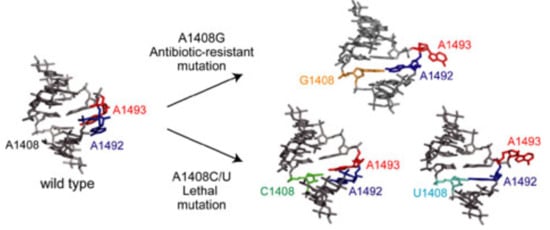
Structural Bases for the Fitness Cost of the Antibiotic-Resistance and Lethal Mutations at Position 1408 of 16S rRNA
Abstract
:1. Introduction
2. Results and Discussion
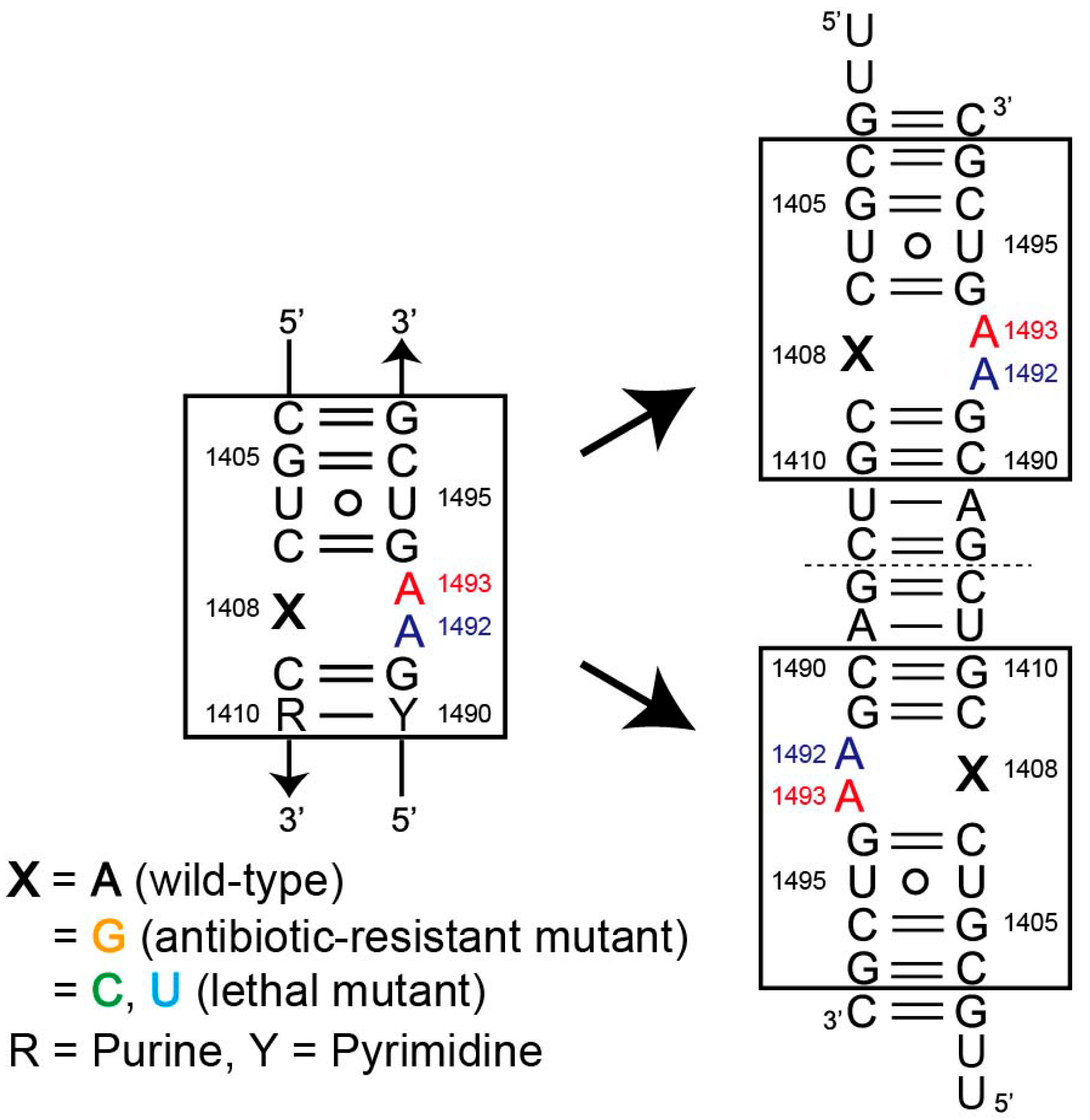
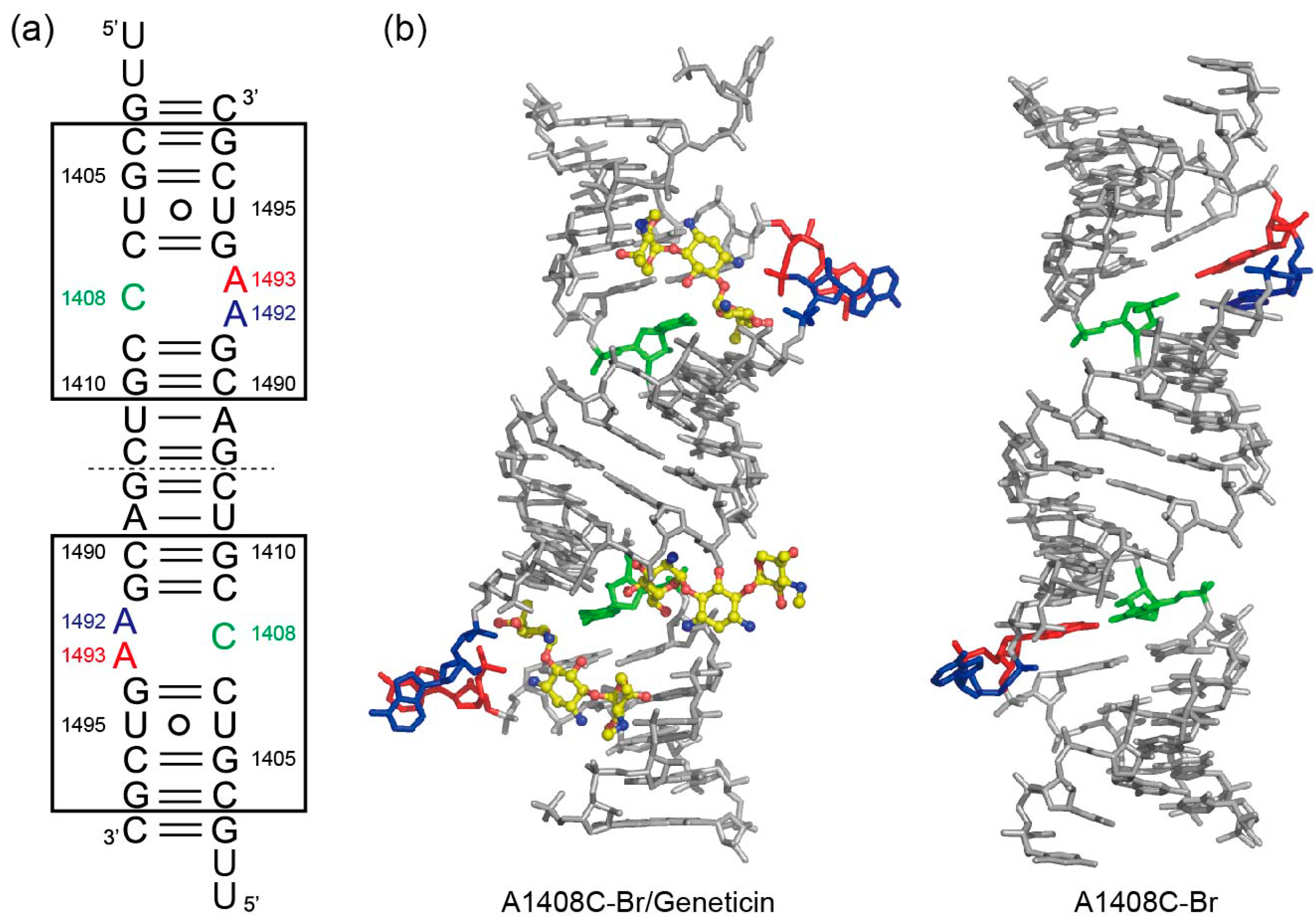
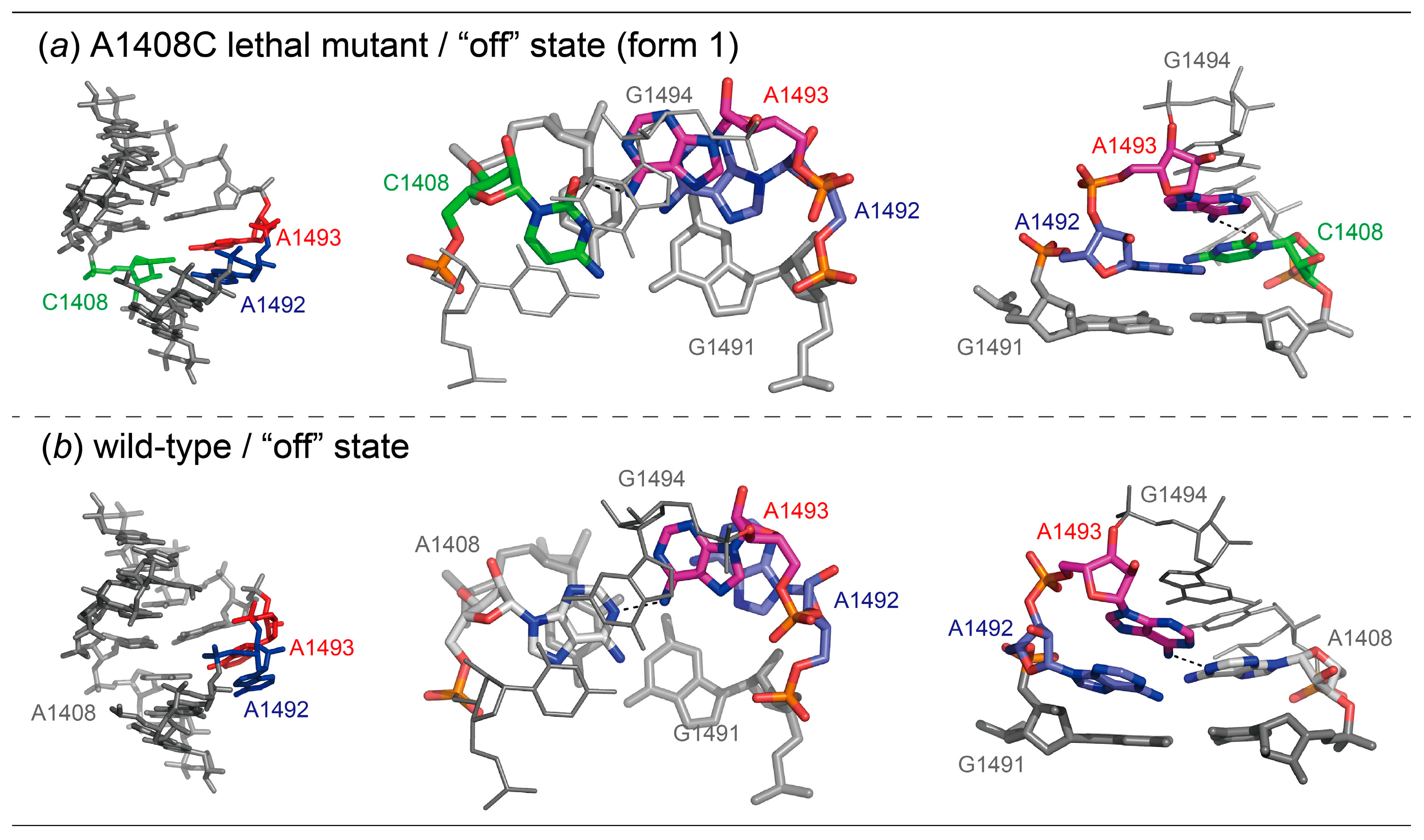
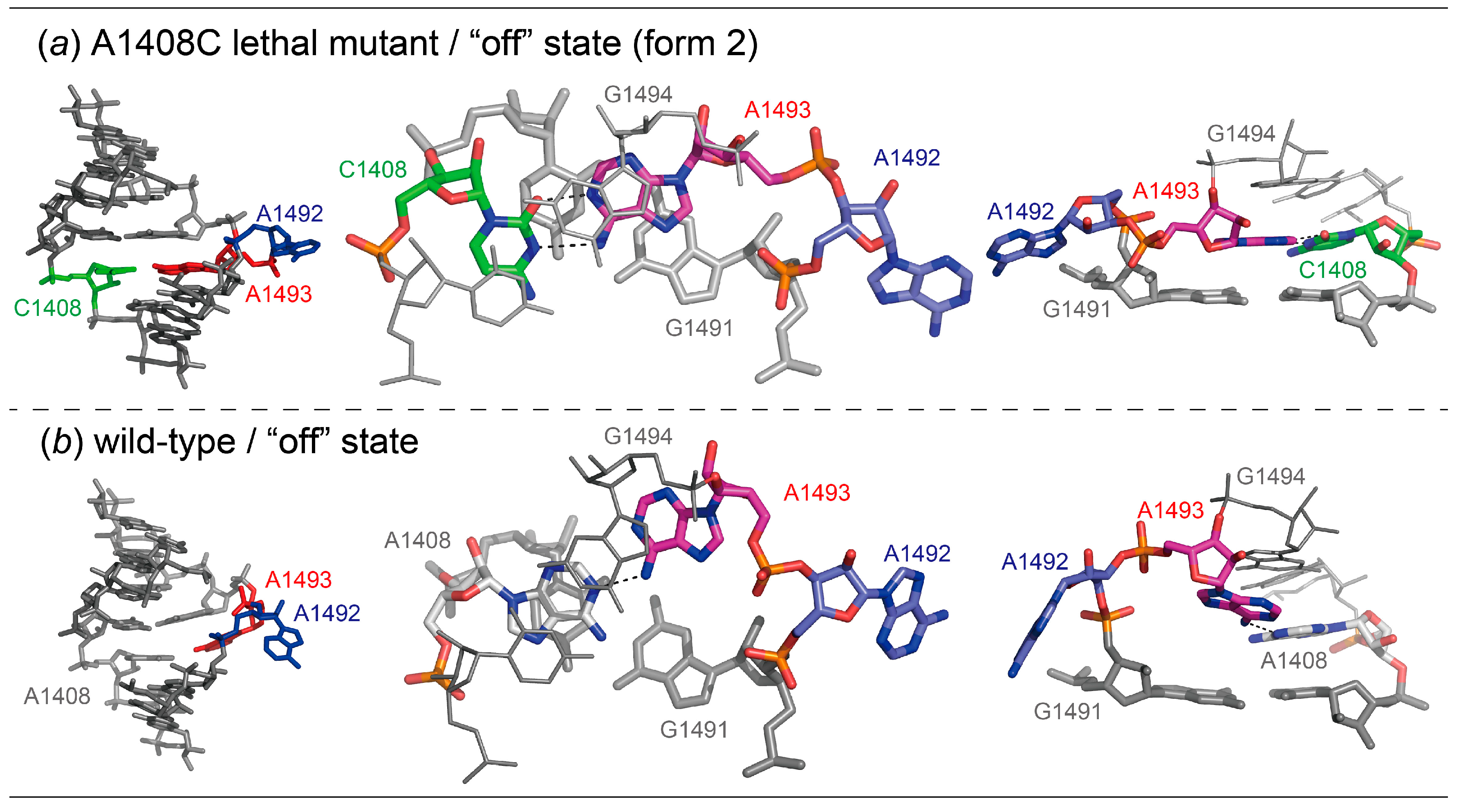
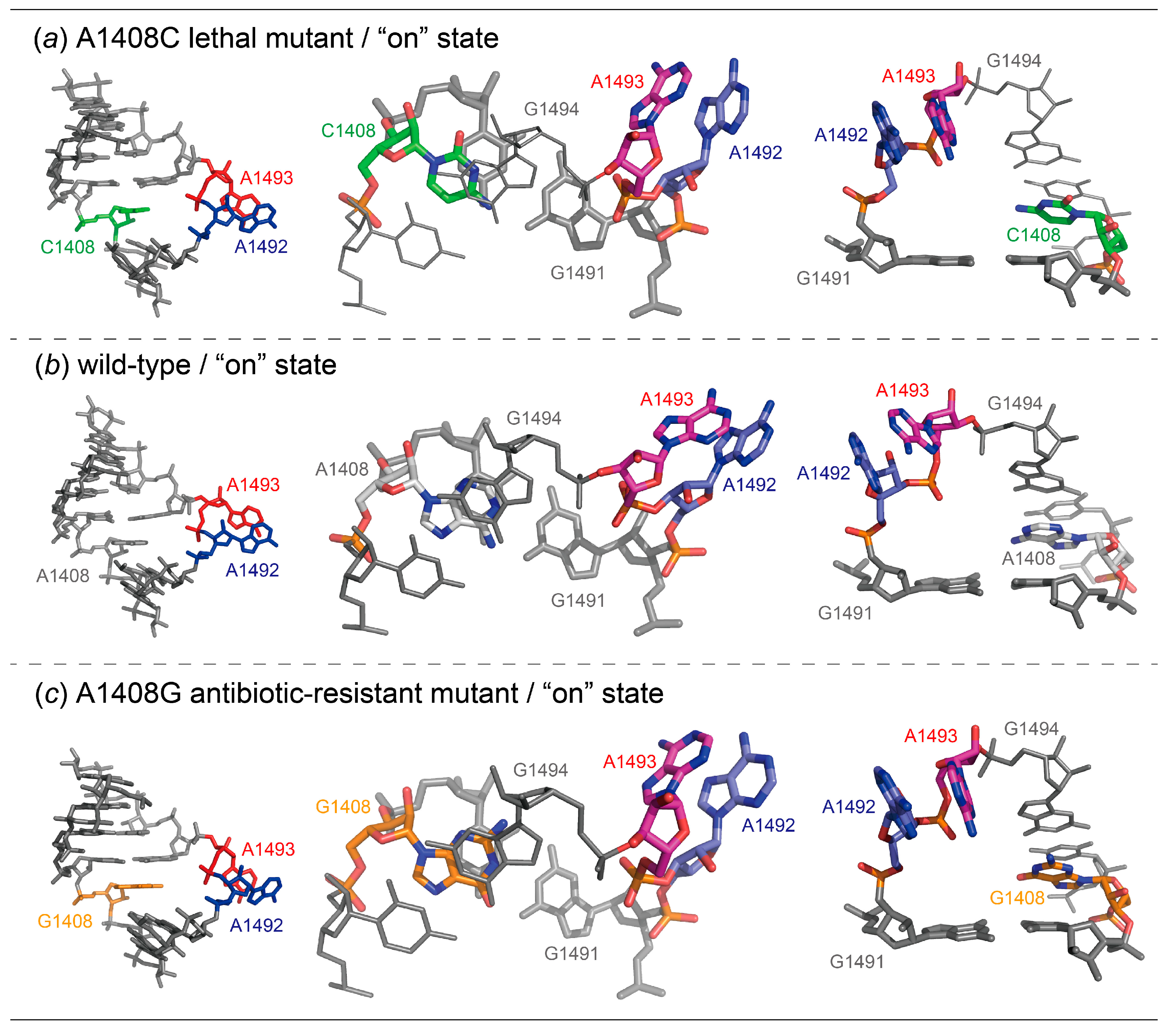
2.1. Structures of the A1408C Lethal Mutant A Site and Its Comparison with the Wild Type and the A1408G Antibiotic-Resistant Mutant
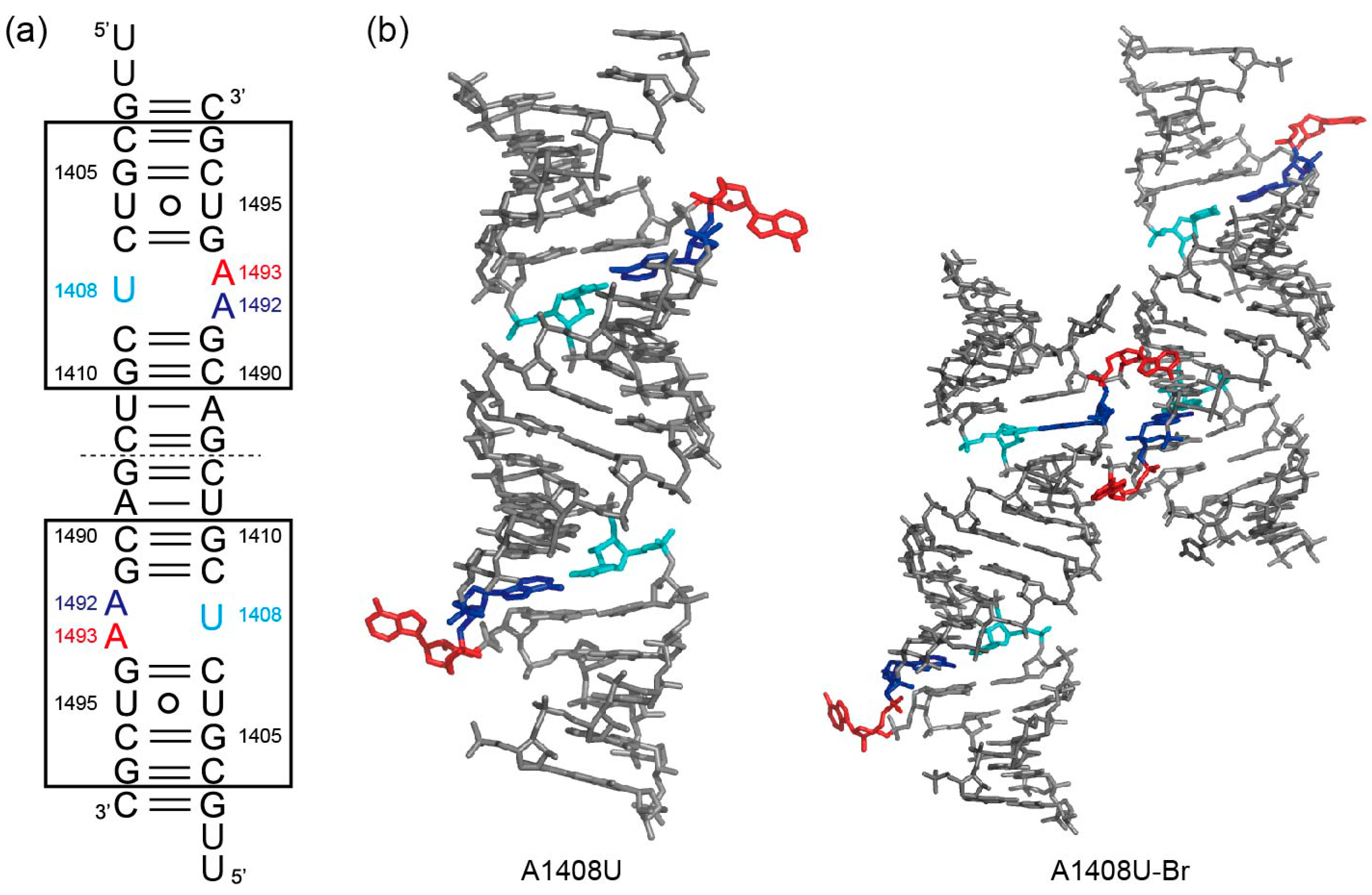
2.2. Structures of the A1408U Lethal Mutant A Site and Its Comparison with the Wild Type and the A1408G Antibiotic-Resistant Mutant
2.3. Conclusions of Discussion
3. Materials and Methods
3.1. Sample Preparations and Crystallizations
3.2. Data Collections, Structure Determinations and Refinements
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ogle, J.M.; Carter, A.P.; Ramakrishnan, V. Insights into the decoding mechanism from recent ribosome structure. Trends Biochem. Sci. 2003, 28, 259–266. [Google Scholar] [CrossRef]
- Ogle, J.M.; Carter, A.P.; Ramakrishnan, V. Structural insights into translation fidelity. Annu. Rev. Biochem. 2005, 74, 129–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noller, H.F. Biochemical characterization of the ribosomal decoding site. Biochimie 2006, 88, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Schmeing, T.M.; Ramakrishnan, V. What recent ribosome structures have revealed about the mechanism of translation. Nature 2009, 461, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Wimberly, B.T.; Brodersen, D.E.; Clemons, W.M., Jr.; Morgan-Warren, R.J.; Carter, A.P.; Vonrhein, C.; Hartsch, T.; Ramakrishnan, V. Structure of the 30S ribosomal subunit. Nature 2000, 407, 327–339. [Google Scholar] [CrossRef]
