Design, Synthesis, and Biological Evaluation of 8-Mercapto-3,7-Dihydro-1H-Purine-2,6-Diones as Potent Inhibitors of SIRT1, SIRT2, SIRT3, and SIRT5
Abstract
:1. Introduction
2. Results and Discussion
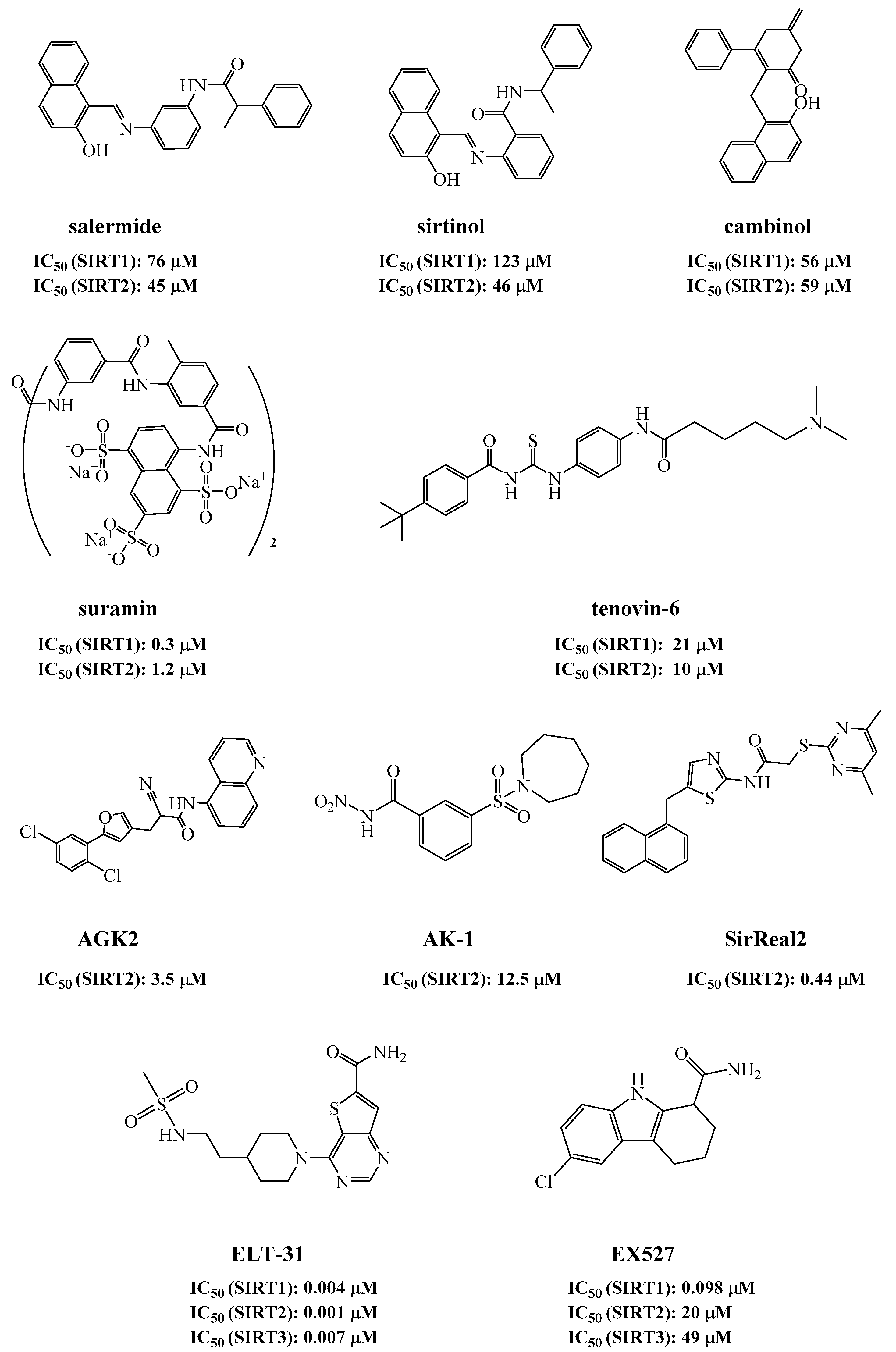
2.1. Discovery of Sirtuin Inhibitors
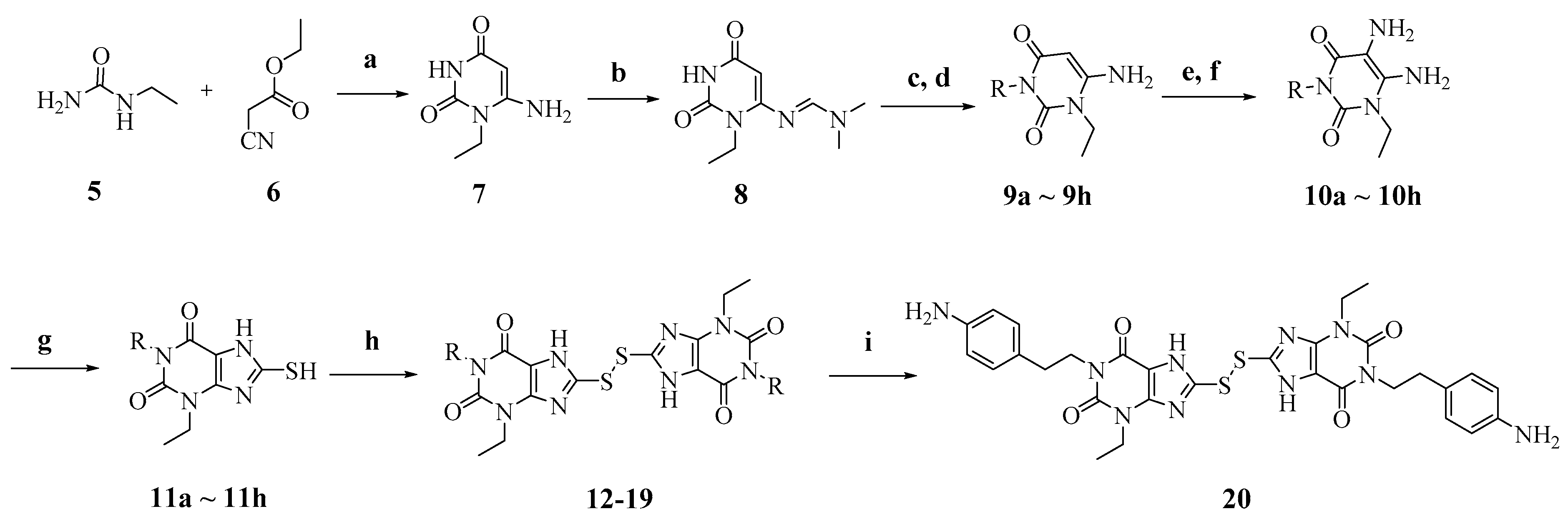
2.2. Chemistry
2.3. Structure–Activity Relationship
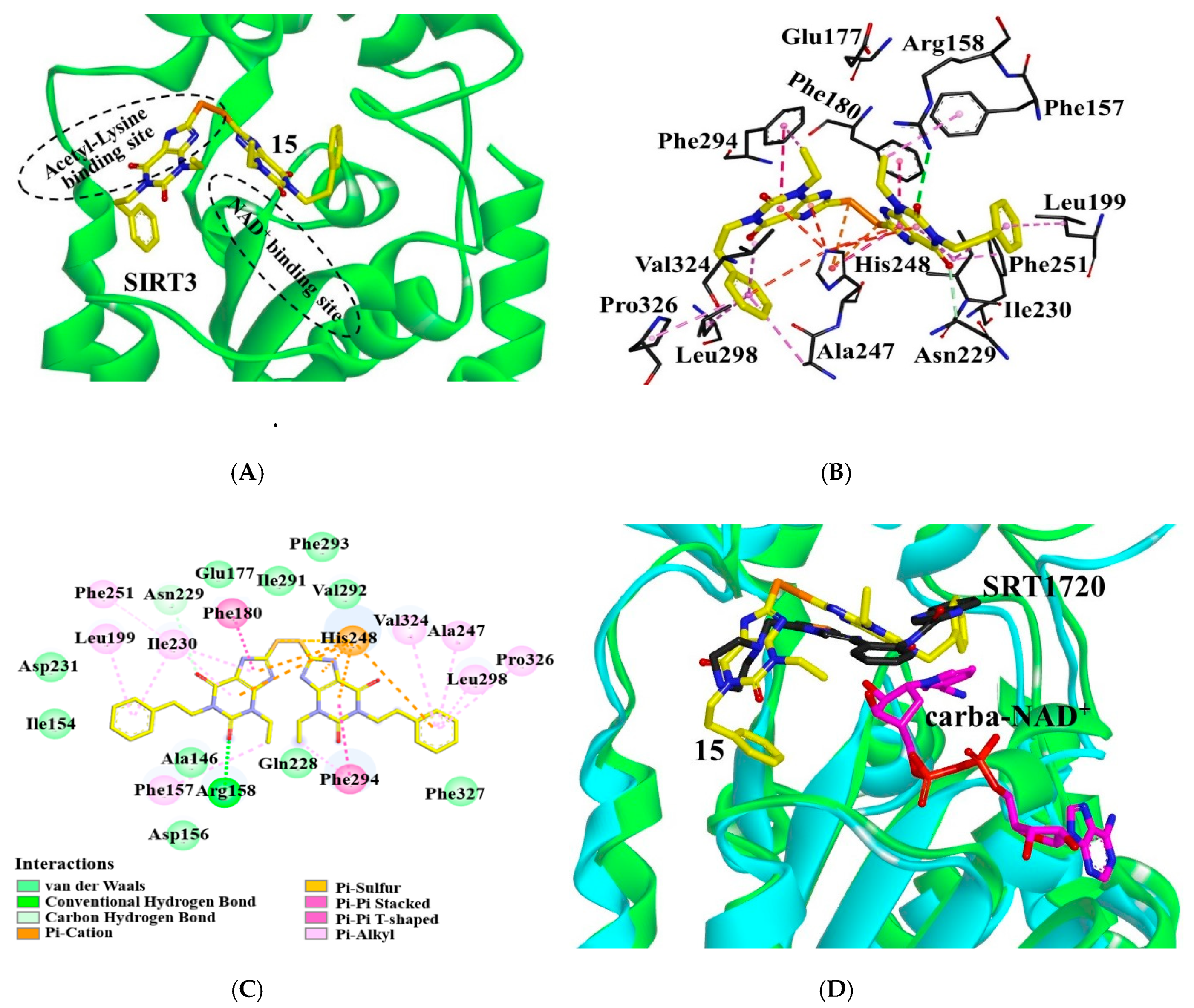
2.4. Binding Mode of the Inhibitor in SIRT3
2.5. Site-Directed Mutagenesis
2.6. Mechanism of Inhibition
2.7. Compounds Are Potent Inhibitors of SIRT1/2/3/5.
3. Conclusions
4. Experimental Section
4.1. Chemicals and General Methods
4.2. Synthesis of Compounds
4.3. Protein Expression and Purification
4.4. Molecular Modeling
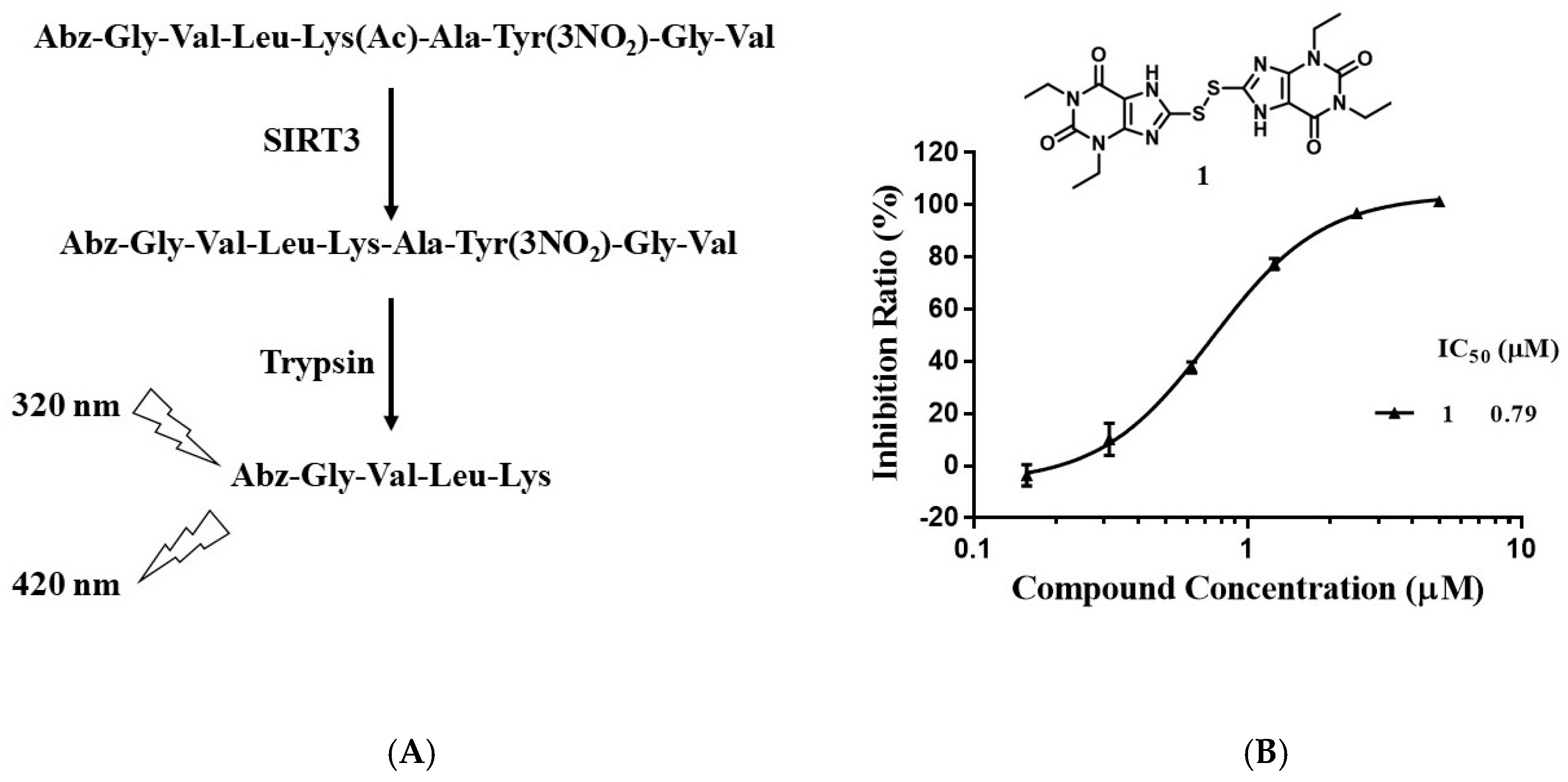
4.5. Screening for Sirtuin Inhibitor
4.6. Determination of the Inhibition Pattern of 15
4.7. Microscale Thermophoresis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NAD+ | nicotinamide adenine dinucleotide |
| ADP | adenosine diphosphate |
| DMSO | dimethyl sulfoxide |
| DCM | dichloromethane |
| DMF | N,N-dimethylformamide |
| DMA | dimethyl acetamide |
| DMF-DMA | N,N-Dimethylformamide dimethyl acetal |
| EA | ethyl acetate |
| TFA | trifluoroacetic acid |
| TEV | tobacco etch virus |
| PMSF | phenylmethanesulfonyl fluoride |
| sumo | small ubiquitin-related modifier |
| RMSD | root-mean-square deviation |
References
- Sauve, A.A. Sirtuin chemical mechanisms. Biochim. Biophys. Acta (BBA) Bioenerg. 2010, 1804, 1591–1603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, S.; Islam, R. Mammalian Sirt1: Insights on its biological functions. Cell Commun. Signal. 