Molecular Spectroscopic Markers of Abnormal Protein Aggregation
Abstract
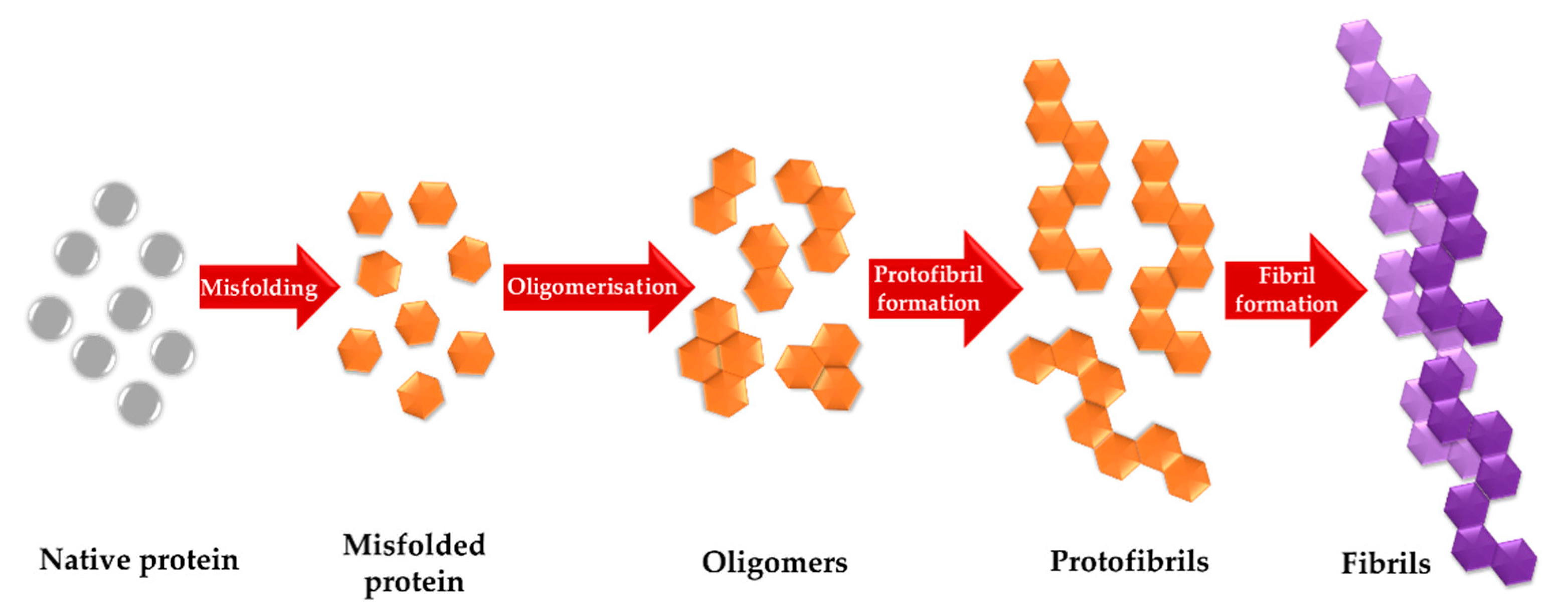
1. Introduction
2. Infrared Spectroscopy Studies of Abnormal Aggregation of Proteins/Peptides
2.1. Infrared Spectroscopy at the Nano Scale in Studies of Abnormal Proteins/Peptide Aggregation
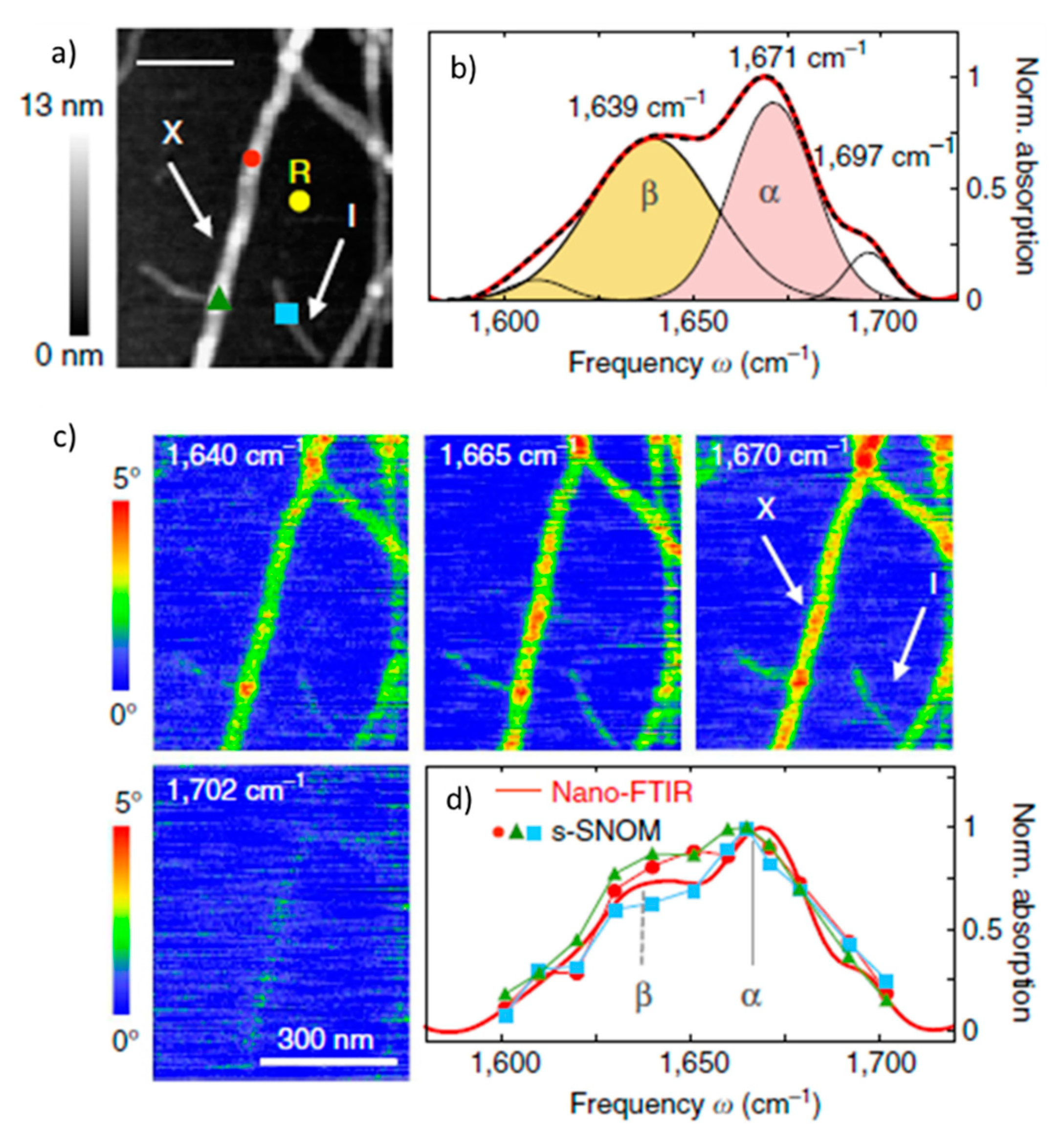
2.1.1. Nano-FTIR In Studies of Abnormal Proteins/Peptide Aggregation
2.1.2. Infrared Spectroscopy Combined with Atomic Force Microscopy (AFM-IR) in Studies of Abnormal Proteins/Peptide Aggregation
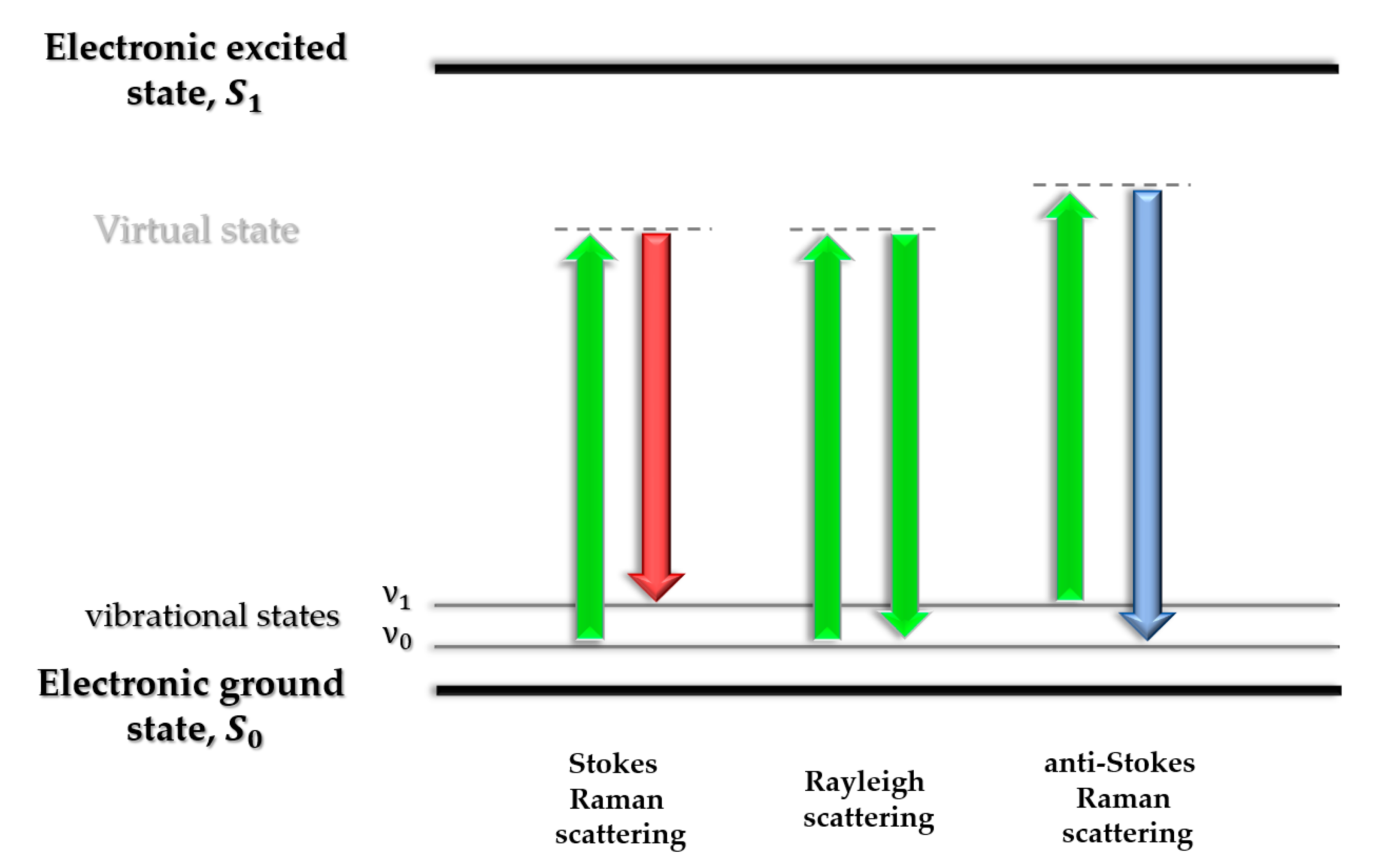
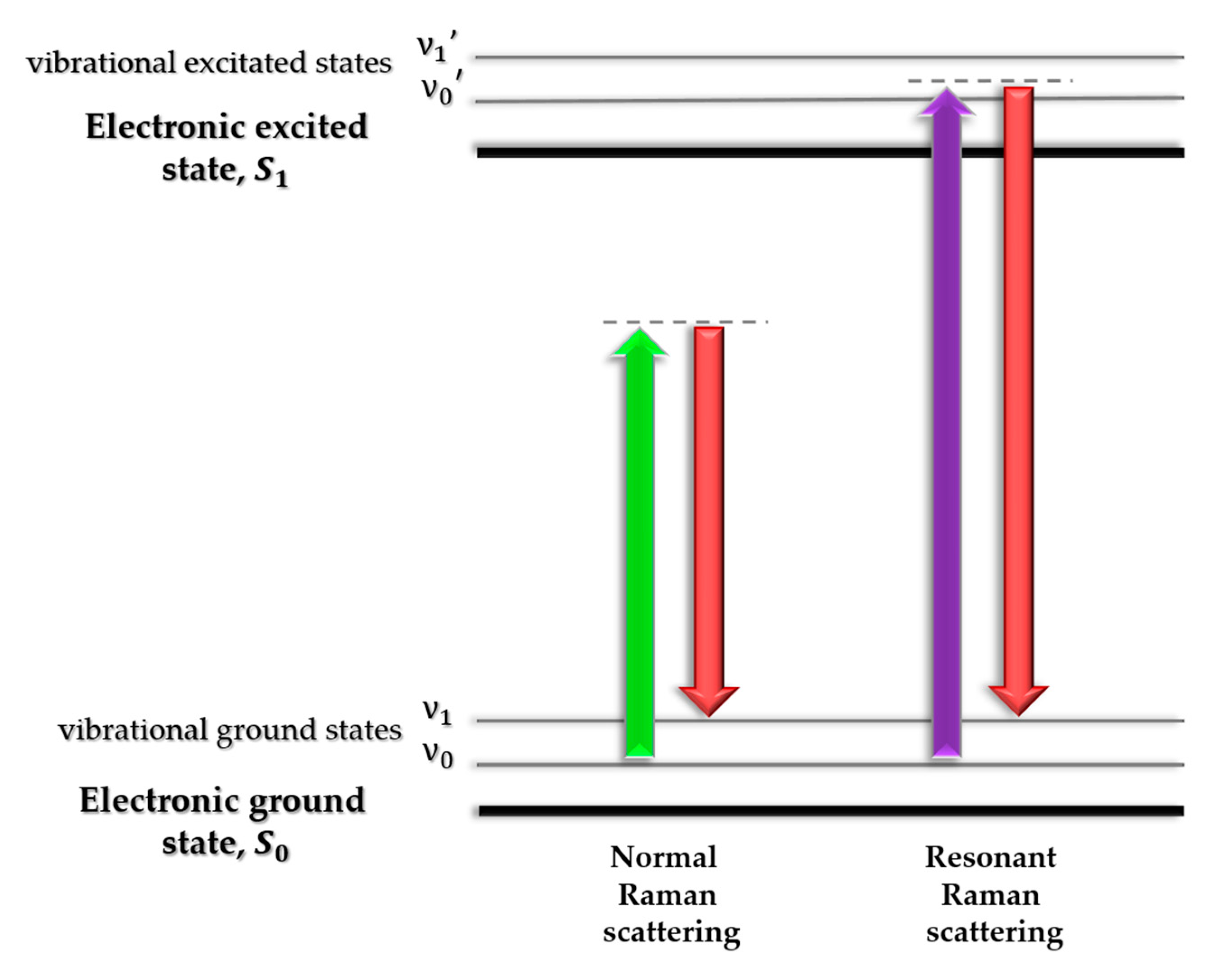
3. Raman Spectroscopy in Studies of Abnormal Proteins/Peptide Aggregation
3.1. SERS In Studies of Abnormal Proteins/Peptide Aggregation
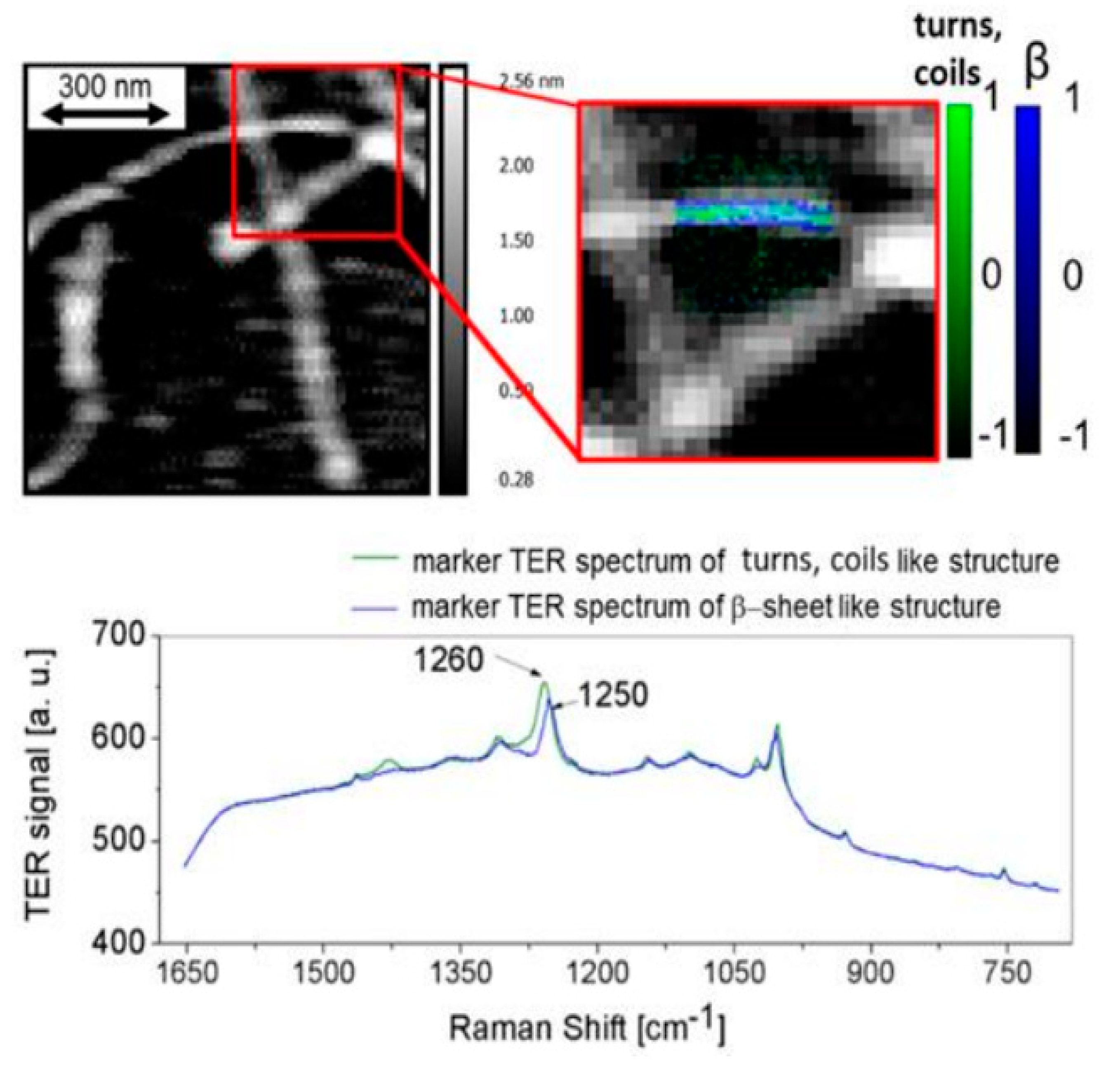
3.2. TERS In Studies of Abnormal Proteins/Peptide Aggregation
4. Multivariate Data Analysis in Studies of Abnormal Proteins/Peptide Aggregation
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Knowles, T.P.J.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Jucker, M.; Walker, L.C. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 2013, 501, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Riek, R.; Eisenberg, D.S. The activities of amyloids from a structural perspective. Nature 2016, 539, 227–235. [Google Scholar] [CrossRef]
- Alzheimer’s Disease International Dementia statistics | Alzheimer’s Disease International. Alzheimer’s Dis. Int. 2016, 1. [CrossRef]
- Stefani, M. Protein misfolding and aggregation: New examples in medicine and biology of the dark side of the protein world. Biochim. Biophys. Acta—Mol. Basis Dis. 2004, 1739, 5–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, W. Protein aggregation and its inhibition in biopharmaceutics. Int. J. Pharm. 2005, 289, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Tutar, L.; Tutar, Y. Heat Shock Proteins; An Overview. Curr. Pharm. Biotechnol. 2010, 11, 216–222. [Google Scholar] [CrossRef]
- Kumar, V.; Sami, N.; Kashav, T.; Islam, A.; Ahmad, F.; Hassan, M.I. Protein aggregation and neurodegenerative diseases: From theory to therapy. Eur. J. Med. Chem. 2016, 124, 1105–1120. [Google Scholar] [CrossRef]
- Chung, C.G.; Lee, H.; Lee, S.B. Mechanisms of protein toxicity in neurodegenerative diseases. Cell. Mol. Life Sci. 2018, 75, 3159–3180. [Google Scholar] [CrossRef]
- Lee, C.; Yu, M.H. Protein folding and diseases. J. Biochem. Mol. Biol. 2005, 38, 275–280. [Google Scholar] [CrossRef]
- Kolarova, M.; García-Sierra, F.; Bartos, A.; Ricny, J.; Ripova, D. Structure and pathology of tau protein in Alzheimer disease. Int. J. Alzheimers Dis. 2012, 2012. [Google Scholar] [CrossRef]
- Buée, L.; Bussière, T.; Buée-Scherrer, V.; Delacourte, A.; Hof, P.R. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Rev. 2000, 33, 95–130. [Google Scholar] [CrossRef]
- Martin, L.; Latypova, X.; Terro, F. Post-translational modifications of tau protein: Implications for Alzheimer’s disease. Neurochem. Int. 2011, 58, 458–471. [Google Scholar] [CrossRef]
- Murphy, M.P.; Levine, H. Alzheimer’s disease and the amyloid-β peptide. J. Alzheimer’s Dis. 2010, 19, 311–323. [Google Scholar] [CrossRef]
- Atwood, C.S.; Obrenovich, M.E.; Liu, T.; Chan, H.; Perry, G.; Smith, M.A.; Martins, R.N. Amyloid-β: A chameleon walking in two worlds: A review of the trophic and toxic properties of amyloid-β. Brain Res. Rev. 2003, 43, 1–16. [Google Scholar] [CrossRef]
- Meng, F.; Bellaiche, M.M.J.; Kim, J.Y.; Zerze, G.H.; Best, R.B.; Chung, H.S. Highly Disordered Amyloid-β Monomer Probed by Single-Molecule FRET and MD Simulation. Biophys. J. 2018, 114, 870–884. [Google Scholar] [CrossRef]
- Vivekanandan, S.; Brender, J.R.; Lee, S.Y.; Ramamoorthy, A. A partially folded structure of amyloid-beta(1-40) in an aqueous environment. Biochem. Biophys. Res. Commun. 2011, 411, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, L.; Lee, D.H.S.; Yu, L.C.; Zhang, Y. The role of intracellular amyloid β in Alzheimer’s disease. Prog. Neurobiol. 2007, 83, 131–139. [Google Scholar] [CrossRef]
- Ma, Q.L.; Chan, P.; Yoshii, M.; Uéda, K. α-synuclein aggregation and neurodegenerative diseases. J. Alzheimer’s Dis. 2003, 5, 139–148. [Google Scholar] [CrossRef]
- Etezadi, D.; Warner, J.B.; Ruggeri, F.S.; Dietler, G.; Lashuel, H.A.; Altug, H. Nanoplasmonic mid-infrared biosensor for in vitro protein secondary structure detection. Light Sci. Appl. 2017, 6, 17029. [Google Scholar] [CrossRef]