- Carter, A.P.; Clemons, W.M.; Brodersen, D.E.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 2000, 407, 340–348. [Google Scholar] [CrossRef]
- Ogle, J.M.; Brodersen, D.E.; Clemons, W.M., Jr.; Tarry, M.J.; Carter, A.P.; Ramakrishnan, V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 2001, 292, 897–902. [Google Scholar] [CrossRef] [Green Version]
- Ogle, J.M.; Murphy, F.V.; Tarry, M.J.; Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 2002, 111, 721–732. [Google Scholar] [CrossRef] [Green Version]
- Murphy, F.V., 4th; Ramakrishnan, V.; Malkiewicz, A.; Agris, P.F. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 2004, 11, 1186–1191. [Google Scholar] [CrossRef]
- Murphy, F.V., 4th; Ramakrishnan, V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol. 2004, 11, 1251–1252. [Google Scholar] [CrossRef]
- Schuwirth, B.S.; Borovinskaya, M.A.; Hau, C.W.; Zhang, W.; Vila-Sanjurjo, A.; Holton, J.M.; Cate, J.H. Structures of the bacterial ribosome at 3.5 A resolution. Science 2005, 310, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Selmer, M.; Dunham, C.M.; Murphy, F.V., 4th; Weixlbaumer, A.; Petry, S.; Kelley, A.C.; Weir, J.R.; Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006, 313, 1935–1942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunham, C.M.; Selmer, M.; Phelps, S.S.; Kelley, A.C.; Suzuki, T.; Joseph, S.; Ramakrishnan, V. Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit. RNA 2007, 13, 817–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weixlbaumer, A.; Murphy, F.V., 4th; Dziergowska, A.; Malkiewicz, A.; Vendeix, F.A.; Agris, P.F.; Ramakrishnan, V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 2007, 14, 498–502. [Google Scholar] [CrossRef] [Green Version]
- Kurata, S.; Weixlbaumer, A.; Ohtsuki, T.; Shimazaki, T.; Wada, T.; Kirino, Y.; Takai, K.; Watanabe, K.; Ramakrishnan, V.; Suzuki, T. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U.G wobble pairing during decoding. J. Biol. Chem. 2008, 283, 18801–18811. [Google Scholar] [CrossRef] [Green Version]