2011, 9, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Zhang, L.; Zhang, W.; Meng, D.; Zhang, H.; Jiang, Y.; Xu, X.; Van Meter, M.; Seluanov, A.; Gorbunova, V.; et al. SIRT6 rescues the age related decline in base excision repair in a PARP1-dependent manner. Cell Cycle 2015, 14, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Ford, E.; Voit, R.; Liszt, G.; Magin, C.; Grummt, I.; Guarente, L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genome Res. 2006, 20, 1075–1080. [Google Scholar] [CrossRef] [Green Version]
- North, B.J.; Marshall, B.L.; Borra, M.T.; Denu, J.M.; Verdin, E. The human Sir2 Ortholog, SIRT2, Is an NAD+-dependent tubulin deacetylase. Mol. Cell 2003, 11, 437–444. [Google Scholar] [CrossRef]
- Giralt, A.; Villarroya, F. SIRT3, a pivotal actor in mitochondrial functions: Metabolism, cell death and aging. Biochem. J. 2012, 444, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haigis, M.C.; Mostoslavsky, R.; Haigis, K.M.; Fahie, K.; Christodoulou, D.C.; Murphy, A.J.; Valenzuela, D.M.; Yancopoulos, G.D.; Karow, M.; Blander, G.; et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell 2006, 126, 941–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gertz, M.; Steegborn, C. Function and regulation of the mitochondrial Sirtuin isoform Sirt5 in Mammalia. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2010, 1804, 1658–1665. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cao, N.; Fenech, M.; Wang, X. Role of sirtuins in maintenance of genomic stability: Relevance to cancer and healthy aging. DNA Cell Biol. 2016, 35, 542–575. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Ma, T.; Zhu, Q.; Xu, Y.; Zha, X. Recent advances in inhibitors of sirtuin1/2: An update and perspective. Futur. Med. Chem. 2018, 10, 907–934. [Google Scholar] [CrossRef] [PubMed]
