Searching for Low Molecular Weight Seleno-Compounds in Sprouts by Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
2.1. Total Selenium Concentration in Se-Enriched Sprouts
2.2. Selenium Species in the Sprouts
2.2.1. Identification of Selenium Compounds in Sprout’s Extracts by HPLC-ICP-MS
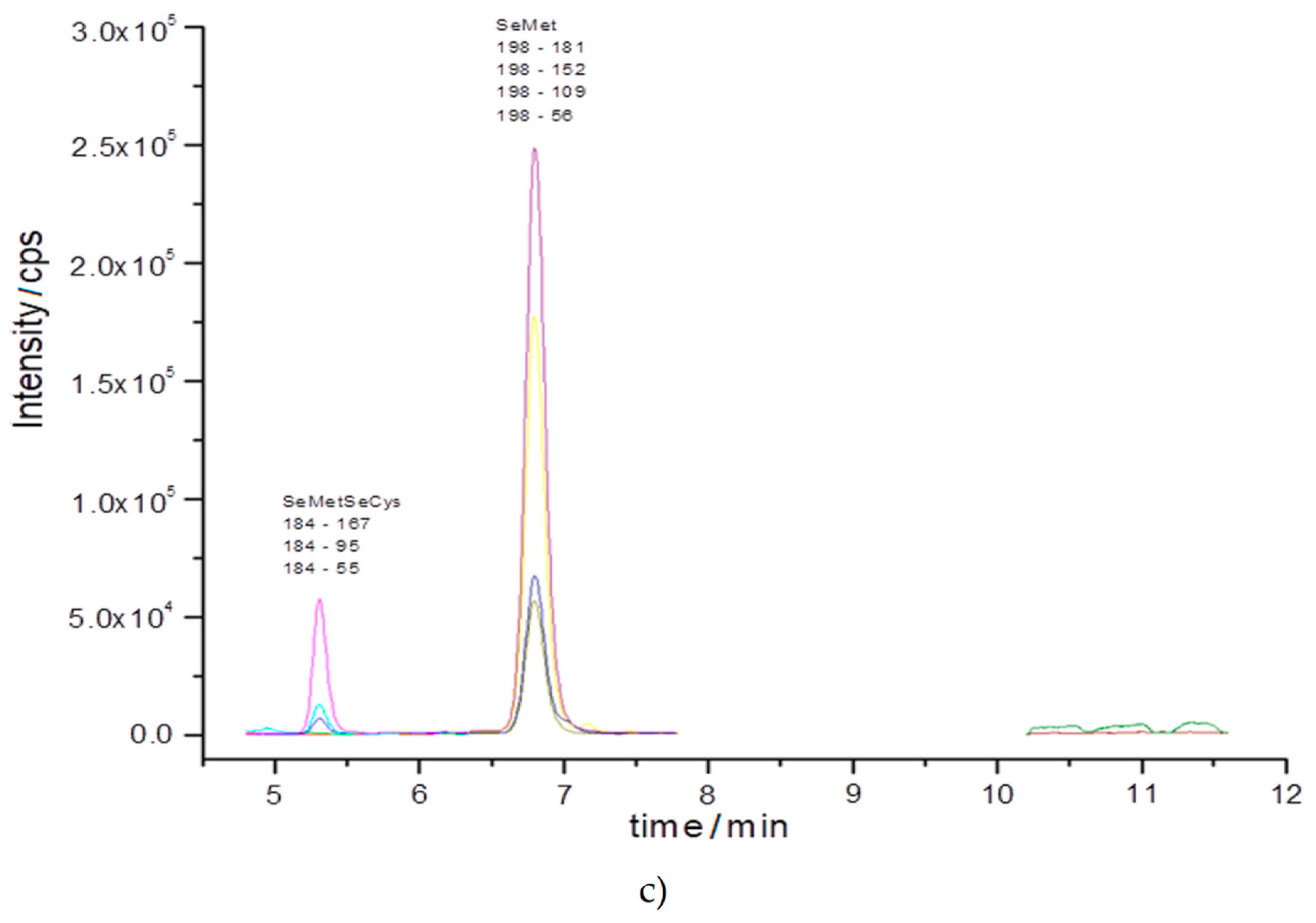
2.2.2. Identification of Selenium Compounds with the Use of Tandem Mass Spectrometry
2.2.3. Identification of Selenium Compounds with the use of UHPLC-ESI-Orbitrap-MS/MS
3. Materials and Methods
3.1. Preparation of Sprout’s Samples and Investigation of Selenium Intake
3.1.1. Se-Enriched Sprouts
3.1.2. Determination of Total Selenium in Sprouts
3.1.3. Extraction of Selenium Species from Sprouts
3.2. Instrumentation
3.2.1. Limit of Quantification and Limit of Detection
3.2.2. Precision
3.3. Reagents
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Szpunar, J.; Łobiński, R. Hyphenated Techniques in Speciation Analysis; Royal Society of Chemistry: Cambridge, UK, 2003. [Google Scholar]
- Bulska, E.; Ruszczyńska, A. Analytical Techniques for Trace Element Determination. Phys. Sci. Rev. 2017, 2, 1–14. [Google Scholar] [CrossRef]
- Wojciechowski, M.; Bulska, E.; Piaścik, M. Noble metal modifiers for antimony determination by graphite furnace atomic absorption spectrometry in biological samples. J. Anal. At. Spectrom. 2001, 16, 99–101. [Google Scholar] [CrossRef]
- Moens, L. Applications of mass spectrometry in the trace element analysis of biological materials. Anal. Bioanal. Chem. 1997, 359, 309–316. [Google Scholar] [CrossRef]
- Czauderna, M.; Kowalczyk, J.; Bulska, E.; Boldižarova, K.; Niedźwiedzka, K.M.; Ruszczyńska, A.; Leng, Ľ. Effect of dietary CLA isomers on selenium, zinc, copper, chromium, magnesium and calcium levels in rat liver. J. Anim. Feed. Sci. 2005, 14, 529–532. [Google Scholar] [CrossRef] [Green Version]
- Wróbel, K.; Wróbel, K.; Kannamkumarath, S.S.; Caruso, J.A.; Wysocka, I.A.; Bulska, E.; Świątek, J.; Wierzbicka, M. HPLC-ICP-MS speciation of selenium in enriched onion leaves—A potential dietary source of Se-methylselenocystine. Food. Chem. 2004, 86, 617–623. [Google Scholar] [CrossRef]
- Auger, J.; Yang, W.; Arnault, I.; Pannier, F.; Potin-Gautier, M. High-performance liquid chromatographic–inductively coupled plasma mass spectrometric evidence for Se-“alliins” in garlic and onion grown in Se-rich soil. J. Chromatogr. A 2004, 1032, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Kápolna, E.; Shah, M.; A Caruso, J.; Fodor, P. Selenium speciation studies in Se-enriched chives (Allium schoenoprasum) by HPLC-ICP–MS. Food Chem. 2007, 101, 1398–1406. [Google Scholar] [CrossRef]
- Pyrzyńska, K. Determination of Selenium Species in Environmental Samples. Microchim. Acta 2002, 140, 55–62. [Google Scholar] [CrossRef]