- Schmeing, T.M.; Voorhees, R.M.; Kelley, A.C.; Gao, Y.G.; Murphy, F.V., 4th; Weir, J.R.; Ramakrishnan, V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 2009, 326, 688–694. [Google Scholar] [CrossRef] [Green Version]
- Schmeing, T.M.; Voorhees, R.M.; Kelley, A.C.; Ramakrishnan, V. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat. Struct. Mol. Biol. 2011, 18, 432–436. [Google Scholar] [CrossRef] [Green Version]
- Neubauer, C.; Gillet, R.; Kelley, A.C.; Ramakrishnan, V. Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science 2012, 335, 1366–1369. [Google Scholar] [CrossRef] [Green Version]
- Demeshkina, N.; Jenner, L.; Westhof, E.; Yusupov, M.; Yusupova, G. A new understanding of the decoding principle on the ribosome. Nature 2012, 484, 256–259. [Google Scholar] [CrossRef]
- Voorhees, R.M.; Mandal, D.; Neubauer, C.; Köhrer, C.; RajBhandary, U.L.; Ramakrishnan, V. The structural basis for specific decoding of AUA by isoleucine tRNA on the ribosome. Nat. Struct. Mol. Biol. 2013, 20, 641–643. [Google Scholar] [CrossRef] [Green Version]
- Fernández, I.S.; Ng, C.L.; Kelley, A.C.; Wu, G.; Yu, Y.T.; Ramakrishnan, V. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature 2013, 500, 107–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demeshkina, N.; Jenner, L.; Westhof, E.; Yusupov, M.; Yusupova, G. New structural insights into the decoding mechanism: Translation infidelity via a G·U pair with Watson-Crick geometry. FEBS Lett. 2013, 587, 1848–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vicens, Q.; Westhof, E. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure 2001, 9, 647–658. [Google Scholar] [CrossRef] [Green Version]
