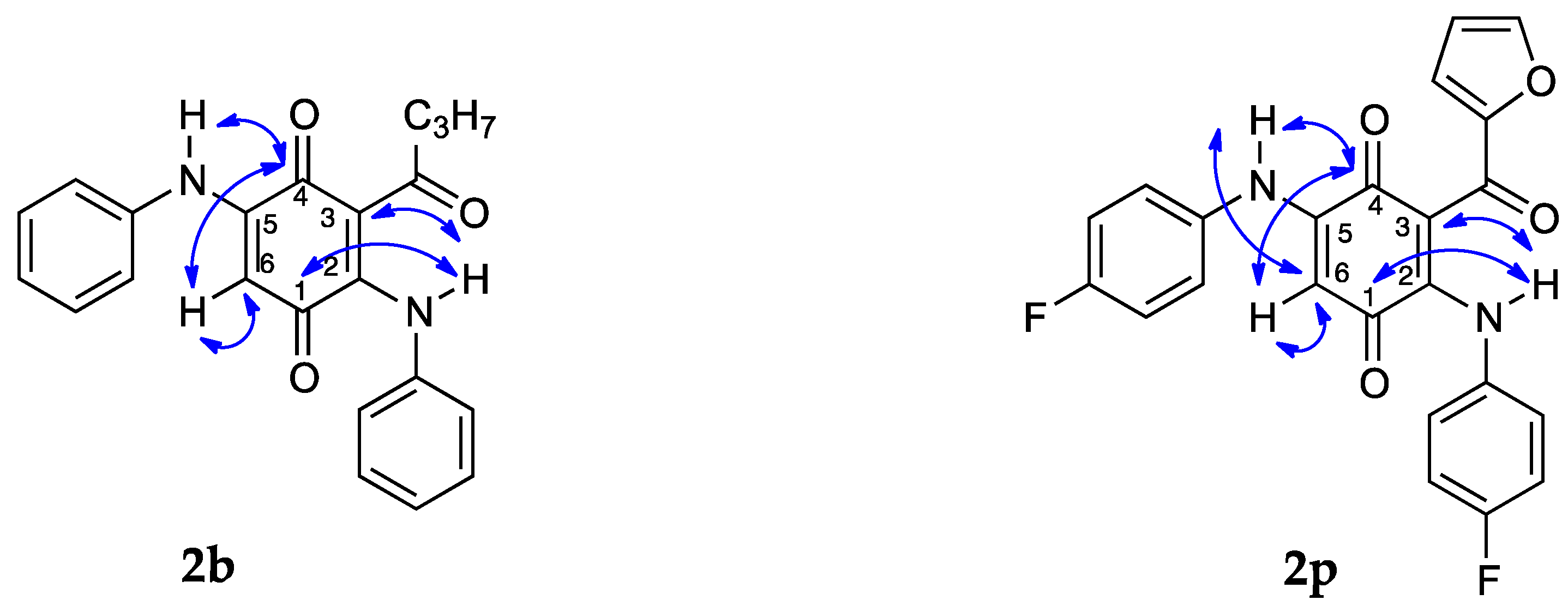
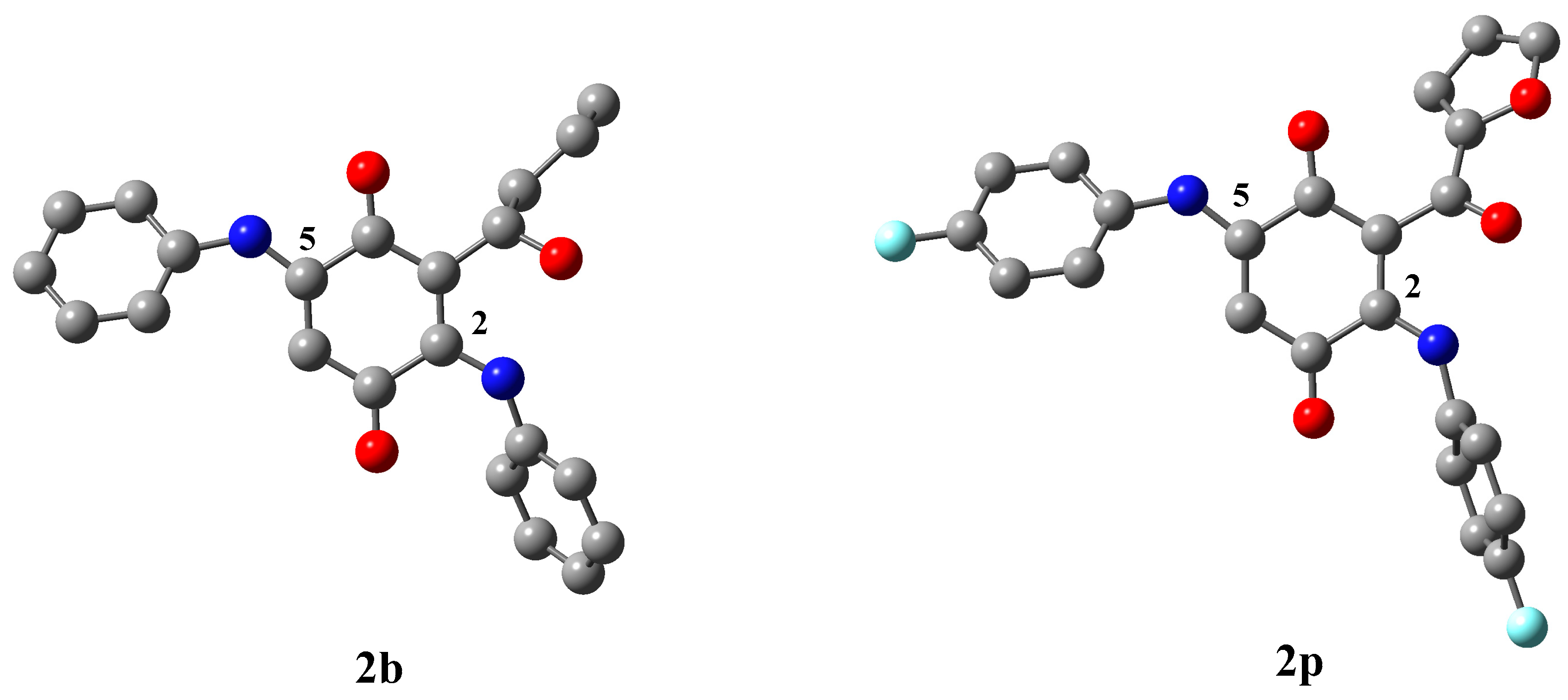
2. Results and Discussion
3. Materials and Methods
3.1. General Information
3.2. Chemistry
Preparation of 3-Acyl-2,5-bis(phenylamino)-1,4-benzoquinones 2a–q, General Procedure.