- Sugihara, S.; Kondo, M.; Chihara, Y.; Yuji, M.; Hattori, H.; Yoshida, M. Preparation of selenium-enriched sprouts and identification of their selenium species by high-performance liquid chromatography-inductively coupled plasma mass spectrometry. Biosci. Biotechnol. Biochem. 2004, 68, 193–199. [Google Scholar] [CrossRef] [Green Version]
- Bianga, J.; Govasmark, E.; Szpunar, J. Characterization of Selenium Incorporation into Wheat Proteins by Two-Dimensional Gel Electrophoresis–Laser Ablation ICP MS followed by capillary HPLC–ICP MS and Electrospray Linear Trap Quadrupole Orbitrap MS. Anal. Chem. 2013, 85, 2037–2043. [Google Scholar] [CrossRef]
- Dernovics, M.; Far, J.; Łobinski, R. Identification of anionic selenium species in Se-rich yeast by electrospray QTOF MS/MS and hybrid linear ion trap/orbitrap MSn. Metallomics 2009, 1, 317. [Google Scholar] [CrossRef] [PubMed]
- Encinar, J.R.; Ouerdane, L.; Buchmann, W.; Tortajada, J.; Łobinski, R.; Szpunar, J. Identification of Water-Soluble Selenium-Containing Proteins in Selenized Yeast by Size-Exclusion-Reversed-Phase HPLC/ICPMS Followed by MALDI-TOF and Electrospray Q-TOF Mass Spectrometry. Anal. Chem. 2003, 75, 3765–3774. [Google Scholar] [CrossRef] [PubMed]
- Far, J.; Preud’Homme, H.; Łobinski, R. Detection and identification of hydrophilic selenium compounds in selenium-rich yeast by size exclusion–microbore normal-phase HPLC with the on-line ICP–MS and electrospray Q-TOF-MS detection. Anal. Chim. Acta 2010, 657, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Bierła, K.; Suzuki, N.; Ogra, Y.; Szpunar, J.; Łobiński, R. Identification and determination of selenohomolanthionine – The major selenium compound in Torula yeast. Food. Chem. 2017, 237, 1196–1201. [Google Scholar] [CrossRef] [PubMed]
- Gil Casal, S.; Far, J.; Bierla, K.; Ouerdane, L.; Szpunar, J. Study of the Se-containing metabolomes in Se-rich yeast by size-exclusion—cation-exchange HPLC with the parallel ICP MS and electrospray orbital ion trap detection. Metallomics 2010. [Google Scholar] [CrossRef] [PubMed]
- Bulska, E.; Wysocka, I.; Wierzbicka, M.H.; Proost, K.; Janssens, K.; Falkenberg, G. In Vivo Investigation of the Distribution and the Local Speciation of Selenium inAlliumcepa L. by Means of Microscopic X-ray Absorption Near-Edge Structure Spectroscopy and Confocal Microscopic X-ray Fluorescence Analysis. Anal. Chem. 2006, 78, 7616–7624. [Google Scholar] [CrossRef] [PubMed]
- Tapiero, H.; Townsend, D.; Tew, K. The antioxidant role of selenium and seleno-compounds. Biomed. Pharmacother. 2003, 57, 134–144. [Google Scholar] [CrossRef]
- Carsella, J.S.; Sánchez-Lombardo, I.; Bonetti, S.J.; Crans, D.C. Selenium Speciation in the Fountain Creek Watershed (Colorado, USA) Correlates with Water Hardness, Ca and Mg Levels. Molecules 2017, 22, 708. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Hu, B.; He, M.; Duan, J. Organic and inorganic selenium speciation in environmental and biological samples by nanometer-sized materials packed dual-column separation/preconcentration on-line coupled with ICP-MS. J. Mass Spectrom. 2008, 43, 336–345. [Google Scholar] [CrossRef]