- Lara, E.; Mai, A.; Calvanese, V.; Altucci, L.; López-Nieva, P.; Martínez-Chantar, M.L.; Varela-Rey, M.; Rotili, D.; Nebbioso, A.; Ropero, S.; et al. Salermide, a Sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene 2008, 28, 781–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawson, M.; Uciechowska, U.; Schemies, J.; Rumpf, T.; Jung, M.; Sippl, W. Inhibitors to understand molecular mechanisms of NAD+-dependent deacetylases (sirtuins). Biochim. Biophys. Acta (BBA)-Bioenerg. 2010, 1799, 726–739. [Google Scholar] [CrossRef] [PubMed]
- Heltweg, B. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006, 66, 4368–4377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- E Voogd, T.; Vansterkenburg, E.L.; Wilting, J.; Janssen, L.H. Recent research on the biological activity of suramin. Pharmacol. Rev. 1993, 45, 177–203. [Google Scholar]
- Lain, S.; Hollick, J.J.; Campbell, J.; Staples, O.D.; Higgins, M.; Aoubala, M.; McCarthy, A.; Appleyard, V.; Murray, K.E.; Baker, L.; et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell 2008, 13, 454–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Outeiro, T.F.; Kontopoulos, E.; Altmann, S.M.; Kufareva, I.; Strathearn, K.E.; Amore, A.M.; Volk, C.B.; Maxwell, M.M.; Rochet, J.C.; McLean, P.J.; et al. Sirtuin 2 Inhibitors Rescue Alpha-Synuclein-Mediated Toxicity in Models of Parkinson’s Disease. Science 2007, 317, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Spires-Jones, T.L.; Fox, L.M.; Rozkalne, A.; Pitstick, R.; Carlson, G.A.; Kazantsev, A.G. Inhibition of Sirtuin 2 with Sulfobenzoic Acid Derivative AK1 is Non-Toxic and Potentially Neuroprotective in a Mouse Model of Frontotemporal Dementia. Front. Pharmacol. 2012, 3, 42. [Google Scholar] [CrossRef] [Green Version]