- Shastry, B.S. Neurodegenerative disorders of protein aggregation. Neurochem. Int. 2003, 43, 1–7. [Google Scholar] [CrossRef]
- Trześniewska, K.; Brzyska, M.; Elbaum, D. Neurodegenerative aspects of protein aggregation. Acta Neurobiol. Exp. (Wars) 2004, 64, 41–52. [Google Scholar]
- Moreno-Gonzalez, I.; Soto, C. Misfolded protein aggregates: Mechanisms, structures and potential for disease transmission. Semin. Cell Dev. Biol. 2011, 22, 482–487. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Bemporad, F.; Chiti, F. Protein misfolded oligomers: Experimental approaches, mechanism of formation, and structure-toxicity relationships. Chem. Biol. 2012, 19, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Sarroukh, R.; Goormaghtigh, E.; Ruysschaert, J.M.; Raussens, V. ATR-FTIR: A “rejuvenated” tool to investigate amyloid proteins. Biochim. Biophys. Acta Biomembr. 2013, 1828, 2328–2338. [Google Scholar] [CrossRef] [PubMed]
- Powers, E.T.; Ferrone, F.A. Kinetic Models for Protein Misfolding and Association. In Protein Misfolding Diseases: Current and Emerging Principles and Therapies; John Wiley & Sons: Hoboken, NJ, USA, 2010; pp. 73–92. [Google Scholar]
- Celej, M.S.; Sarroukh, R.; Goormaghtigh, E.; Fidelio, G.D.; Ruysschaert, J.M.; Raussens, V. Toxic prefibrillar α-synuclein amyloid oligomers adopt a distinctive antiparallel β-sheet structure. Biochem. J. 2012, 443, 719–726. [Google Scholar] [CrossRef]
- Frare, E.; Mossuto, M.F.; de Laureto, P.P.; Tolin, S.; Menzer, L.; Dumoulin, M.; Dobson, C.M.; Fontana, A. Characterization of Oligomeric Species on the Aggregation Pathway of Human Lysozyme. J. Mol. Biol. 2009, 387, 17–27. [Google Scholar] [CrossRef]
- Ruggeri, F.S.; Flagmeier, P.; Kumita, J.R.; Meisl, G.; Chirgadze, D.Y.; Bongiovanni, M.N.; Knowles, T.P.J.; Dobson, C.M. The Influence of Pathogenic Mutations in α-Synuclein on Biophysical and Structural Characteristics of Amyloid Fibrils. ACS Nano 2020. [Google Scholar] [CrossRef]
- Juszczyk, P.; Kolodziejczyk, A.S.; Grzonka, Z. FTIR spectroscopic studies on aggregation process of the β-amyloid 11-28 fragment and its variants. J. Pept. Sci. 2009, 15, 23–29. [Google Scholar] [CrossRef]
- Sandberg, A.; Luheshi, L.M.; Söllvander, S.; De Barros, T.P.; Macao, B.; Knowles, T.P.J.; Biverstål, H.; Lendel, C.; Ekholm-Petterson, F.; Dubnovitsky, A.; et al. Stabilization of neurotoxic Alzheimer amyloid-β oligomers by protein engineering. Proc. Natl. Acad. Sci. USA 2010, 107, 15595–15600. [Google Scholar] [CrossRef]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman Spectroscopy of Proteins: A Review; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; Volume 44, pp. 1061–1076. [Google Scholar]
- Stuart, B.H. Infrared Spectroscopy: Fundamentals and Applications; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2005. [Google Scholar]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [PubMed]
- Sofińska, K.; Wilkosz, N.; Szymoński, M.; Lipiec, E. Molecular spectroscopic markers of DNA damage. Molecules 2020, 25, 561. [Google Scholar] [CrossRef] [PubMed]
- Haris, P.I.; Chapman, D. Does Fourier-transform infrared spectroscopy provide useful information on protein structures? Trends Biochem. Sci. 1992, 17, 328–333. [Google Scholar] [CrossRef]
- Shivu, B.; Seshadri, S.; Li, J.; Oberg, K.A.; Uversky, V.N.; Fink, A.L. Distinct β-sheet structure in protein aggregates determined by ATR-FTIR spectroscopy. Biochemistry 2013, 52, 5176–5183. [Google Scholar] [CrossRef]
- Bouchard, M.; Zurdo, J.; Nettleton, E.J.; Dobson, C.M.; Robinson, C.V. Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci. 2000, 9, 1960–1967. [Google Scholar] [CrossRef]
- Cerf, E.; Sarroukh, R.; Tamamizu-Kato, S.; Breydo, L.; Derclayes, S.; Dufrênes, Y.F.; Narayanaswami, V.; Goormaghtigh, E.; Ruysschaert, J.M.; Raussens, V. Antiparallel β-sheet: A signature structure of the oligomeric amyloid β-peptide. Biochem. J. 2009, 421, 415–423. [Google Scholar] [CrossRef]
- Eckert, A.; Hauptmann, S.; Scherping, I.; Meinhardt, J.; Rhein, V.; Dröse, S.; Brandt, U.; Fändrich, M.; Müller, W.E.; Götz, J. Oligomeric and fibrillar species of β-amyloid (Aβ42) both impair mitochondrial function in P301L tau transgenic mice. J. Mol. Med. 2008, 86, 1255–1267. [Google Scholar] [CrossRef]
- Sarroukh, R.; Cerf, E.; Derclaye, S.; Dufrêne, Y.F.; Goormaghtigh, E.; Ruysschaert, J.M.; Raussens, V. Transformation of amyloid β(1-40) oligomers into fibrils is characterized by a major change in secondary structure. Cell. Mol. Life Sci. 2011, 68, 1429–1438. [Google Scholar] [CrossRef]