- Umysová, D.; Vítová, M.; Doušková, I.; Bišová, K.; Slavková, M.; Čížková, M.; Machát, J.; Doucha, J.; Zachleder, V. Bioaccumulation and toxicity of selenium compounds in the green alga Scenedesmus quadricauda. BMC Plant Boil. 2009, 9, 58. [Google Scholar] [CrossRef] [Green Version]
- Guerrero, B.; Llugany, M.; Palacios, O.; Valiente, M. Dual effects of different selenium species on wheat. Plant Physiol. Biochem. 2014, 83, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Maseko, T.; Callahan, D.; Dunshea, F.R.; Doronila, A.; Kolev, S.D.; Ng, K. Chemical characterisation and speciation of organic selenium in cultivated selenium-enriched Agaricus bisporus. Food Chem. 2013, 141, 3681–3687. [Google Scholar] [CrossRef] [PubMed]
- Kieliszek, M.; Błażejak, S. Selenium: Significance, and outlook for supplementation. Nutrition 2013, 29, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Kouba, A.; Velisek, J.; Stara, A.; Masojidek, J.; Kozák, P. Supplementation with Sodium Selenite and Selenium-Enriched Microalgae Biomass Show Varying Effects on Blood Enzymes Activities, Antioxidant Response, and Accumulation in Common Barbel (Barbus barbus). BioMed Res. Int. 2014, 2014, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avila, F.W.; Faquin, V.; Yang, Y.; Ramos, S.J.; Guilherme, L.R.G.; Thannhauser, T.W.; Li, L. Assessment of the anticancer compounds Se-Methylselenocysteine and Glucosinolates in Se-biofortified broccoli (Brassica oleracea L. var. italica) sprouts and florets. J. Agric. Food Chem. 2013, 61, 6216–6223. [Google Scholar] [CrossRef] [PubMed]
- Finley, J.W.; Davis, C.D.; Feng, Y. Selenium from high selenium broccoli protects rats from colon cancer. J. Nutr. 2000, 130, 2384–2389. [Google Scholar] [CrossRef] [PubMed]
- Finley, J.W.; Ip, C.; Lisk, D.J.; Davis, C.D.; Hintze, K.J.; Whanger, P.D. Cancer-protective properties of high-selenium broccoli. J. Agric. Food Chem. 2001, 49, 2679–2683. [Google Scholar] [CrossRef]
- Shao, S.; Mi, X.; Ouerdane, L.; Lobinski, R.; Garcia-Reyes, J.F.; Molina-Diaz, A.; Vass, A.; Dernovics, M. Quantification of Se-Methylselenocysteine and Its γ-Glutamyl Derivative from Naturally Se-Enriched Green Bean (Phaseolus vulgaris vulgaris) After HPLC-ESI-TOF-MS and Orbitrap MSn-Based Identification. Food Anal. Methods 2014, 7, 1147–1157. [Google Scholar] [CrossRef]
- Ip, C.; Birringer, M.; Block, E.; Kotrebai, M.; Tyson, J.F.; Udén, P.; Lisk, D.J. Chemical speciation influences comparative activity of selenium-enriched garlic and yeast in mammary cancer prevention. J. Agric. Food Chem. 2000, 48, 2062–2070. [Google Scholar] [CrossRef] [Green Version]
- Ip, C.; Thompson, H.J.; Zhu, Z.; E Ganther, H. In vitro and in vivo studies of methylseleninic acid: Evidence that a monomethylated selenium metabolite is critical for cancer chemoprevention. Cancer Res. 2000, 60, 2882–2886. [Google Scholar] [PubMed]