- Cheon, M.G.; Kim, W.; Choi, M.; Kim, J.-E. AK-1, a specific SIRT2 inhibitor, induces cell cycle arrest by downregulating Snail in HCT116 human colon carcinoma cells. Cancer Lett. 2015, 356, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Mellini, P.; Itoh, Y.; Tsumoto, H.; Li, Y.; Suzuki, M.; Tokuda, N.; Kakizawa, T.; Miura, Y.; Takeuchi, J.; Lahtela-Kakkonen, M.; et al. Potent mechanism-based sirtuin-2-selective inhibition by an in situ-generated occupant of the substrate-binding site. Chem. Sci. 2017, 8, 6400–6408. [Google Scholar] [CrossRef] [Green Version]
- Schiedel, M.; Robaa, D.; Rumpf, T.; Sippl, W.; Jung, M. The current state of NAD+-Dependent histone deacetylases (sirtuins) as novel therapeutic targets. Med. Res. Rev. 2017, 38, 147–200. [Google Scholar] [CrossRef] [PubMed]
- Disch, J.S.; Evindar, G.; Chiu, C.H.; Blum, C.A.; Dai, H.; Jin, L.; Schuman, E.; Lind, K.E.; Belyanskaya, S.L.; Deng, J.; et al. Discovery of Thieno[3,2-d]pyrimidine-6-carboxamides as Potent Inhibitors of SIRT1, SIRT2, and SIRT3. J. Med. Chem. 2013, 56, 3666–3679. [Google Scholar] [CrossRef] [PubMed]
- Sussmuth, S.D.; Haider, S.; Landwehrmeyer, G.B.; Farmer, R.; Frost, C.; Tripepi, G.; Andersen, C.A.; Di Bacco, M.; Lamanna, C.; Diodato, E.; et al. An exploratory double-blind, randomized clinical trial with selisistat, a SirT1 inhibitor, in patients with Huntington’s disease. Br. J. Clin. Pharmacol. 2015, 79, 465–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roessler, C.; Tüting, C.; Meleshin, M.; Steegborn, C.; Schutkowski, M. A Novel continuous assay for the deacylase sirtuin 5 and other deacetylases. J. Med. Chem. 2015, 58, 7217–7223. [Google Scholar] [CrossRef] [PubMed]