- Shandrick, S.; Zhao, Q.; Han, Q.; Ayida, B.K.; Takahashi, M.; Winters, G.C.; Simonsen, K.B.; Vourloumis, D.; Hermann, T. Monitoring molecular recognition of the ribosomal decoding site. Angew. Chem. Int. Ed. 2004, 43, 3177–3182. [Google Scholar] [CrossRef]
- Kondo, J.; Urzhumtsev, A.; Westhof, E. Two conformational states in the crystal structure of the Homo sapiens cytoplasmic ribosomal decoding A site. Nucleic Acids Res. 2006, 34, 676–685. [Google Scholar] [CrossRef] [Green Version]
- Kondo, J.; François, B.; Urzhumtsev, A.; Westhof, E. Crystal structure of the Homo sapiens cytoplasmic ribosomal decoding site complexed with apramycin. Angew. Chem. Int. Ed. Engl. 2006, 45, 3310–3314. [Google Scholar] [CrossRef]
- Kondo, J.; Westhof, E. Structural comparisons between prokaryotic and eukaryotic ribosomal decoding A sites free and complexed with aminoglycosides. In Aminoglycoside Antibiotics, from Chemical Biology to Drug Discovery; Arya, D.P., Ed.; Wiley-Interscience: Hoboken, NJ, USA, 2007; pp. 209–223. [Google Scholar]
- Kondo, J.; Westhof, E. The bacterial and mitochondrial ribosomal A-site molecular switches possess different conformational substates. Nucleic Acids Res. 2008, 36, 2654–2666. [Google Scholar] [CrossRef]
- Kondo, J. A structural basis for the antibiotic resistance conferred by an A1408G mutation in 16S rRNA and for the antiprotozoal activity of aminoglycosides. Angew. Chem. Int. Ed. Engl. 2012, 51, 465–468. [Google Scholar] [CrossRef]
- Kanazawa, H.; Baba, F.; Koganei, M.; Kondo, J. A structural basis of the antibiotic resistance conferred by an N1-methylation of A1408 in 16S rRNA. Nucleic Acids Res. 2017, 45, 12529–12535. [Google Scholar] [CrossRef] [Green Version]
- Cannone, J.J.; Subramanian, S.; Schnare, M.N.; Collett, J.R.; D’Souza, L.M.; Du, Y.; Feng, B.; Lin, N.; Madabusi, L.V.; Müller, K.M.; et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 2002, 3, 2. [Google Scholar]