- Clark, L.C.; Combs, G.F.; Turnbull, B.W.; Slate, E.H.; Chalker, D.K.; Chow, J.; Davis, L.S.; Glover, R.A.; Graham, G.F.; Gross, E.G.; et al. Effects of Selenium Supplementation for Cancer Prevention in Patients with Carcinoma of the Skin. JAMA 1996, 276, 1957. [Google Scholar] [CrossRef] [PubMed]
- Lippman, S.M.; Klein, E.A.; Goodman, P.J.; Lucia, M.S.; Thompson, I.M.; Ford, L.G.; Parnes, H.L.; Minasian, L.M.; Gaziano, J.M.; Hartline, J.A.; et al. Effect of selenium and vitamin e on risk of prostate cancer and other cancers the selenium and vitamin e cancer prevention trial (select). JAMA 2009, 301, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michalska-Kacymirow, M.; Kurek, E.; Smolis, A.; Wierzbicka, M.; Bulska, E. Biological and chemical investigation of Allium cepa L. response to selenium inorganic compounds. Anal. Bioanal. Chem. 2014, 406, 3717–3722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, X.-J.; Block, E.; Udén, P.; Zhang, X.; Quimby, B.D.; Sullivan, J.J. Allium Chemistry: Identification of Selenoamino Acids in Ordinary and Selenium-Enriched Garlic, Onion, and Broccoli Using Gas Chromatography with Atomic Emission Detection. J. Agric. Food Chem. 1995, 43, 1754–1757. [Google Scholar] [CrossRef]
- Ogra, Y.; Ishiwata, K.; Iwashita, Y.; Suzuki, K.T. Simultaneous speciation of selenium and sulfur species in selenized odorless garlic (Allium sativum L. Shiro) and shallot (Allium ascalonicum) by HPLC–inductively coupled plasma-(octopole reaction system)-mass spectrometry and electrospray ionization-tandem mass spectrometry. J. Chromatogr. A 2005, 1093, 118–125. [Google Scholar] [CrossRef]
- Whanger, P.D.; Ip, C.; Polan, C.E.; Uden, P.C.; Welbaum, G. Tumorigenesis, metabolism, speciation, bioavailability, and tissue deposition of selenium in selenium-enriched ramps (Allium tricoccum). J. Agric. Food Chem. 2000, 48, 5723–5730. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Lisk, D.; Block, E.; Ip, C. Characterization of the Biological Activity of g-Glutamyl-Semethylselenocysteine: A Novel, Naturally Occurring Anticancer Agent from Garlic. Cancer Res. 2001, 61, 2923–2928. [Google Scholar] [PubMed]
- Ogra, Y.; Ogihara, Y.; Anan, Y. Comparison of the metabolism of inorganic and organic selenium species between two selenium accumulator plants, garlic and Indian mustard. Metallomics 2017, 9, 61–68. [Google Scholar] [CrossRef]
- Kitajima, T.; Chiba, Y. Selenomethionine metabolism and its toxicity in yeast. Biomol. Concepts 2013, 4, 611–616. [Google Scholar] [CrossRef]
- Rao, Y.; McCooeye, M.; Windust, A.; Bramanti, E.; D’Ulivo, A.; Mester, Z. Mapping of Selenium Metabolic Pathway in Yeast by Liquid Chromatography−Orbitrap Mass Spectrometry. Anal. Chem. 2010, 82, 8121–8130. [Google Scholar] [CrossRef] [Green Version]
- Pilon-Smits, E.; Quinn, C.F. Selenium Metabolism in Plants. Plant Cell Monogr. 2010, 17, 225–241. [Google Scholar] [CrossRef]