3.3. Biological Assays
3.3.1. Cell Lines and Cell Cultures
3.3.2. Cytotoxic Assays
3.3.3. Quantitative real-time PCR (qPCR) Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kong, D.; Yamori, T.; Kobayashi, M.; Duan, H. Antiproliferative and antiangiogenic activities of Smenospongine, a marine sponge sesquiterpene aminoquinone. Mar. Drugs 2011, 9, 154–161. [Google Scholar] [CrossRef]
- Gould, S.J.; Weimeb, S.M. Streptonigrin. Fortschr. Chem. Org. Naturst. 1982, 41, 77–111. [Google Scholar]
- Hawas, U.W.; Shaaban, M.; Shaaban, K.A.; Speitling, M.; Maier, A.; Kelter, G.; Fiebig, H.H.; Meiners, M.; Helmke, E.; Laatsch, H. Mansouramycins A–D, cytotoxic isoquinolinequinones from a marine streptomycete. J. Nat. Prod. 2009, 72, 2120–2124. [Google Scholar] [CrossRef] [PubMed]
- Valderrama, J.A.; Cabrera, M.; Benites, J.; Ríos, D.; Inostroza-Rivera, R.; Muccioli, G.G. Synthetic approaches and in vitro cytotoxic evaluation of 2-acyl-3-(3,4,5-trimethoxyanilino)-1,4-naphthoquinones. RSC Adv. 2017, 7, 24813–24821. [Google Scholar] [CrossRef]
- Valderrama, J.A.; Garrido, J.; Delgado, V.; Benites, J.; Theoduloz, C. Access to New Cytotoxic Quinone-Amino Acid Conjugates Linked through A Vinylic Spacer from 2-Acylnaphthoquinones and Methyl 3-Aminocrotonate. Molecules 2017, 22, 2281. [Google Scholar] [CrossRef]
- Benites, J.; Valderrama, J.A.; Bettega, K.; Curi Pedrosa, R.; Buc Calderon, P.; Verrax, J. Biological evaluation of donor-aceptor aminonaphthoquinones as antitumor agents. Eur. J. Med. Chem. 2010, 45, 6052–6057. [Google Scholar] [CrossRef]
- Sarma, M.D.; Ghosh, R.; Patra, A.; Hazra, B. Synthesis of novel aminoquinonoid analogues of diospyrin and evaluation of their inhibitory activity against murine and human cancer cells. Eur. J. Med. Chem. 2008, 43, 1878–1888. [Google Scholar] [CrossRef] [PubMed]
- Miguel del Corral, J.M.; Castro, M.A.; Gordaliza, M.; Martín, M.L.; Gamito, A.M.; Cuevas, C.; San Feliciano, A. Synthesis and cytotoxicity of new heterocyclic terpenylnaphthoquinones. Bioorg. Med. Chem. 2006, 14, 2816–2827. [Google Scholar] [CrossRef] [PubMed]
- Ibacache, J.A.; Faundes, J.; Montoya, M.; Mejías, S.; Valderrama, J.A. Preparation of Novel Homodimers Derived from Cytotoxic Isoquinolinequinones. A Twin Drug Approach. Molecules 2018, 23, 439. [Google Scholar] [CrossRef] [PubMed]
- Pathania, D.; Kuang, Y.; Sechi, M.; Neamati, N. Mechanisms underlying the cytotoxicity of a novel quinazolinedione-based redox modulator, QD232, in pancreatic cancer cells. Br. J. Pharmacol. 2015, 172, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, P.H.; Khanijou, J.K.; Lam, T.H.; Tong, C.T.; Chai, C.L.L. Synthesis and potent cytotoxic activity of 8-and 9-anilinophenanthridine-7,10-diones. Tetrahedron Lett. 2011, 52, 92–94. [Google Scholar] [CrossRef]