- Pradeepkiran, J.A.; Reddy, P.H.; Reddy, P.H.; Adi, P.J. Pharmacophore-based models for therapeutic drugs against phosphorylated tau in Alzheimer’s disease. Drug Discov. Today 2019, 24, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Adi, P.J.; Reddy, P.H. Structure based design and molecular docking studies for phosphorylated tau inhibitors in Alzheimer’s Disease. Cells 2019, 8, 260. [Google Scholar] [CrossRef] [Green Version]
- Adi, P.J.; Reddy, A.P.; Yin, X.; Manczak, M.; Reddy, P.H. Protective effects of BACE1 inhibitory ligand molecules against amyloid beta-induced synaptic and mitochondrial toxicities in Alzheimer’s disease. Hum. Mol. Genet. 2019, 29, 49–69. [Google Scholar] [CrossRef]
- Sanders, B.D.; Jackson, B.; Marmorstein, R. Structural basis for sirtuin function: What we know and what we don’t. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2009, 1804, 1604–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, G.T.T.; Schaefer, S.; Gertz, M.; Weyand, M.; Steegborn, C. Structures of human sirtuin 3 complexes with ADP-ribose and with carba-NAD+and SRT1720: Binding details and inhibition mechanism. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Parenti, M.D.; Bruzzone, S.; Nencioni, A.; Del Rio, A. Selectivity hot-spots of sirtuin catalytic cores. Mol. BioSyst. 2015, 11, 2263–2272. [Google Scholar] [CrossRef] [PubMed]