- Itkin, A.; Dupres, V.; Dufrêne, Y.F.; Bechinger, B.; Ruysschaert, J.M.; Raussens, V. Calcium ions promote formation of amyloid β-peptide (1-40) oligomers causally implicated in neuronal toxicity of Alzheimer’s disease. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Habicht, G.; Haupt, C.; Friedrich, R.P.; Hortschansky, P.; Sachse, C.; Meinhardt, J.; Wieligmann, K.; Gellermann, G.P.; Brodhun, M.; Götz, J.; et al. Directed selection of a conformational antibody domain that prevents mature amyloid fibril formation by stabilizing Aβ protofibrils. Proc. Natl. Acad. Sci. USA 2007, 104, 19232–19237. [Google Scholar] [CrossRef]
- Lashuel, H.A.; Hartley, D.; Petre, B.M.; Walz, T.; Lansbury, P.T. Neurodegenerative disease: Amyloid pores from pathogenic mutations. Nature 2002, 418. [Google Scholar] [CrossRef]
- Habchi, J.; Arosio, P.; Perni, M.; Costa, A.R.; Yagi-Utsumi, M.; Joshi, P.; Chia, S.; Cohen, S.I.A.; Müller, M.B.D.; Linse, S.; et al. Neuroscience: An anticancer drug suppresses the primary nucleation reaction that initiates the production of the toxic Ab42 aggregates linked with Alzheimer’s disease. Sci. Adv. 2016, 2. [Google Scholar] [CrossRef]
- Hong, D.P.; Fink, A.L.; Uversky, V.N. Structural Characteristics of α-Synuclein Oligomers Stabilized by the Flavonoid Baicalein. J. Mol. Biol. 2008, 383, 214–223. [Google Scholar] [CrossRef]
- Uversky, V.N.; Li, J.; Fink, A.L. Evidence for a Partially Folded Intermediate in α-Synuclein Fibril Formation. J. Biol. Chem. 2001, 276, 10737–10744. [Google Scholar] [CrossRef]
- Roeters, S.J.; Iyer, A.; Pletikapiä, G.; Kogan, V.; Subramaniam, V.; Woutersen, S. Evidence for Intramolecular Antiparallel Beta-Sheet Structure in Alpha-Synuclein Fibrils from a Combination of Two-Dimensional Infrared Spectroscopy and Atomic Force Microscopy. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Middleton, C.T.; Marek, P.; Cao, P.; Chiu, C.C.; Singh, S.; Woys, A.M.; De Pablo, J.J.; Raleigh, D.P.; Zanni, M.T. Two-dimensional infrared spectroscopy reveals the complex behaviour of an amyloid fibril inhibitor. Nat. Chem. 2012, 4, 355–360. [Google Scholar] [CrossRef]
- Natalello, A.; Prokorov, V.V.; Tagliavini, F.; Morbin, M.; Forloni, G.; Beeg, M.; Manzoni, C.; Colombo, L.; Gobbi, M.; Salmona, M.; et al. Conformational Plasticity of the Gerstmann-Sträussler-Scheinker Disease Peptide as Indicated by Its Multiple Aggregation Pathways. J. Mol. Biol. 2008, 381, 1349–1361. [Google Scholar] [CrossRef]
- Cordeiro, Y.; Kraineva, J.; Suarez, M.C.; Tempesta, A.G.; Kelly, J.W.; Silva, J.L.; Winter, R.; Foguel, D. Fourier transform infrared spectroscopy provides a fingerprint for the tetramer and for the aggregates of transthyretin. Biophys. J. 2006, 91, 957–967. [Google Scholar] [CrossRef]
- Zandomeneghi, G.; Krebs, M.R.H.; McCammon, M.G.; Fändrich, M. FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Sci. 2009, 13, 3314–3321. [Google Scholar] [CrossRef]
- Fabian, H.; Gast, K.; Laue, M.; Misselwitz, R.; Uchanska-Ziegler, B.; Ziegler, A.; Naumann, D. Early stages of misfolding and association of β2- microglobulin: Insights from infrared spectroscopy and dynamic light scattering. Biochemistry 2008, 47, 6895–6906. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, K.; Ta, H.P.; Géan, J.; Lecomte, S.; Cullin, C. In vivo and in vitro analyses of toxic mutants of HET-s: FTIR antiparallel signature correlates with amyloid toxicity. J. Mol. Biol. 2011, 412, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Li, Y.; Hao, W.; Hu, X.; Ma, G. Parallel β-sheet fibril and antiparallel β-sheet oligomer: New insights into amyloid formation of hen egg white lysozyme under heat and acidic condition from FTIR spectroscopy. J. Phys. Chem. B 2013, 117, 4003–4013. [Google Scholar] [CrossRef] [PubMed]
- Zurdo, J.; Guijarro, J.I.; Dobson, C.M. Preparation and characterization of purified amyloid fibrils. J. Am. Chem. Soc. 2001, 123, 8141–8142. [Google Scholar] [CrossRef] [PubMed]
- Balbirnie, M.; Grothe, R.; Eisenberg, D.S. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated β-sheet structure for amyloid. Proc. Natl. Acad. Sci. USA 2001, 98, 2375–2380. [Google Scholar] [CrossRef]
- Shim, S.H.; Gupta, R.; Ling, Y.L.; Strasfeld, D.B.; Raleigh, D.P.; Zanni, M.T. Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. Proc. Natl. Acad. Sci. USA 2009, 106, 6614–6619. [Google Scholar] [CrossRef]