- Vásquez, D.; Rodríguez, J.A.; Theoduloz, C.; Buc Calderon, P.; Valderrama, J.A. Studies on quinones. Part 46. Synthesis and in vitro antitumor evaluation of Aminopyrimidoisoquinolinequinones. Eur J. Med. Chem. 2010, 45, 5234–5242. [Google Scholar]
- Valderrama, J.A.; Ibacache, J.A.; Arancibia, V.; Rodriguez, J.; Theoduloz, T. Studies on quinones. Part 45: Novel 7-aminoisoquinoline-5,8 -quinone derivatives with antitumor properties on cancer cell lines. Bioorg. Med. Chem. 2009, 17, 2894–2901. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.S.; Chen, C.; Lee, P.T.; Chiu, S.J.; Liu, H.F.; Tsai, C.C.; Chao, J.I. 7-Chloro-6-piperidin-1-yl-quinoline-5,8-dione (PT-262), a novel synthetic compound induces lung carcinoma cell death associated with inhibiting ERK and CDC2 phosphorylation via a p53-independent pathway. Cancer Chemother. Pharmacol. 2008, 62, 799–808. [Google Scholar] [CrossRef]
- Lodygin, D.; Menssen, A.; Hermeking, H. Induction of the Cdk inhibitor p21 by LY83583 inhibits tumor cell proliferation in a p53-independent manner. J. Clin. Invest. 2002, 110, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Ryu, C.K.; Jeong, H.J.; Lee, S.K.; You, H.J.; Choi, K.U.; Shim, J.Y.; Heo, Y.H.; Lee, C.O. Effects of 6-arylamino-5,8-quinolinediones and 6-chloro-7-arylamino-5,8-isoquinolinediones on NAD(P)H: Quinone oxidoreductase (NQO1) activity and their cytotoxic potential. Arch. Pharm. Res. 2001, 24, 390–896. [Google Scholar] [CrossRef]
- Lee, Y.S.; Wurster, R.D. Mechanism of potentiation of LY83583-induced growth inhibition by sodium nitroprusside in human brain tumor cells. Cancer Chemother. Pharmacol. 1995, 36, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Verter, H.S.; Rogers, J. Preparation of 2,5-Bis(alkylamino)-3,6-dimethoxy-p-benzoquinones. J. Org. Chem. 1966, 31, 987–988. [Google Scholar] [CrossRef]
- Burton, J.F.; Cain, H.F. Antileukaemic activity of polyporic acid. Nature 1959, 184, 1326–1327. [Google Scholar] [CrossRef]
- Mathew, A.E.; Zee-Cheng, R.K.Y.; Cheng, C.C. Amino-substituted p-benzoquinones. J. Med. Chem. 1986, 29, 1792–1795. [Google Scholar] [CrossRef]
- Schafer, W.; Moore, H.W.; Aguado, A. Synthesen des 2,1-Benzoxazol-4,7-chinon-Systems. Synthesis 1974, 30–32. [Google Scholar] [CrossRef]
- Suto, M.J.; Stier, M.A.; Winters, R.T.; Turner, W.R.; Pinter, C.D.; Elliot, W.E.; Sebolt-Leopold, J.S. Synthesis and evaluation of a series 3,5-disubstituted benzisoxazole-4,7-diones. Potent radiosensitizers in vitro. J. Med. Chem. 1991, 34, 3290–3294. [Google Scholar] [CrossRef]
