Unravelling the Potential of Salivary Volatile Metabolites in Oral Diseases. A Review
Abstract
:1. Introduction
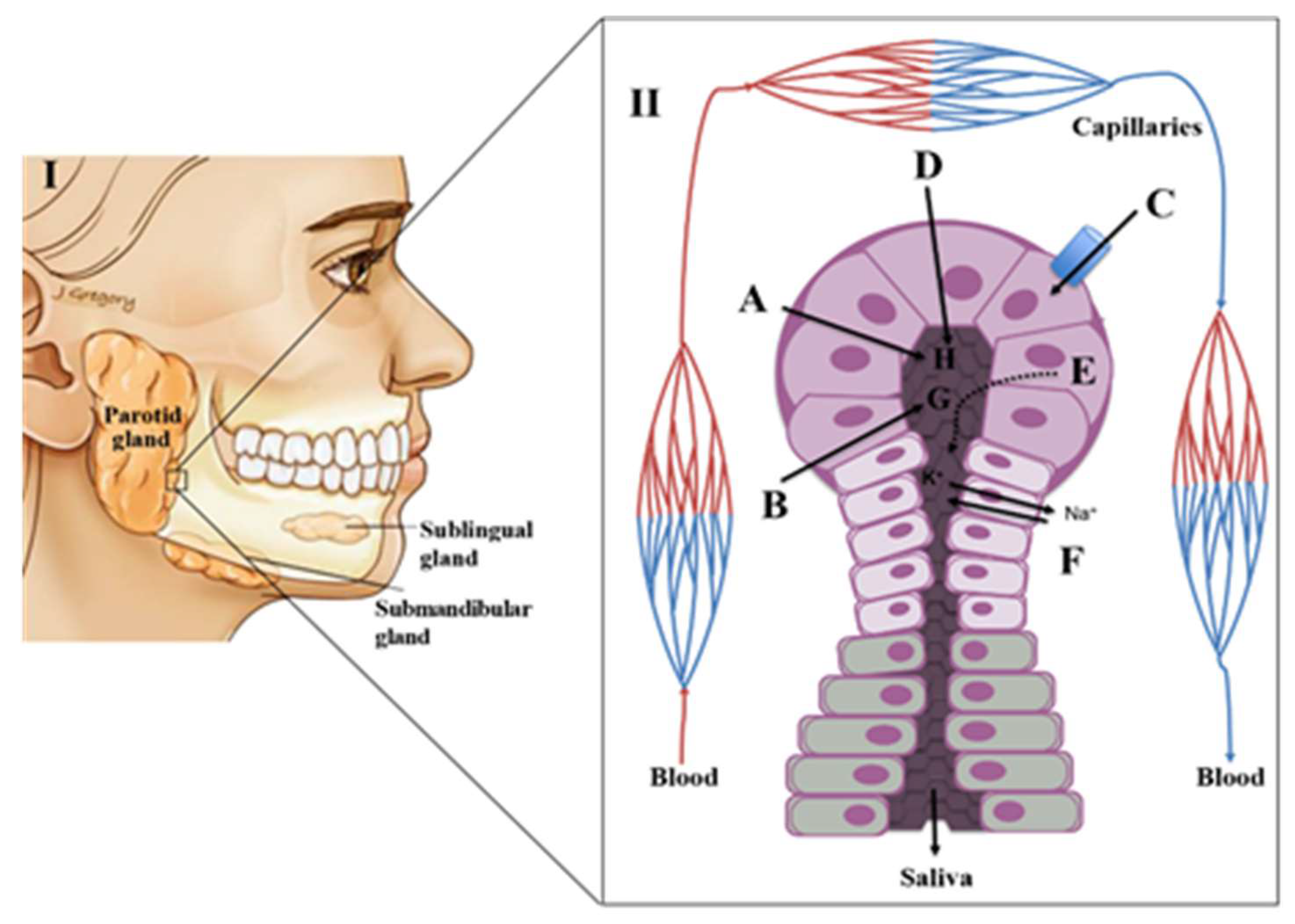
2. Physiology of Saliva
Saliva Composition and Production
3. Putative Salivary Biomarkers for Oral Diseases
3.1. Gingivitis and Periodontal Disease
3.2. Dental Caries
3.3. Oral Cancer
3.4. Oral Potentially Malignant Disorders (OPMD)
3.5. Burning Mouth Syndrome
3.6. Recurrent Aphthous Ulceration (RAS)
4. Salivary Volatomics
5. Analytical Platforms Used in the Volatomic Analysis of Saliva
5.1. GC-MS
5.1.1. Solid Phase Micro Extraction (SPME)
5.1.2. Needle Trap Micro Extraction (NTME)
5.1.3. Thin Film Micro Extraction (TFME)
5.1.4. Stir Bar Sorptive Extraction (SBSE)
5.2. Direct Injection Mass Spectrometry
5.3. eNOSE
6. Data Analysis
7. Current Challenges and Future Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADH | antidiuretic hormone |
| ANNs | artificial neural networks |
| CE-TOF-MS | capillary electrophoresis time-of-flight mass spectrometry |
| GC-MS | gas chromatography-mass spectrometry |
| GFR | glomerular filtration rate |
| 1H-NMR | proton nuclear magnetic resonance |
| HS-trap/GC-MS | headspace-trap gas chromatography-mass spectrometry |
| IL | interleukin |
| LDA | linear discriminant analysis |
| LC-MS/MS | liquid chromatography tandem mass spectrometry |
| lncRNA | long non-coding RNA |
| LMICs | low- and middle-income countries |
| MALDI-TOF-MS | matrix-assisted laser desorption/ionization time-of-flight mass spectrometry |
| MMPs | matrix metalloproteinases |
| MVSA | multivariate statistical analysis |
| NTME | Needle Trap Micro Extraction |
| OD | oral diseases |
| OLP | oral leukoplakia |
| OPMDs | oral potentially malignant disorders |
| OSCC | oral squamous cell carcinoma |
| PCA | Principal component analysis |
| PLS-DA | discriminant analysis of partial least squares |
| POCT | point of care device |
| pptV | parts per trillion by volume |
| PTR-MS | Proton Transfer Reaction Mass Spectrometry |
| RAS | recurrent aphthous ulceration |
| SBSE | stir bar sorptive extraction |
| SESI-MS | Secondary Electrospray Ionization-Mass Spectrometry |
| SIFT-MS | Selected Ion Flow Tube-Mass Spectrometry |
| SPME | Solid Phase Micro Extraction |
| TFME | Thin Film Microextraction |
| TNF-α | tumour necrosis factor α |
| UHPLC-MS/MS | ultra-high pressure liquid chromatography tandem mass spectrometry |
| VOCs | volatile organic metabolites |
| VSCs | volatile sulphur containing compounds |
| 2DE | two-dimensional gel electrophoresis |
References