- Li, H.-F.; McGrath, S.P.; Zhao, F.-J. Selenium uptake, translocation and speciation in wheat supplied with selenate or selenite. New Phytol. 2008, 178, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Cuderman, P.; Ožbolt, L.; Kreft, I.; Stibilj, V. Extraction of Se species in buckwheat sprouts grown from seeds soaked in various Se solutions. Food Chem. 2010, 123, 941–948. [Google Scholar] [CrossRef]
- Funes-Collado, V.; Morell-Garcia, A.; Rubio, R.; López-Sánchez, J.F. Study of selenocompounds from selenium-enriched culture of edible sprouts. Food Chem. 2013, 141, 3738–3743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ávila, F.; Yang, Y.; Faquin, V.; Ramos, S.J.; Guilherme, L.R.G.; Thannhauser, T.W.; Li, L. Impact of selenium supply on Se-methylselenocysteine and glucosinolate accumulation in selenium-biofortified Brassica sprouts. Food Chem. 2014, 165, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Diowksz, A.; Kordialik-Bogacka, E.; Ambroziak, W. Se-enriched sprouted seeds as functional additives in sourdough fermentation. LWT 2014, 56, 524–528. [Google Scholar] [CrossRef]
- Ruszczyńska, A.; Konopka, A.; Kurek, E.; Elguera, J.C.T.; Bulska, E. Investigation of biotransformation of selenium in plants using spectrometric methods. Spectrochim. Acta Part B At. Spectrosc. 2017, 130, 7–16. [Google Scholar] [CrossRef]
- Preud’Homme, H.; Far, J.; Gil-Casal, S.; Łobinski, R. Large-scale identification of selenium metabolites by online size-exclusion-reversed phase liquid chromatography with combined inductively coupled plasma (ICP-MS) and electrospray ionization linear trap-Orbitrap mass spectrometry (ESI-MSn). Metallomics 2012, 4, 422. [Google Scholar] [CrossRef]
- Larsen, E.H.; Lobinski, R.; Burger-Meÿer, K.; Hansen, M.; Ruzik, R.; Mazurowska, L.; Rasmussen, P.H.; Sloth, J.J.; Scholten, O.; Kik, C. Uptake and speciation of selenium in garlic cultivated in soil amended with symbiotic fungi (mycorrhiza) and selenite. Anal. Bioanal. Chem. 2006, 385, 1098–1108. [Google Scholar] [CrossRef]
- Arnaudguilhem, C.; Bierla, K.; Ouerdane, L.; Preud’Homme, H.; Yiannikouris, A.; Łobinski, R. Selenium metabolomics in yeast using complementary reversed-phase/hydrophilic ion interaction (HILIC) liquid chromatography–electrospray hybrid quadrupole trap/Orbitrap mass spectrometry. Anal. Chim. Acta 2012, 757, 26–38. [Google Scholar] [CrossRef] [PubMed]
- McSheehy, S.; Szpunar, J.; Haldys, V.; Tortajada, J. Identification of selenocompounds in yeast by electrospray quadrupole-time of flight mass spectrometry. J. Anal. At. Spectrom. 2002, 17, 507–514. [Google Scholar] [CrossRef]
- Dernovics, M.; Lobinski, R. Characterization of the selenocysteine-containing metabolome in selenium-rich yeast. J. Anal. At. Spectrom. 2008, 72, 72–83. [Google Scholar] [CrossRef]
Sample Availability: Samples are available from the authors. |






| Selenium in Nutrient Solution (mg/L Se) | Total Se Content (mg/kg Dried Weight) * | ||
|---|---|---|---|