- Ami, D.; Lavatelli, F.; Rognoni, P.; Palladini, G.; Raimondi, S.; Giorgetti, S.; Monti, L.; Doglia, S.M.; Natalello, A.; Merlini, G. In situ characterization of protein aggregates in human tissues affected by light chain amyloidosis: A FTIR microspectroscopy study. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Amenabar, I.; Poly, S.; Nuansing, W.; Hubrich, E.H.; Govyadinov, A.A.; Huth, F.; Krutokhvostov, R.; Zhang, L.; Knez, M.; Heberle, J.; et al. Structural analysis and mapping of individual protein complexes by infrared nanospectroscopy. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Dazzi, A.; Prazeres, R.; Glotin, F.; Ortega, J.M. Local infrared microspectroscopy with subwavelength spatial resolution with an atomic force microscope tip used as a photothermal sensor. Opt. Lett. 2005, 30. [Google Scholar] [CrossRef]
- Unger, M.; Marcott, C. Recent Advances and Applications of Nanoscale Infrared Spectroscopy and Imaging. In Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 1–26. [Google Scholar]
- Dazzi, A.; Prazeres, R.; Glotin, F.; Ortega, J.M. Subwavelength infrared spectromicroscopy using an AFM as a local absorption sensor. Infrared Phys. Technol. 2006, 49, 113–121. [Google Scholar] [CrossRef]
- Henry, S.; Bercu, N.B.; Bobo, C.; Cullin, C.; Molinari, M.; Lecomte, S. Interaction of Aβ1-42 peptide or their variant with model membrane of different composition probed by infrared nanospectroscopy. Nanoscale 2018, 10, 936–940. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, F.S.; Longo, G.; Faggiano, S.; Lipiec, E.; Pastore, A.; Dietler, G. Infrared nanospectroscopy characterization of oligomeric and fibrillar aggregates during amyloid formation. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, F.S.; Vieweg, S.; Cendrowska, U.; Longo, G.; Chiki, A.; Lashuel, H.A.; Dietler, G. Nanoscale studies link amyloid maturity with polyglutamine diseases onset. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rizevsky, S.; Kurouski, D. Nanoscale Structural Organization of Insulin Fibril Polymorphs Revealed by Atomic Force Microscopy–Infrared Spectroscopy (AFM-IR). ChemBioChem 2020, 21, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Galante, D.; Ruggeri, F.S.; Dietler, G.; Pellistri, F.; Gatta, E.; Corsaro, A.; Florio, T.; Perico, A.; D’Arrigo, C. A critical concentration of N-terminal pyroglutamylated amyloid beta drives the misfolding of Ab1-42 into more toxic aggregates. Int. J. Biochem. Cell Biol. 2016, 79, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Kumosinski, T.F.; Unruh, J.J. Quantitation of the global secondary structure of globular proteins by FTIR spectroscopy: Comparison with X-ray crystallographic structure. Talanta 1996, 43, 199–219. [Google Scholar] [CrossRef]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. (Shanghai) 2007, 39, 549–559. [Google Scholar] [CrossRef]
- Ramer, G.; Ruggeri, F.S.; Levin, A.; Knowles, T.P.J.; Centrone, A. Determination of Polypeptide Conformation with Nanoscale Resolution in Water. ACS Nano 2018, 12, 6612–6619. [Google Scholar] [CrossRef]
- Creasey, R.C.G.; Louzao, I.; Arnon, Z.A.; Marco, P.; Adler-Abramovich, L.; Roberts, C.J.; Gazit, E.; Tendler, S.J.B. Disruption of diphenylalanine assembly by a Boc-modified variant. Soft Matter 2016, 12, 9451–9457. [Google Scholar] [CrossRef]
- Sivanandam, V.N.; Jayaraman, M.; Hoop, C.L.; Kodali, R.; Wetzel, R.; Van Der Wel, P.C.A. The aggregation-enhancing huntingtin N-terminus is helical in amyloid fibrils. J. Am. Chem. Soc. 2011, 133, 4558–4566. [Google Scholar] [CrossRef]
- Haris, P.I.; Severcan, F. FTIR spectroscopic characterization of protein structure in aqueous and non-aqueous media. Proc. J. Mol. Catal. B Enzym. 1999, 7, 207–221. [Google Scholar] [CrossRef]
- Vignaud, H.; Bobo, C.; Lascu, I.; Sörgjerd, K.M.; Zako, T.; Maeda, M.; Salin, B.; Lecomte, S.; Cullin, C. A structure-toxicity study of Aß42 reveals a new anti-parallel aggregation pathway. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- McCreery, R.L. Raman Spectroscopy for Chemical Analysis. Meas. Sci. Technol. 2001, 12, 653–654. [Google Scholar] [CrossRef]
- Long, D.A. The Raman Effect: A Unified Treatment of the Theory of Raman Scattering by Molecules; John Wiley & Sons, Ltd.: Chichester, UK, 2002; Volume 8. [Google Scholar]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Choo-Smith, L.P.; Edwards, H.G.M.; Endtz, H.P.; Kros, J.M.; Heule, F.; Barr, H.; Robinson, J.S.; Bruining, H.A.; Puppels, G.J. Medical applications of Raman spectroscopy: From proof of principle to clinical implementation. Biopolym. Biospectrosc. Sect. 2002, 67, 1–9. [Google Scholar] [CrossRef]