- Benites, J.; Valderrama, J.A.; Ramos, M.; Muccioli, G.G.; Buc Calderon, P. Targeting Akt as strategy to kill cancer cells using 3-substituted 5-anilinobenzo[c]isoxazolequinones: A preliminary study. Biomed. Pharmacother. 2018, 97, 778–783. [Google Scholar] [CrossRef]
- Benites, J.; Rios, D.; Díaz, P.; Valderrama, J.A. The solar-chemical photo-Friedel-Crafts heteroacylation of 1,4-quinones. Tetrahedron Lett. 2011, 52, 609–611. [Google Scholar] [CrossRef]
- Schäfer, W.; Aguado, A. Oxidative Amination of Hydroquinones. Angew. Chem. Internat. Edit. 1971, 10, 405–406. [Google Scholar] [CrossRef]
- Ibacache, J.A.; Delgado, V.; Benites, J.; Theoduloz, C.; Arancibia, V.; Muccioli, G.; Valderrama, J.A. Synthesis, half wave potentials and antiproliferative activity of 1-aryl-substituted Aminoisoquinolinequinones. Molecules 2014, 19, 726–739. [Google Scholar] [CrossRef]
- De Abreu, F.C.; de Ferraz, P.A.; Goulart, M.O.F. Some applications of electrochemistry in biomedical chemistry. Emphasis on the correlation of electrochemical and bioactive properties. J. Braz. Chem. Soc. 2002, 13, 19–35. [Google Scholar] [CrossRef]
- Aguilar-Martinez, M.; Cuevas, G.; Jimenez-Estrada, M.; González, I.; Lotina-Hennsen, B.; Macias-Ruvalcaba, N. An experimental and theoretical study of the substituent effects on the redox properties of 2-[(R-phenyl)amine]-1,4-naphthalenediones in acetonitrile. J. Org. Chem. 1999, 64, 3684–3694. [Google Scholar] [CrossRef] [PubMed]
- Scudiero, D.A.; Shoemaker, R.H.; Paull, K.D.; Monks, A.; Tierney, S.; Nofziger, T.H.; Currens, M.J.; Seniff, D.; Boyd, M.R. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 1988, 48, 4827–4833. [Google Scholar]
- Showalter, A.; Limaye, A.; Oyer, J.L.; Igarashi, R.; Kittipatarin, Ch.; Copik, A.J.; Khaled, A.R. Cytokines in immunogenic cell death: Applications for cancer immunotherapy. Cytokine 2017, 97, 123–132. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Guerrero-Castilla, A.; Olivero-Verbel, J. Altered gene expression in HepG2 cells exposed to a methanolic coal dust extract. Environ. Toxicol. Pharmacol. 2014, 38, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.; Gaiteiro, C.; Peixoto, A.; Soares, J.; Neves, M.; Santos, L.L.; Ferreira, J.A. Reference Genes for Addressing Gene Expression of Bladder Cancer Cell Models under Hypoxia: A Step Towards Transcriptomic Studies. PLoS ONE 2016, 11, e0166120. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 2b, 2g, 2h, 2j, 2m, 2ñ, 2p are available from the authors. |

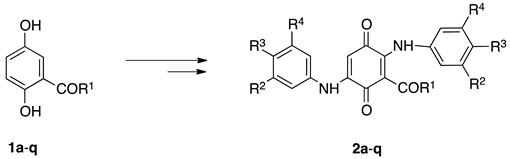
| Product Nº | R1 | R2 | R3 | R4 | Yield (%) a | ClogP | −EI 1/2 (mV) | −EII 1/2 (mV) |
|---|---|---|---|---|---|---|---|---|
| 2a | CH3 | H | H | H | 64 | 0.14 | 565 | 1170 |
| 2b | C3H7 | H | H | H | 56 | 1.21 | 570 | 1150 |
| 2c | C5H11 | H | H | H | 57 | 2.05 | 570 | 1090 |