- Prammananan, T.; Sander, P.; Brown, B.A.; Frischkorn, K.; Onyi, G.O.; Zhang, Y.; Böttger, E.C.; Wallace, R.J., Jr. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 1998, 177, 1573–1581. [Google Scholar] [CrossRef] [Green Version]
- Sander, P.; Prammananan, T.; Böttger, E.C. Introducing mutations into a chromosomal rRNA gene using a genetically modified eubacterial host with a single rRNA operon. Mol. Microbiol. 1996, 22, 841–848. [Google Scholar] [CrossRef]
- Suzuki, Y.; Katsukawa, C.; Tamaru, A.; Abe, C.; Makino, M.; Mizuguchi, Y.; Taniguchi, H. Detection of kanamycin-resistant Mycobacterium tuberculosis by identifying mutations in the 16S rRNA gene. J. Clin. Microbiol. 1998, 36, 1220–1225. [Google Scholar] [PubMed]
- Alangaden, G.J.; Kreiswirth, B.N.; Aouad, A.; Khetarpal, M.; Igno, F.R.; Moghazeh, S.L.; Manavathu, E.K.; Lerner, S.A. Mechanism of resistance to amikacin and kanamycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 1998, 42, 1295–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krüüner, A.; Jureen, P.; Levina, K.; Ghebremichael, S.; Hoffner, S. Discordant resistance to kanamycin and amikacin in drug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2003, 47, 2971–2973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maus, C.E.; Plikaytis, B.B.; Shinnick, T.M. Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2005, 49, 3192–3197. [Google Scholar] [CrossRef] [Green Version]
- Feuerriegel, S.; Cox, H.S.; Zarkua, N.; Karimovich, H.A.; Braker, K.; Rüsch-Gerdes, S.; Niemann, S. Sequence analyses of just four genes to detect extensively drug-resistant Mycobacterium tuberculosis strains in multidrug-resistant tuberculosis patients undergoing treatment. Antimicrob. Agents Chemother. 2009, 53, 3353–3356. [Google Scholar] [CrossRef] [Green Version]