- Peres, M.A.; Macpherson, L.M.D.; Weyant, R.J.; Daly, B.; Venturelli, R.; Mathur, M.R.; Listl, S.; Celeste, R.K.; Guarnizo-Herreno, C.C.; Kearns, C.; et al. Oral diseases: A global public health challenge. Lancet 2019, 394, 249–260. [Google Scholar] [CrossRef]
- Dawes, C.; Wong, D.T.W. Role of Saliva and Salivary Diagnostics in the Advancement of Oral Health. J. Dent. Res. 2019, 98, 133–141. [Google Scholar] [CrossRef]
- Deepa, T.; Thirrunavukkarasu, N. Saliva as a potential diagnostic tool. Indian J. Med. Sci. 2010, 64, 293–306. [Google Scholar] [CrossRef]
- Katsani, K.R.; Sakellari, D. Saliva proteomics updates in biomedicine. J. Biol. Res. 2019, 26, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellagambi, F.G.; Lomonaco, T.; Salvo, P.; Vivaldi, F.; Hangouët, M.; Ghimenti, S.; Biagini, D.; Di Francesco, F.; Fuoco, R.; Errachid, A. Saliva sampling: Methods and devices. An overview. Tractrends Anal. Chem. 2020, 124, 115781. [Google Scholar] [CrossRef]
- Farnaud, S.J.; Kosti, O.; Getting, S.J.; Renshaw, D. Saliva: Physiology and diagnostic potential in health and disease. Sci. World J. 2010, 10, 434–456. [Google Scholar] [CrossRef]
- Gardner, A.; Carpenter, G.; So, P.W. Salivary metabolomics: From diagnostic biomarker discovery to investigating biological function. Metabolites 2020, 10, 47. [Google Scholar] [CrossRef] [Green Version]
- Aps, J.K.; Martens, L.C. Review: The physiology of saliva and transfer of drugs into saliva. Forensic Sci. Int. 2005, 150, 119–131. [Google Scholar] [CrossRef]
- Papacosta, E.; Nassis, G.P. Saliva as a tool for monitoring steroid, peptide and immune markers in sport and exercise science. J. Sci. Med. Sport 2011, 14, 424–434. [Google Scholar] [CrossRef]
- Edgar, W.; O’Mullane, D.; Dawes, C. Saliva and Oral Health; British Dental Association: London, UK, 2004; Volume 146. [Google Scholar]
- Sugimoto, M.; Saruta, J.; Matsuki, C.; To, M.; Onuma, H.; Kaneko, M.; Soga, T.; Tomita, M.; Tsukinoki, K. Physiological and environmental parameters associated with mass spectrometry-based salivary metabolomic profiles. Metabolomics 2013, 9, 454–463. [Google Scholar] [CrossRef]
- Ishikawa, S.; Sugimoto, M.; Kitabatake, K.; Tu, M.; Sugano, A.; Yamamori, I.; Iba, A.; Yusa, K.; Kaneko, M.; Ota, S.; et al. Effect of timing of collection of salivary metabolomic biomarkers on oral cancer detection. Amino Acids 2017, 49, 761–770. [Google Scholar] [CrossRef]
- Lamy, E.; Mau, M. Saliva proteomics as an emerging, non-invasive tool to study livestock physiology, nutrition and diseases. J. Proteom. 2012, 75, 4251–4258. [Google Scholar] [CrossRef] [PubMed]