- Nabiev, I.; Chourpa, I.; Manfait, M. Applications of Raman and surface-enhanced Raman scattering spectroscopy in medicine. J. Raman Spectrosc. 1994, 25, 13–23. [Google Scholar] [CrossRef]
- Shashilov, V.A.; Sikirzhytski, V.; Popova, L.A.; Lednev, I.K. Quantitative methods for structural characterization of proteins based on deep UV resonance Raman spectroscopy. Methods 2010, 52, 23–37. [Google Scholar] [CrossRef]
- Kurouski, D.; Van Duyne, R.P.; Lednev, I.K. Exploring the structure and formation mechanism of amyloid fibrils by Raman spectroscopy: A review. Analyst 2015, 140, 4967–4980. [Google Scholar] [CrossRef]
- Kurouski, D.; Lednev, I.K. The impact of protein disulfide bonds on the amyloid fibril morphology. Int. J. Biomed. Nanosci. Nanotechnol. 2011, 2, 167–176. [Google Scholar] [CrossRef]
- Rosario-Alomar, M.F.; Quiñones-Ruiz, T.; Kurouski, D.; Sereda, V.; Ferreira, E.B.; De Jesús-Kim, L.; Hernández-Rivera, S.; Zagorevski, D.V.; López-Garriga, J.; Lednev, I.K. Hydrogen sulfide inhibits amyloid formation. J. Phys. Chem. B 2015, 119, 1265–1274. [Google Scholar] [CrossRef]
- Kurouski, D.; Washington, J.; Ozbil, M.; Prabhakar, R.; Shekhtman, A.; Lednev, I.K. Disulfide bridges remain intact while native insulin converts into amyloid fibrils. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Ermolenkov, V.V.; He, W.; Uversky, V.N.; Fredriksen, L.; Lednev, I.K. Lysozyme fibrillation: Deep UV Raman spectroscopic characterization of protein structural transformation. Biopolymers 2005, 79, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Ermolenkov, V.V.; Uversky, V.N.; Lednev, I.K. Hen egg white lysozyme fibrillation: A deep-UV resonance Raman spectroscopic study. J. Biophotonics 2008, 1, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Stiles, P.L.; Dieringer, J.A.; Shah, N.C.; Van Duyne, R.P. Surface-Enhanced Raman Spectroscopy SERS: Surface-enhanced Raman spectroscopy Raman scattering: Inelastic scattering of a photon from a molecule in which the frequency change precisely matches the difference in vibrational energy levels. Annu. Rev. Anal. Chem. 2008, 1, 601–626. [Google Scholar] [CrossRef]
- Willets, K.A.; Van Duyne, R.P. Localized Surface Plasmon Resonance Spectroscopy and Sensing. Annu. Rev. Phys. Chem. 2007, 58, 267–297. [Google Scholar] [CrossRef]
- Kneipp, K.; Kneipp, H.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Ultrasensitive Chemical Analysis by Raman Spectroscopy. Chem. Rev. 1999, 99, 2957–2975. [Google Scholar] [CrossRef]
- Xie, W.; Schlücker, S. Medical applications of surface-enhanced Raman scattering. Phys. Chem. Chem. Phys. 2013, 15, 5329–5344. [Google Scholar] [CrossRef]
- Cialla, D.; März, A.; Böhme, R.; Theil, F.; Weber, K.; Schmitt, M.; Popp, J. Surface-enhanced Raman spectroscopy (SERS): Progress and trends. Anal. Bioanal. Chem. 2012, 403, 27–54. [Google Scholar] [CrossRef]
- Han, X.X.; Zhao, B.; Ozaki, Y. Surface-enhanced Raman scattering for protein detection. Anal. Bioanal. Chem. 2009, 394, 1719–1727. [Google Scholar] [CrossRef]
- Beier, H.T.; Cowan, C.B.; Chou, I.H.; Pallikal, J.; Henry, J.E.; Benford, M.E.; Jackson, J.B.; Good, T.A.; Coté, G.L. Application of surface-enhanced raman spectroscopy for detection of beta amyloid using nanoshells. Plasmonics 2007, 2, 55–64. [Google Scholar] [CrossRef]
- Choi, I.; Huh, Y.S.; Erickson, D. Ultra-sensitive, label-free probing of the conformational characteristics of amyloid beta aggregates with a SERS active nanofluidic device. Microfluid. Nanofluid. 2012, 12, 663–669. [Google Scholar] [CrossRef]
- Chou, I.H.; Benford, M.; Beier, H.T.; Coté, G.L.; Wang, M.; Jing, N.; Kameoka, J.; Good, T.A. Nanofluidic biosensing for β-amyloid detection using surface enhanced raman spectroscopy. Nano Lett. 2008, 8, 1729–1735. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, D.; Mote, K.R.; MacLaughlin, C.M.; Biswas, N.; Chandra, B.; Basu, J.K.; Walker, G.C.; Madhu, P.K.; Maiti, S. Cell-Membrane-Mimicking Lipid-Coated Nanoparticles Confer Raman Enhancement to Membrane Proteins and Reveal Membrane-Attached Amyloid-β Conformation. ACS Nano 2015, 9, 9070–9077. [Google Scholar] [CrossRef] [PubMed]
- Kurouski, D.; Sorci, M.; Postiglione, T.; Belfort, G.; Lednev, I.K. Detection and structural characterization of insulin prefibrilar oligomers using surface enhanced Raman spectroscopy. Biotechnol. Prog. 2014, 30, 488–495. [Google Scholar] [CrossRef]