- Jugheli, L.; Bzekalava, N.; de Rijk, P.; Fissette, K.; Portaels, F.; Rigouts, L. High level of cross-resistance between kanamycin, amikacin, and capreomycin among Mycobacterium tuberculosis isolates from Georgia and a close relation with mutations in the RRS gene. Antimicrob. Agents Chemother. 2009, 53, 5064–5068. [Google Scholar] [CrossRef] [Green Version]
- Kogure, T.; Shimada, R.; Ishikawa, J.; Yazawa, K.; Brown, J.M.; Mikami, Y.; Gonoi, T. Homozygous triplicate mutations in three 16S rRNA genes responsible for high-level aminoglycoside resistance in Nocardia farcinica clinical isolates from a Canada-wide bovine mastitis epizootic. Antimicrob. Agents Chemother. 2010, 54, 2385–2390. [Google Scholar] [CrossRef] [Green Version]
- Biswas, S.; Raoult, D.; Rolain, J.M. Molecular mechanism of gentamicin resistance in Bartonella henselae. Clin. Microbiol. Infect. 2009, 15, 98–99. [Google Scholar] [CrossRef] [Green Version]
- Recht, M.I.; Douthwaite, S.; Puglisi, J.D. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 1999, 18, 3133–3138. [Google Scholar] [CrossRef] [PubMed]
- Pfister, P.; Hobbie, S.; Vicens, Q.; Böttger, E.C.; Westhof, E. The molecular basis for A-site mutations conferring aminoglycoside resistance: Relationship between ribosomal susceptibility and X-ray crystal structures. ChemBioChem 2003, 4, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Gregory, S.T.; Carr, J.F.; Rodriguez-Correa, D.; Dahlberg, A.E. Mutational analysis of 16S and 23S rRNA genes of Thermus thermophilus. J. Bacteriol. 2005, 187, 4804–4812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobbie, S.N.; Pfister, P.; Brüll, C.; Westhof, E.; Böttger, E.C. Analysis of the contribution of individual substituents in 4,6-aminoglycoside-ribosome interaction. Antimicrob. Agents Chemother. 