- Gröschl, M. Saliva: A reliable sample matrix in bioanalytics. Bioanalysis 2017, 9, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Proctor, G.B. The physiology of salivary secretion. Periodontol. 2000 2016, 70, 11–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, J.A.M.; Taware, R.; Porto-Figueira, P.; Rapole, S.; Câmara, J.S. Chapter 29–The salivary volatome in breast cancer. In Precision Medicine for Investigators, Practitioners and Providers; Faintuch, J., Faintuch, S., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 301–307. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amado, F.M.L.; Ferreira, R.P.; Vitorino, R. One decade of salivary proteomics: Current approaches and outstanding challenges. Clin. Biochem. 2013, 46, 506–517. [Google Scholar] [CrossRef]
- Amado, F.M.L.; Vitorino, R.M.P.; Domingues, P.M.D.N.; Lobo, M.J.C.; Duarte, J.A.R. Analysis of the human saliva proteome. Expert Rev. Proteomics 2005, 2, 521–539. [Google Scholar] [CrossRef]
- Cong, X.; Zhang, Y.; He, Q.H.; Wei, T.; Zhang, X.M.; Zhang, J.Z.; Xiang, R.L.; Yu, G.Y.; Wu, L.L. Endothelial Tight Junctions Are Opened in Cholinergic-Evoked Salivation in Vivo. J. Dent. Res. 2017, 96, 562–570. [Google Scholar] [CrossRef]
- Simón-Soro, Á.; Tomás, I.; Cabrera-Rubio, R.; Catalan, M.D.; Nyvad, B.; Mira, A. Microbial geography of the oral cavity. J. Dent. Res. 2013, 92, 616–621. [Google Scholar] [CrossRef]
- Azen, E.A.; Goodman, P.A.; Lalley, P.A. Human salivary proline-rich protein genes on chromosome 12. Am. J. Hum. Genet. 1985, 37, 418–424. [Google Scholar]
- Castagnola, M.; Cabras, T.; Vitali, A.; Sanna, M.T.; Messana, I. Biotechnological implications of the salivary proteome. Trends Biotechnol. 2011, 29, 409–418. [Google Scholar] [CrossRef]
- Pihlstrom, B.L.; Michalowicz, B.S.; Johnson, N.W. Periodontal diseases. Lancet 2005, 366, 1809–1820. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, L.D.R.; Soares, M.R.; Nogueira, F.C.S.; Garcia, C.H.S.; Camisasca, D.R.; Domont, G.; Feitosa, A.C.R.; Pereira, D.A.; Zingali, R.B.; Alves, G. Analysis of the salivary proteome in gingivitis patients. J. Periodontal Res. 2011, 46, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.H.; Rahim, Z.H.A.; Jessie, K.; Hashim, O.H.; Taiyeb-Ali, T.B. Salivary proteins associated with periodontitis in patients with type 2 diabetes mellitus. Int. J. Mol. Sci. 2012, 13, 4652–4654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, L.D.R.; Soares, M.R.; Nogueira, F.C.S.; Garcia, C.; Camisasca, D.R.; Domont, G.; Feitosa, A.C.R.; Pereira, D.d.A.; Zingali, R.B.; Alves, G. Comparative proteomic analysis of whole saliva from chronic periodontitis patients. J. Proteom. 2010, 73, 1334–1341. [Google Scholar] [CrossRef]