| 2d | C7H15 | H | H | H | 59 | 2.88 | 590 | 910 |
| 2e | 3,4-(OMe)2Ph | H | H | H | 55 | 1.69 | 560 | 1140 |
| 2f | 3,4,5-(OMe)3Ph | H | H | H | 58 | 1.57 | 560 | 1120 |
| 2g | Furan-2-yl | H | H | H | 56 | 0.56 | 570 | 870 |
| 2h | Thiophen-2-yl | H | H | H | 60 | 1.93 | 500 | 1030 |
| 2i | Ph | MeO | MeO | MeO | 74 | 1.19 | 800 | 950 |
| 2j | 4-MeOPh | MeO | MeO | MeO | 57 | 1.06 | 740 | 950 |
| 2k | 3-OMe-4-HOPh | MeO | MeO | MeO | 63 | 0.67 | 610 | 1010 |
| 2l | Furan-2-yl | MeO | MeO | MeO | 67 | −0.20 | 790 | 930 |
| 2m | Thiophen-2-yl | MeO | MeO | MeO | 64 | 1.17 | 850 | 1010 |
| 2n | Ph | H | F | H | 47 | 2.26 | 560 | 1040 |
| 2ñ | 4-MeOPh | H | F | H | 62 | 2.13 | 710 | 860 |
| 2o | 3-MeO-4-HOPh | H | F | H | 52 | 1.74 | 550 | 920 |
| 2p | Furan-2-yl | H | F | H | 49 | 0.88 | 680 | 950 |
| 2q | Thiophen-2-yl | H | F | H | 55 | 2.24 | 730 | 980 |
| Quinone | T24 | DU-145 | Mean value | HEK-293 |
|---|---|---|---|---|
| 2a | >100 | 96.0 ± 7.9 | 98.0 | >100 |
| 2b | >100 | 92.2 ± 8.7 | 96.1 | >100 |
| 2c | 16.3 ± 1.7 | 45.2 ± 4.5 | 30.7 | >100 |
| 2d | 34.0 ± 3.7 | 23.5 ± 2.1 | 28.7 | >100 |
| 2e | 58.2 ± 7.1 | 47.7 ± 4.2 | 52.9 | >100 |
| 2f | 51.8 ± 6.2 | 20.4 ± 2.1 | 36.1 | 85.8 ± 0.9 |
| 2g | 40.2 ± 4.0 | 39.9 ± 3.6 | 40.0 | 61.7 ± 4.2 |
| 2h | 75.6 ± 9.1 | 80.9 ± 9.7 | 78.2 | >100 |
| 2i | >100 | 92.9 ± 1.4 | 96.4 | >100 |
| 2j | 85.3 ± 1.9 | 72.4 ± 2.6 | 78.8 | 56.9 ± 5.2 |
| 2k | >100 | 45.9 ± 3.2 | 72.9 | 28.3 ± 1.5 |
| 2l | 47.1 ± 2.0 | 23.0 ± 2.3 | 35.0 | 33.0 ± 4.8 |
| 2m | 63.2 ± 0.6 | 48.6 ± 3.0 | 55.9 | 30.7 ± 3.1 |
| 2n | 74.5 ± 2.2 | >100 | 87.2 | >100 |
| 2ñ | >100 | >100 | 100.0 | >100 |
| 2o | >100 | >100 | 100.0 | >100 |
| 2p | >100 | 90.0 ± 6.4 | 95.0 | 72.8 ± 7.0 |
| 2q | >100 | >100 | 100 | >100 |
| DOX | 0.46 ± 0.08 | 0.93 ± 0.06 | 0.69 | 4.27 ± 0.34 |
| Gene Name | Gene Symbol | Relative Expression Levels in T24 Cells (μM) | ||
|---|---|---|---|---|
| Vehicle | 2c | 2d | ||
| Apoptosis regulator (BCL2), transcript variant alpha | BCL2 | 1.00 ± 0.05 | 1.28 ± 0.15 | 0.80 ± 0.04 |
| Mechanistic target of rapamycin kinase | mTOR | 1.00 ± 0.04 | 0.99 ± 0.13 | 0.95 ± 0.02 |
| Glutathione S-transferase pi 1 | GSTP1 | 1.00 ± 0.06 | 0.90 ± 0.08 | 1.21 ± 0.04 * |
| Glutathione-disulfide reductase | GSR | 1.00 ± 0.05 | 0.96 ± 0.05 | 1.07 ± 0.04 |
| Cell division cycle 25A | CDC25A | 1.00 ± 0.06 | 1.54 ± 0.13 *** | 1.07 ± 0.03 |
| Tumor protein p53 | TP53 | 1.00 ± 0.04 | 1.06 ± 0.11 | 1.01 ± 0.04 |
| v-Ha-ras Harvey rat sarcoma viral oncogene homolog | HRAS | 1.00 ± 0.06 | 1.07 ± 0.13 | 1.33 ± 0.06 * |
| Histone deacetylase 3 | HDAC3 | 1.00 ± 0.06 | 1.03 ± 0.15 | 1.02 ± 0.05 |
| Histone deacetylase 4 | HDAC4 | 1.00 ± 0.05 | 1.08 ± 0.13 | 1.01 ± 0.02 |