- Li, Z.; Wang, H.; Chen, Z. Monitoring and modulation of insulin fibers by a protein isomerization targeting dye bromophenol blue. Sens. Actuators B Chem. 2019, 287, 496–502. [Google Scholar] [CrossRef]
- Karaballi, R.A.; Merchant, S.; Power, S.R.; Brosseau, C.L. Electrochemical surface-enhanced Raman spectroscopy (EC-SERS) study of the interaction between protein aggregates and biomimetic membranes. Phys. Chem. Chem. Phys. 2018, 20, 4513–4526. [Google Scholar] [CrossRef]
- Yu, X.; Hayden, E.Y.; Xia, M.; Liang, O.; Cheah, L.; Teplow, D.B.; Xie, Y.H. Surface enhanced Raman spectroscopy distinguishes amyloid Β-protein isoforms and conformational states. Protein Sci. 2018, 27, 1427–1438. [Google Scholar] [CrossRef]
- Stöckle, R.M.; Suh, Y.D.; Deckert, V.; Zenobi, R. Nanoscale chemical analysis by tip-enhanced Raman spectroscopy. Chem. Phys. Lett. 2000, 318, 131–136. [Google Scholar] [CrossRef]
- Lipiec, E.W.; Wood, B.R. Tip-Enhanced Raman Scattering: Principles, Instrumentation, and the Application toe Biological Systems. In Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 1–26. [Google Scholar]
- Novotny, L.; Van Hulst, N. Antennas for light. Nat. Photonics 2011, 5, 83–90. [Google Scholar] [CrossRef]
- Chen, C.; Hayazawa, N.; Kawata, S. A 1.7 nm resolution chemical analysis of carbon nanotubes by tip-enhanced Raman imaging in the ambient. Nat. Commun. 2014, 5, 1–5. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, Y.; Dong, Z.C.; Jiang, S.; Zhang, C.; Chen, L.G.; Zhang, L.; Liao, Y.; Aizpurua, J.; Luo, Y.; et al. Chemical mapping of a single molecule by plasmon-enhanced Raman scattering. Nature 2013, 498, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Jiang, S.; Hu, C.; Zhang, R.; Kuang, Y.; Zhu, J.; Zhang, Y.; Dong, Z. Tip-enhanced raman spectroscopic imaging of individual carbon nanotubes with subnanometer resolution. Nano Lett. 2016, 16, 4040–4046. [Google Scholar] [CrossRef] [PubMed]
- Domke, K.F.; Zhang, D.; Pettinger, B. Enhanced Raman spectroscopy: Single molecules or carbon? J. Phys. Chem. C 2007, 111, 8611–8616. [Google Scholar] [CrossRef]
- Blum, C.; Schmid, T.; Opilik, L.; Metanis, N.; Weidmann, S.; Zenobi, R. Missing amide i mode in gap-mode tip-enhanced raman spectra of proteins. J. Phys. Chem. C 2012, 116, 23061–23066. [Google Scholar] [CrossRef]
- Kurouski, D.; Postiglione, T.; Deckert-Gaudig, T.; Deckert, V.; Lednev, I.K. Amide i vibrational mode suppression in surface (SERS) and tip (TERS) enhanced Raman spectra of protein specimens. Analyst 2013, 138, 1665–1673. [Google Scholar] [CrossRef]
- Szczerbiński, J.; Metternich, J.B.; Goubert, G.; Zenobi, R. How Peptides Dissociate in Plasmonic Hot Spots. Small 2020, 16. [Google Scholar] [CrossRef]
- Lipiec, E.; Perez-Guaita, D.; Kaderli, J.; Wood, B.R.; Zenobi, R. Direct Nanospectroscopic Verification of the Amyloid Aggregation Pathway. Angew. Chem. 2018, 130, 8655–8660. [Google Scholar] [CrossRef]
- Hermann, P.; Fabian, H.; Naumann, D.; Hermelink, A. Comparative study of far-field and near-field raman spectra from silicon-based samples and biological nanostructures. J. Phys. Chem. C 2011, 115, 24512–24520. [Google Scholar] [CrossRef]
- Kurouski, D.; Deckert-Gaudig, T.; Deckert, V.; Lednev, I.K. Structure and composition of insulin fibril surfaces probed by TERS. J. Am. Chem. Soc. 2012, 134, 13323–13329. [Google Scholar] [CrossRef]
- Deckert-Gaudig, T.; Deckert, V. High resolution spectroscopy reveals fibrillation inhibition pathways of insulin. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Deckert-Gaudig, T.; Kurouski, D.; Hedegaard, M.A.B.; Singh, P.; Lednev, I.K.; Deckert, V. Spatially resolved spectroscopic differentiation of hydrophilic and hydrophobic domains on individual insulin amyloid fibrils. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- vandenAkker, C.C.; Deckert-Gaudig, T.; Schleeger, M.; Velikov, K.P.; Deckert, V.; Bonn, M.; Koenderink, G.H. Nanoscale Heterogeneity of the Molecular Structure of Individual hIAPP Amyloid Fibrils Revealed with Tip-Enhanced Raman Spectroscopy. Small 2015, 11, 4131–4139. [Google Scholar] [CrossRef] [PubMed]
- Paulite, M.; Blum, C.; Schmid, T.; Opilik, L.; Eyer, K.; Walker, G.C.; Zenobi, R. Full spectroscopic tip-enhanced raman imaging of single nanotapes formed from β-Amyloid(1-40) peptide fragments. ACS Nano 2013, 7, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Pearson, K. LIII. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Hotelling, H. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 1933, 24, 417–441. [Google Scholar] [CrossRef]