- Sultani, H.N.; Ghazal, R.A.; Hayallah, A.M.; Abdulrahman, L.K.; Abu-Hammour, K.; Abuhammad, S.; Taha, M.O.; Al-Eitan, L. Inhibitory effects of new mercapto xanthine derivatives in human mcf7 and k562 cancer cell lines. J. Heterocycl. Chem. 2016, 54, 450–456. [Google Scholar] [CrossRef]
- Famulok, M.; Hayallah, A.M. Synthesis of New 1,3,8-Trisubstituted Purine-2,6-diones and 1,3,6-Trisubstituted Thiazolo[2,3-f]purine-2,4-diones. HETEROCYCLES 2007, 74, 369. [Google Scholar] [CrossRef]
- ElZein, E.; Kalla, R.V.; Li, X.; Perry, T.; Gimbel, A.; Zeng, D.; Lustig, D.; Leung, K.; Zablocki, J. Discovery of a novel A2BAdenosine receptor antagonist as a clinical candidate for chronic inflammatory airway diseases. J. Med. Chem. 2008, 51, 2267–2278. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Case, A.W.; Riera, T.; Considine, T.; Lee, J.E.; Hamuro, Y.; Zhao, H.; Jiang, Y.; Sweitzer, S.M.; Pietrak, B.; et al. Crystallographic structure of a small molecule SIRT1 activator-enzyme complex. Nat. Commun. 2015, 6, 7645. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, D.; Chen, L.; Li, J.; Wang, J.; Ning, C.; Yu, N.; Zhao, F.; Chen, D.; Chen, X.; et al. Discovery and mechanism study of SIRT1 activators that promote the deacetylation of fluorophore-labeled substrate. J. Med. Chem. 2013, 56, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Gertz, M.; Nguyen, G.T.T.; Fischer, F.; Suenkel, B.; Schlicker, C.; Fränzel, B.; Tomaschewski, J.; Aladini, F.; Becker, C.; Wolters, D.; et al. A molecular mechanism for direct Sirtuin activation by resveratrol. PLoS ONE 2012, 7, e49761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, P.W.; Feldman, J.L.; Devries, M.K.; Dong, A.; Edwards, A.M.; Denu, J.M. Structure and biochemical functions of SIRT6*. J. Biol. Chem. 2011, 