| Tumor necrosis factor | TNF | 1.00 ± 0.03 | 2.04 ± 0.11 *** | 1.44 ± 0.10 ** |
| Gene Name | Gene Symbol | Entrez Gene ID | Forward (5′ → 3′) | Reverse (5′ → 3′) | Amplicon Size (pb) | ||
|---|---|---|---|---|---|---|---|
| Apoptosis | |||||||
| Apoptosis regulator (BCL2), transcript variant alpha | BCL2 | NM_000633.2 | ATGTGTGTGGAGAGCGTCAA | GAGACAGCCAGGAGAAATCAA | 181 | ||
| PI3 Kinases & Phosphatases | |||||||
| Mechanistic target of rapamycin kinase | mTOR | NM_004958.3 | TCCGAGAGATGAGTCAAGAGG | CACCTTCCACTCCTATGAGGC | 141 | ||
| Drug Metabolism/Oxidative Stress | |||||||
| Glutathione S-transferase pi 1 | GSTP1 | NM_000852.3 | CATCTACACCAACTATGAGGCG | AGCAGGGTCTCAAAAGGCTTC | 81 | ||
| Glutathione-disulfide reductase | GSR | NM_000637.4 | CACTTGCGTGAATGTTGGATG | TGGGATCACTCGTGAAGGCT | 242 | ||
| Cell Cycle | |||||||
| Cell division cycle 25A | CDC25A | NM_001789.2 | TGGGCCATTGGACAGTAAAG | TCCCAACAGCTTCTGAGGTA | 76 | ||
| Tumor protein p53 | TP53 | NM_000546.5 | ACAGCTTTGAGGTGCGTGTTT | CCCTTTCTTGCGGAGATTCTCT | 77 | ||
| Ras Signaling | |||||||
| V-Ha-Ras Harvey rat sarcoma viral oncogene homolog | HRAS | NM_005343 | GACGTGCCTGTTGGACATC | CTTCACCCGTTTGATCTGCTC | 166 | ||
| Histone Deacetylases | |||||||
| Histone deacetylase 3 | HDAC3 | NM_003883.3 | TCTGGCTTCTGCTATGTCAACG | CCCGGTCAGTGAGGTAGAAAG | 136 | ||
| Histone deacetylase 4 | HDAC4 | NM_006037.3 | AGCGTCCGTTGGATGTCAC | CCTTCTCGTGCCACAAGTCT | 169 | ||
| Inflammation | |||||||
| Tumor necrosis factor | TNF | NM_000594.3 | AGAACTCACTGGGGCCTACA | GCTCCGTGTCTCAAGGAAGT | 177 | ||
| Housekeeping | |||||||
| Beta-2-microglobulin | B2M | NM_004048.2 | ATGAGTATGCCTGCCGTGTGA | GGCATCTTCAAACCTCCATG | 97 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benites, J.; Valderrama, J.A.; Ramos, M.; Valenzuela, M.; Guerrero-Castilla, A.; Muccioli, G.G.; Buc Calderon, P. Half-Wave Potentials and In Vitro Cytotoxic Evaluation of 3-Acylated 2,5-Bis(phenylamino)-1,4-benzoquinones on Cancer Cells. Molecules 2019, 24, 1780. https://doi.org/10.3390/molecules24091780
Benites J, Valderrama JA, Ramos M, Valenzuela M, Guerrero-Castilla A, Muccioli GG, Buc Calderon P. Half-Wave Potentials and In Vitro Cytotoxic Evaluation of 3-Acylated 2,5-Bis(phenylamino)-1,4-benzoquinones on Cancer Cells. Molecules. 2019; 24(9):1780. https://doi.org/10.3390/molecules24091780
Chicago/Turabian StyleBenites, Julio, Jaime A. Valderrama, Maryan Ramos, Maudy Valenzuela, Angélica Guerrero-Castilla, Giulio G. Muccioli, and Pedro Buc Calderon. 2019. "Half-Wave Potentials and In Vitro Cytotoxic Evaluation of 3-Acylated 2,5-Bis(phenylamino)-1,4-benzoquinones on Cancer Cells" Molecules 24, no. 9: 1780. https://doi.org/10.3390/molecules24091780