- Lastovicka, J.L.; Jackson, J.E. A User’s Guide to Principal Components. J. Mark. Res. 1992, 29. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis, Second Edition. Encycl. Stat. Behav. Sci. 2002, 30. [Google Scholar] [CrossRef]
- Halkidi, M.; Batistakis, Y.; Vazirgiannis, M. On clustering validation techniques. J. Intell. Inf. Syst. 2001, 17, 107–145. [Google Scholar] [CrossRef]


| Amide Bands | Wavenumber [cm−1] | Assigned Vibration 1 |
|---|---|---|
| A | 3500 | ν(N-H) |
| B | 3100 | ν(N-H) |
| I | 1700–1600 | 80% ν(C=O), 10% ν(C-N), 10% δ(N-H) |
| II | 1580–1480 | 40% ν(C-N), 60% δ(N-H) |
| III | 1300–1230 | 30% ν(C-N), 30% δ(N-H), 10% ν(CH3-C), 10% δ(O=C-N), 20% other |
| IV | 770–625 | 40% δ(O=C-N), 60% other |
| V | 800–640 | γ(N-H) |
| VI | 600–540 | γ(C=O) |
| VII | 200 | skeletal mode |
| Marker Band of the Aggregation [cm−1] | Assignment | Peptide | Reference | |
|---|---|---|---|---|
| amide I (1700-1600 cm−1) | ||||
| 1695-1685/1633-1623 | antiparallel β-sheet | amyloid β1-42 | oligomers | [40,41] |
| amyloid β1-40 | [42,43,44] | |||
| α-synuclein | [28,47,48] | |||
| PrP82–146 | [51] | |||
| 1693-1685/1623–1613 | antiparallel β-sheet | amyloid β11-28 fragment and its mutants in 21-23 position | [31] | |
| 1692/1620 | antiparallel β-sheet | HET218–289 | [55] | |
| 1691/1630 | antiparallel β-sheet | Aβ 42CC oligomers/protofibrils | [32] | |
| 1688/1620 | antiparallel β-sheet | human lysozyme, oligomers | [29] | |
| 1686/1616 | antiparallel β-sheet | transthyretin (TTR) soluble aggregates | [52] | |
| 1686/1614 | antiparallel β-sheet | hen egg white lysozyme (HEWL) | [56] | |
| 1684/1616 | β2-microglobulin (short curved structures) | [54] | ||
| 1684/1612 | antiparallel β-sheet | SH3 domain, amorphous aggregates, non-fibrilar | [57] | |
| 1683/1612 ↑→ | antiparallel β-sheet | insulin | oligomer | [39] |
| 1678-1670 | β-turns | amyloid β11-28 fragment and its mutants in 21-23 position | [31] | |
| 1670 | β-turns | amyloid β1-42 | oligomers and fibrils | [40] |
| α-synuclein | [28] | |||
| 1669 | β-turns | HET218–289 | [55] | |
| 1667-1661 | 310-helix | E22K and A21G mutants of Aβ(11-28) fragment | [31] | |
| 1664 | β-turns | SH3 fibrils/pepsin digested | [57] | |
| 1660-1650 | random coil and/or helical structures | amyloid β1-42 | oligomers and fibrils | [40] |
| α-synuclein | [28] | |||
| 1659-1652 | α-helix | amyloid β11-28 fragment and its mutants in 21-23 position | [31] | |
| 1658 | Turns | human lysozyme | monomers, oligomers, fibrils | [29] |
| 1655 | random coil | HET218–289 | [55] | |
| 1649 | unstructured | SH3 amorphous aggregates | [57] | |
| 1648-1639 | random coil | amyloid β11-28 fragment and its mutants in 21-23 position | [31] | |
| 1648 | random coil | PrP82–146 | [51] | |
| 1644-1641 | disordered/loops | human lysozyme | oligomers, fibrils | [29] |
| 1641 | disordered structures | SH3 fibrils/pepsin digested | [57] | |
| 1635-1624 | β-sheet | amyloid β11-28 fragment and its mutants in 21-23 position | [31] | |
| 1633 | parallel β-sheet | Sup35 crystals, prion-like | [58] | |
| 1630 | parallel β-sheets | HET218–289 | [55] | |
| 1630-1623 | parallel β-sheet | amyloid β1-42 | fibrils | [40,41] |
| amyloid β1-40 | [42,43,44] | |||
| 1630-1614 | parallel β-sheet | human lysozyme | fibrils | [29] |
| 1628 | parallel β-sheet | α-synuclein | fibrils | [28] |
| 1626 ↑ | parallel β-sheet | PrP82–146 | fibrils | [51] |
| 1626 ↑→ | parallel β-sheet | insulin | fibrils | [39] |
| 1625 | parallel β-sheet | transthyretin (TTR) | fibrils | [52] |
| 1620-1618 | β2-microglobulin, fibrils | [54] | ||
| 1620-1600 | β-sheets | hen egg white lysozyme (HEWL) | [56] | |
| 1618 ← | parallel β-sheet | SH3 fibrils/pepsin digested | [57] | |
| Marker Band of the Aggregation [cm−1] | Assignment | Peptide | Reference |
|---|---|---|---|
| amide I (1730–1600 cm−1) | |||
| 1730 | C=O | Boc-FF, FF | [72,73] |
| 1700–1690 | anti-parallel β-sheet | amyloid β (AβpE3-42) | [69] |
| 1700–1600 | ν(C=O) bk | unexpanded Exon1 (22Q) | [26,34,35,67,74] |
| 1696–1690 | anti-parallel β-sheet, carbamate group | Boc-FF, FF | [72,73] |
| 1695–1665 | β-turn and antiparallel β-sheets | ataxin-3 | [38,66,70,71,75] |
| 1695,1684 | β-turn, anti-parallel β-sheet | amyloid β (AβpE3-42) | [69] |
| 1695 | β-turn | insulin | [68] |
| 1692 | anti-parallel β-sheet | amyloid β with 5% pyroglutamylated peptide | [69] |
| 1692 | anti-parallel β-sheet | unexpanded Exon1 (22Q) | [26,34,35,67,74] |