286, 14575–14587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- E Lohning, A.; Levonis, S.M.; Williams-Noonan, B.; Schweiker, S.S. A practical guide to molecular docking and homology modelling for medicinal chemists. Curr. Top. Med. Chem. 2017, 17, 2023–2040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jerabek-Willemsen, M.; Wienken, C.J.; Braun, D.; Baaske, P.; Duhr, S. Molecular interaction studies using microscale thermophoresis. ASSAY Drug Dev. Technol. 2011, 9, 342–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]



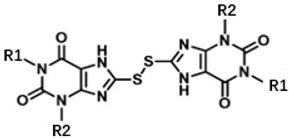
| Cmpd | R1 | R2 | IC50 (μM) | ||||
|---|---|---|---|---|---|---|---|
| SIRT3 | SIRT1 | SIRT2 | SIRT5 | SIRT6 | |||
| 1 | –CH2CH3 | –CH2CH3 | 0.79 ± 0.06 | 0.10 ± 0.01 | 1.17 ± 0.04 | 0.42 ± 0.01 | 116.0 ± 6.2 |
| 4 | –CH2CH2CH3 | –CH2CH2CH3 | 0.54 ± 0.05 | 0.12 ± 0.01 | 1.19 ± 0.06 | 0.39 ± 0.03 | 128.7 ± 16.1 |
| 12 | –CH2CH(CH3)2 | –CH2CH3 | 1.77 ± 0.05 | 0.43 ± 0.03 | 4.86 ± 0.08 | 1.51 ± 0.10 | 296.6 ± 11.3 |
| 13 |  | –CH2CH3 | 0.72 ± 0.06 | 0.17 ± 0.01 | 2.25 ± 0.12 | 0.50 ± 0.01 | 155.8 ± 6.5 |
| 14 | 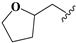 | –CH2CH3 | 0.69 ± 0.12 | 0.15 ± 0.02 | 1.62 ± 0.05 | 0.54 ± 0.02 | 126.5 ± 10.2 |
| 15 | 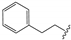 | –CH2CH3 | 0.37 ± 0.05 | 0.17 ± 0.01 | 1.35 ± 0.05 | 0.45 ± 0.01 | 103.3 ± 6.3 |
| 16 | 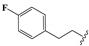 | –CH2CH3 | 0.46 ± 0.04 | 0.19 ± 0.01 | 2.03 ± 0.04 | 0.84 ± 0.08 | 110.9 ± 2.6 |
| 17 |  | –CH2CH3 | 1.15 ± 0.08 | 0.41 ± 0.04 | 2.19 ± 0.08 | 1.00 ± 0.03 | 128.3 ± 4.1 |
| 18 | 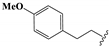 | –CH2CH3 | 0.61 ± 0.03 | 0.21 ± 0.02 | 2.16 ± 0.08 | 0.61 ± 0.02 | 102.3 ± 9.1 |