- Rangé, H.; Léger, T.; Huchon, C.; Ciangura, C.; Diallo, D.; Poitou, C.; Meilhac, O.; Bouchard, P.; Chaussain, C. Salivary proteome modifications associated with periodontitis in obese patients. J. Clin. Periodontol. 2012, 39, 799–806. [Google Scholar] [CrossRef]
- Wu, Y.; Shu, R.; Luo, L.J.; Ge, L.H.; Xie, Y.F. Initial comparison of proteomic profiles of whole unstimulated saliva obtained from generalized aggressive periodontitis patients and healthy control subjects. J. Periodontal Res. 2009, 44, 636–644. [Google Scholar] [CrossRef]
- Bostanci, N.; Selevsek, N.; Wolski, W.; Grossmann, J.; Bao, K.; Wahlander, A.; Trachsel, C.; Schlapbach, R.; Oztürk, V.Ö.; Afacan, B.; et al. Targeted proteomics guided by label-free quantitative proteome analysis in saliva reveal transition signatures from health to periodontal disease. Mol. Cell. Proteom. 2018, 17, 1392–1409. [Google Scholar] [CrossRef] [Green Version]
- Sorsa, T.; Gursoy, U.K.; Nwhator, S.; Hernandez, M.; Tervahartiala, T.; Leppilahti, J.; Gursoy, M.; Könönen, E.; Emingil, G.; Pussinen, P.J.; et al. Analysis of matrix metalloproteinases, especially MMP-8, in gingival creviclular fluid, mouthrinse and saliva for monitoring periodontal diseases. Periodontol. 2000 2016, 70, 142–163. [Google Scholar] [CrossRef]
- Emecen-Huja, P.; Hasan, I.; Miller, C.S. Biologic markers of failing implants. Dent. Clin. North Am. 2015, 59, 179–194. [Google Scholar] [CrossRef]
- Gursoy, U.K.; Kononen, E.; Pussinen, P.J.; Tervahartiala, T.; Hyvarinen, K.; Suominen, A.L.; Uitto, V.J.; Paju, S.; Sorsa, T. Use of host- and bacteria-derived salivary markers in detection of periodontitis: A cumulative approach. Dis. Markers 2011, 30, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Kaczynski, T.; Wronski, J.; Gluszko, P.; Kryczka, T.; Miskiewicz, A.; Gorski, B.; Radkowski, M.; Strzemecki, D.; Grieb, P.; Gorska, R. Salivary interleukin 6, interleukin 8, interleukin 17A, and tumour necrosis factor alpha levels in patients with periodontitis and rheumatoid arthritis. Cent. Eur. J. Immunol. 2019, 44, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Aimetti, M.; Cacciatore, S.; Graziano, A.; Tenori, L. Metabonomic analysis of saliva reveals generalized chronic periodontitis signature. Metabolomics 2012, 8, 465–474. [Google Scholar] [CrossRef]
- Gawron, K.; Wojtowicz, W.; Łazarz-Bartyzel, K.; Łamasz, A.; Qasem, B.; Mydel, P.; Chomyszyn-Gajewska, M.; Potempa, J.; Mlynarz, P. Metabolomic status of the oral cavity in chronic periodontitis. Vivo 2019, 33, 1165–1174. [Google Scholar] [CrossRef] [Green Version]