| Sunflower Sprouts | Radish Sprouts | Onion Sprouts | |
| control sample | <LOQ | <LOQ | 1.51 ± 0.08 |
| 10 | 20 ± 1 | 64 ± 3 | 91 ± 5 |
| 20 | 46 ± 2 | 145 ± 7 | 198 ± 10 |
| 40 | 87 ± 4 | 304 ± 15 | 574 ± 29 |
| 60 | 132 ± 6 | 388 ± 19 | 825 ± 41 |
| Compound | MRM Transition Precursor Ion m/z → Product Ion m/z | Fragmentor (V) | Collision Energy (V) |
|---|---|---|---|
| SeMet | 198 → 181 | 80 | 5 |
| 198 → 152 | 80 | 9 | |
| 198 → 109 | 80 | 25 | |
| 198 → 56 | 80 | 21 | |
| SeMetSeCys | 184 → 167 | 50 | 5 |
| 184 → 95 | 50 | 29 | |
| 184 → 55 | 50 | 21 | |
| SeCys | 337 → 248 | 80 | 9 |
| 337 → 88 | 80 | 21 | |
| 337 → 74 | 80 | 29 | |
| gamma-Glu-MetSeCys | 313 → 167 | 70 | 13 |
| 313 → 84 | 100 | 29 |
| Sample/Fraction | tR * | Seleno-Compound Identified by HPLC-ICP-MS | m/zteor | m/zexp | Chemical Formula (M+H+) | Seleno-Compound Identified by UHPLC-ESI-Orbitrap-MS/MS | References | |
|---|---|---|---|---|---|---|---|---|
| HPLC-ICP-MS | UHPLC-ESI-Orbitrap-MS/MS | |||||||
| RF1 | 1.9–2.5 | 7.84 | U4 | 332.0256 | 332.0242 | C10H14O3N5Se+ | 5′-seleno adenosine | [49] |
| RF2a | 2.5–3.0 | 5.17 | SeMetCys | 166.9606 | 166.9607 | C4H7O2Se+ | SeMetCys –NH3 | [29] |
| RF2b | 3.0–3.4 | 5.17 | 166.9606 | 166.9607 | C4H7O2Se+ | SeMetCys–NH3 | [49] | |
| RF2c | 3.4–3.8 | 5.17 | 166.9606 | 166.9607 | C4H7O2Se+ | SeMetCys–NH3 | ||
| RF3a | 4.0–4.3 | SeMet | [50] | |||||
| RF3b | 4.3–5.0 | 6.57 | SeMet | 198.0028 | 198.0027 | C5H12O2NSe+ | SeMet | [16,51,52] |
| RF3b | 4.3–5.0 | 6.57 | 180.9762 | 180.9764 | C5H9O2Se+ | SeMet-NH3 | [49] | |
| RF3c | 5.0–5.7 | |||||||
| RF4 | 6.0–7.8 | 10.49 | U5 | 345.0196 | 345.0193 | C9H17O7N2Se+ | 2,3-DHP-selenolanthionine | [14,16] |
| RF5 | 7.8–8.9 | 10.05 | Se (IV) | 475.0396 | 475.0393 | C13H23O8N4SSe+ | Se-S conjugate of cysteino-selenoglutathione | [16,51] |
| RF6 | 8.9–9.8 | 12.05 | U6 | 376.9916 | 376.9912 | C9H17O7N2SSe+ | 2,3-DHP-selenocysteine-cysteine | [16,51] |
| RF7 | 9.8–10.8 | U7 | ||||||
| RF8 | 10.8–11,6 | 11.22 | gamma-Glu-MetSeCys | 313.0297 | 313.0292 | C9H17O5N2Se+ | gamma-Glu-MetSeCys | [12,16] |
| RF9 | 11.6–13.0 | U8 | ||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurek, E.; Michalska-Kacymirow, M.; Konopka, A.; Kościuczuk, O.; Tomiak, A.; Bulska, E. Searching for Low Molecular Weight Seleno-Compounds in Sprouts by Mass Spectrometry. Molecules 2020, 25, 2870. https://doi.org/10.3390/molecules25122870
Kurek E, Michalska-Kacymirow M, Konopka A, Kościuczuk O, Tomiak A, Bulska E. Searching for Low Molecular Weight Seleno-Compounds in Sprouts by Mass Spectrometry. Molecules. 2020; 25(12):2870. https://doi.org/10.3390/molecules25122870
Chicago/Turabian StyleKurek, Eliza, Magdalena Michalska-Kacymirow, Anna Konopka, Olga Kościuczuk, Anna Tomiak, and Ewa Bulska. 2020. "Searching for Low Molecular Weight Seleno-Compounds in Sprouts by Mass Spectrometry" Molecules 25, no. 12: 2870. https://doi.org/10.3390/molecules25122870