| 1689,1625 | anti-parallel β-sheet | mutant oligomer G37C | [65,76] |
| 1684 | glutamine side chain vibrations and β-turn | expanded Exon1 (42Q) | [26,34,35,67,74] |
| 1664,1655 | α-helix, 3-helix | Boc-FF, FF | [72,73] |
| 1662 | β-turn | amyloid β | [65,76] |
| 1660–1650 | α- helix | ataxin-3 | [38,66,70,71,75] |
| 1660 | α-helix/unordered protein secondary structures | Insulin | [68] |
| 1658 | poor α-helix | amyloid β (AβpE3-42) | [69] |
| 1658 | α-helix | expanded Exon1 (42Q) | [26,34,35,67,74] |
| 1645–1630 | random coil | ataxin-3 | [38,66,70,71,75] |
| 1645–1635 | random coil | unexpanded Exon1 (22Q) | [26,34,35,67,74] |
| 1640–1600 | residual water absorption, NH3+ group | Boc-FF, FF | [72,73] |
| 1638 | random coil | amyloid β (AβpE3-42), amyloid β with 5% pyroglutamylated peptide | [69] |
| 1635–1610 | low density native/high density amyloid β-sheets | ataxin-3 | [38,66,70,71,75] |
| 1635 | β-sheet | unexpanded Exon1 (22Q), expanded Exon1 (42Q) | [26,34,35,67,74] |
| 1631 | parallel β-sheet secondary structure | amyloid β | [65,76] |
| 1623 | high β-sheet | amyloid β (AβpE3-42) | [69] |
| 1620 | β-sheet | Insulin | [68] |
| amide II (1600–1500 cm−1) | |||
| 1580–1510 | bk δ(N-H), ν(C-N) | ataxin-3 | [38,66,70,71,75] |
| 1555,1520 | NH vibrations | Boc-FF, FF | [72,73] |
| C-C ring vibrations | |||
| 1605,1495,1452,1430 | C-C ring vibrations | FF | [72,73] |
| amide III (1400–1200 cm−1) | |||
| 1350–1200 | ν(C-N), δ(N-H),ν(C-C), δ(C=O) | ataxin-3 | [38,66,70,71,75] |
| Marker Band of the Aggregation 1 | Assignment | Peptide | Reference |
|---|---|---|---|
| amide I (1700–1600 cm−1) | |||
| 1690–1600 ↑1672→ | β-sheets formation | insulin, hen egg white lysozyme (HEWL) | [85,86,88] |
| amide II (1580–1480 cm−1) | |||
| 1580–1480 ↑ | β-sheets formation | HEWL | [84,85,86] |
| 1550→ | 1-SS-carboxymethyl lactalbulin (1-SS-LA), HEWL | ||
| Cα H | |||
| 1390 ↑ | insulin, HEWL | [85,86,88] | |
| amide III (1283–1218 cm−1) | |||
| 1320–1270 ↑1270–1230 ↓ | HEWL | [88] | |
| disulfide (S-S) (550–450 cm−1) | |||
| 523, 507 ↓490 | HEWL | [84,85] | |
| 540, 510 ↓508 | apo-α-lactabulin (LA) | ||
| phenylalanine (Phe) | |||
| 1000 ↓ | HEWL | [87,88] | |
| Marker Band of the Aggregation | Assignment | Peptide | Reference |
|---|---|---|---|
| amide I (1700–1600 cm−1) | |||
| 1678–1664↑ | β-sheets | insulin | [99] |
| 1664–1640↓ | α-helics, unordered | ||
| amide III (1283−1218 cm−1) | |||
| 1244 ↑ | β-sheets | amyloid β1–42 | [95,96] |
| 1266 ↓ | α-helics | ||
| CCN stretching | |||
| 1144 ↓ | amyloid β1–42 | [96] | |
| C-C stretching | |||
| 960 ↑ | amyloid β1–42 | [95] | |
| Marker Band of the Aggregation [cm−1] | Assignment | Peptide | Reference |
|---|---|---|---|
| amide I (1700–1600 cm−1) | |||
| 1680–1660 ↑ | β-sheets | amyloid β1-42, insulin, | [111,113,115,116,117,118] |
| β2-microglobulin | |||
| 1674 ↑ | β-sheets | hIAPP (Amylin) | [112] |
| 1655–1630↓ | random coil | amyloid β1-42 | [117] |
| 1640↓,1664–1640 ↓ | α-helics, unordered | hIAPP (amylin), insulin | [112,113,115,118] |
| Cα H/N-H (1370–1360 cm−1) | |||
| 1364 ↑ | β-sheets | amyloid β1-42 | [116] |
| amide III (1283–1218 cm−1) | |||
| 1228–1218 ↑ | parallel β-sheets | amyloid β1-42 | [117] |
| 1242–1233, 1250↑ | antiparallel β-sheets | insulin | [116,117] |
| 1235–1230↑ | β-sheets | amyloid β1-42, | [111,115] |
| 1261–1248, 1258 ↓ | random coil | β2-microglobulin | [116,117] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilkosz, N.; Czaja, M.; Seweryn, S.; Skirlińska-Nosek, K.; Szymonski, M.; Lipiec, E.; Sofińska, K. Molecular Spectroscopic Markers of Abnormal Protein Aggregation. Molecules 2020, 25, 2498. https://doi.org/10.3390/molecules25112498
Wilkosz N, Czaja M, Seweryn S, Skirlińska-Nosek K, Szymonski M, Lipiec E, Sofińska K. Molecular Spectroscopic Markers of Abnormal Protein Aggregation. Molecules. 2020; 25(11):2498. https://doi.org/10.3390/molecules25112498
Chicago/Turabian StyleWilkosz, Natalia, Michał Czaja, Sara Seweryn, Katarzyna Skirlińska-Nosek, Marek Szymonski, Ewelina Lipiec, and Kamila Sofińska. 2020. "Molecular Spectroscopic Markers of Abnormal Protein Aggregation" Molecules 25, no. 11: 2498. https://doi.org/10.3390/molecules25112498
APA StyleWilkosz, N., Czaja, M., Seweryn, S., Skirlińska-Nosek, K., Szymonski, M., Lipiec, E., & Sofińska, K. (2020). Molecular Spectroscopic Markers of Abnormal Protein Aggregation. Molecules, 25(11), 2498. https://doi.org/10.3390/molecules25112498