| 19 | 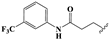 | –CH2CH3 | 8.52 ± 0.22 | 1.87 ± 0.04 | 9.44 ± 0.19 | 26.12 ± 3.15 | 452.0 ± 9.8 |
| 20 | 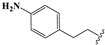 | –CH2CH3 | 0.97 ± 0.06 | 0.49 ± 0.01 | 4.61 ± 0.13 | 0.92 ± 0.04 | 209.6 ± 5.9 |
| ELT-31 a | 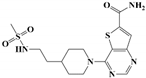 | 0.35 ± 0.02 | 0.27 ± 0.02 | 0.12 ± 0.01 | >500 | 462.0 ± 14.9 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, H.; Li, C.; Li, M.; Yang, L.; Zhao, S.; Wang, Z.; Liu, H.; Liu, D. Design, Synthesis, and Biological Evaluation of 8-Mercapto-3,7-Dihydro-1H-Purine-2,6-Diones as Potent Inhibitors of SIRT1, SIRT2, SIRT3, and SIRT5. Molecules 2020, 25, 2755. https://doi.org/10.3390/molecules25122755
Han H, Li C, Li M, Yang L, Zhao S, Wang Z, Liu H, Liu D. Design, Synthesis, and Biological Evaluation of 8-Mercapto-3,7-Dihydro-1H-Purine-2,6-Diones as Potent Inhibitors of SIRT1, SIRT2, SIRT3, and SIRT5. Molecules. 2020; 25(12):2755. https://doi.org/10.3390/molecules25122755
Chicago/Turabian StyleHan, Haozhen, Chunpu Li, Man Li, Lisheng Yang, Sen Zhao, Zhifei Wang, Hong Liu, and Dongxiang Liu. 2020. "Design, Synthesis, and Biological Evaluation of 8-Mercapto-3,7-Dihydro-1H-Purine-2,6-Diones as Potent Inhibitors of SIRT1, SIRT2, SIRT3, and SIRT5" Molecules 25, no. 12: 2755. https://doi.org/10.3390/molecules25122755
APA StyleHan, H., Li, C., Li, M., Yang, L., Zhao, S., Wang, Z., Liu, H., & Liu, D. (2020). Design, Synthesis, and Biological Evaluation of 8-Mercapto-3,7-Dihydro-1H-Purine-2,6-Diones as Potent Inhibitors of SIRT1, SIRT2, SIRT3, and SIRT5. Molecules, 25(12), 2755. https://doi.org/10.3390/molecules25122755