2005, 49, 5112–5118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobbie, S.N.; Pfister, P.; Bruell, C.; Sander, P.; François, B.; Westhof, E.; Böttger, E.C. Binding of neomycin-class aminoglycoside antibiotics to mutant ribosomes with alterations in the A site of 16S rRNA. Antimicrob. Agents Chemother. 2006, 50, 1489–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobbie, S.N.; Bruell, C.; Kalapala, S.; Akshay, S.; Schmidt, S.; Pfister, P.; Böttger, E.C. A genetic model to investigate drug-target interactions at the ribosomal decoding site. Biochimie 2006, 88, 1033–1043. [Google Scholar] [CrossRef]
- Andersson, D.I. The biological cost of mutational antibiotic resistance: Any practical conclusions? Curr. Opin. Microbiol. 2006, 9, 461–465. [Google Scholar] [CrossRef]
- Sander, P.; Springer, B.; Prammananan, T.; Sturmfels, A.; Kappler, M.; Pletschette, M.; Böttger, E.C. Fitness cost of chromosomal drug resistance-conferring mutations. Antimicrob. Agents Chemother. 2002, 46, 1204–1211. [Google Scholar] [CrossRef] [Green Version]
- Shcherbakov, D.; Akbergenov, R.; Matt, T.; Sander, P.; Andersson, D.I.; Böttger, E.C. Directed mutagenesis of Mycobacterium smegmatis 16S rRNA to reconstruct the in-vivo evolution of aminoglycoside resistance in Mycobacterium tuberculosis. Mol. Microbiol. 2010, 77, 830–840. [Google Scholar] [CrossRef]
- Taliaferro, D.L.; Farabaugh, P.J. Testing constraints on rRNA bases that make nonsequence-specific contacts with the codon-anticodon complex in the ribosomal A site. RNA 2007, 13, 1279–1286. [Google Scholar] [CrossRef] [Green Version]
- Leontis, N.B.; Stombaugh, J.; Westhof, E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002, 30, 3497–3531. [Google Scholar] [CrossRef] [PubMed]
- Auffinger, P.; Hashem, Y. SwS: A solvation web service for nucleic acids. Bioinformatics 2007, 23, 1035–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, W.K.; Bansal, M.; Burley, S.K.; Dickerson, R.E.; Gerstein, M.; Harvey, S.C.; Heinemann, U.; Lu, X.J.; Neidle, S.; Shakked, Z.; et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001, 313, 229–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994, 50, 760–763. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Grosse-Kunstleve, R.W.; Adams, P.D. On symmetries of substructures. Acta Crystallogr. D Biol. Crystallogr. 2003, 59, 1974–1977. [Google Scholar] [CrossRef] [Green Version]
- Terwilliger, T.C.; Adams, P.D.; Read, R.J.; McCoy, A.J.; Moriarty, N.W.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Zwart, P.H.; Hung, L.W. Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 2009, 65, 582–601. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Brünger, A.T.; Adams, P.D.; Clore, G.M.; DeLano, W.L.; Gros, P.; Grosse-Kunstleve, R.W.; Jiang, J.S.; Kuszewski, J.; Nilges, M.; Pannu, N.S.; et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998, 54, 905–921. [Google Scholar] [PubMed]
- Brünger, A.T. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature 1992, 355, 472–475. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific LLC: Palo Alto, CA, USA, 2008. [Google Scholar]
Sample Availability: Samples of the compounds are available from the authors. |

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondo, J.; Koganei, M. Structural Bases for the Fitness Cost of the Antibiotic-Resistance and Lethal Mutations at Position 1408 of 16S rRNA. Molecules 2020, 25, 159. https://doi.org/10.3390/molecules25010159
Kondo J, Koganei M. Structural Bases for the Fitness Cost of the Antibiotic-Resistance and Lethal Mutations at Position 1408 of 16S rRNA. Molecules. 2020; 25(1):159. https://doi.org/10.3390/molecules25010159
Chicago/Turabian StyleKondo, Jiro, and Mai Koganei. 2020. "Structural Bases for the Fitness Cost of the Antibiotic-Resistance and Lethal Mutations at Position 1408 of 16S rRNA" Molecules 25, no. 1: 159. https://doi.org/10.3390/molecules25010159