- Romano, F.; Meoni, G.; Manavella, V.; Baima, G.; Tenori, L.; Cacciatore, S.; Aimetti, M. Analysis of salivary phenotypes of generalized aggressive and chronic periodontitis through nuclear magnetic resonance-based metabolomics. J. Periodontol. 2018, 89, 1452–1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rzeznik, M.; Triba, M.N.; Levy, P.; Jungo, S.; Botosoa, E.; Duchemann, B.; Le Moyec, L.; Bernaudin, J.F.; Savarin, P.; Guez, D. Identification of a discriminative metabolomic fingerprint of potential clinical relevance in saliva of patients with periodontitis using 1H nuclear magnetic resonance (NMR) spectroscopy. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.P.; Saxena, M.; Saimbi, C.S.; Arif, J.M.; Roy, R. Metabolic profiling by 1H NMR spectroscopy of saliva shows clear distinction between control and diseased case of periodontitis. Metabolomics 2017, 13. [Google Scholar] [CrossRef]
- Liebsch, C.; Pitchika, V.; Pink, C.; Samietz, S.; Kastenmüller, G.; Artati, A.; Suhre, K.; Adamski, J.; Nauck, M.; Völzke, H.; et al. The Saliva Metabolome in Association to Oral Health Status. J. Dent. Res. 2019, 98, 642–651. [Google Scholar] [CrossRef]
- Huang, Y.; Zhu, M.; Li, Z.; Sa, R.; Chu, Q.; Zhang, Q.; Zhang, H.; Tang, W.; Zhang, M.; Yin, H. Mass spectrometry-based metabolomic profiling identifies alterations in salivary redox status and fatty acid metabolism in response to inflammation and oxidative stress in periodontal disease. Free Radic. Biol. Med. 2014, 70, 223–232. [Google Scholar] [CrossRef]
- Ebersole, J.L.; Nagarajan, R.; Akers, D.; Miller, C.S. Targeted salivary biomarkers for discrimination of periodontal health and disease(s). Front. Cell. Infect. Microbiol. 2015, 5, 62. [Google Scholar] [CrossRef] [Green Version]
- Hussain, Q.A.; McKay, I.J.; Gonzales-Marin, C.; Allaker, R.P. Detection of adrenomedullin and nitric oxide in different forms of periodontal disease. J. Periodontal Res. 2016, 51, 16–25. [Google Scholar] [CrossRef] [PubMed]
- O’Brien-Simpson, N.M.; Burgess, K.; Brammar, G.C.; Darby, I.B.; Reynolds, E.C. Development and evaluation of a saliva-based chair-side diagnostic for the detection of Porphyromonas gingivalis. J. Oral Microbiol. 2015, 7, 29129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitts, N.B.; Zero, D.T.; Marsh, P.D.; Ekstrand, K.; Weintraub, J.A.; Ramos-Gomez, F.; Tagami, J.; Twetman, S.; Tsakos, G.; Ismail, A. Dental caries. Nat. Rev. Dis. Primers 2017, 3, 17030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.Z.; Cheng, X.Q.; Li, J.Y.; Zhang, P.; Yi, P.; Xu, X.; Zhou, X.D. Saliva in the diagnosis of diseases. Int. J. Oral Sci. 2016, 8, 133–137. [Google Scholar] [CrossRef] [Green Version]
- Fiorillo, L.; Cervino, G.; Laino, L.; D’Amico, C.; Mauceri, R.; Tozum, T.F.; Gaeta, M.; Cicciu, M. Porphyromonas gingivalis, Periodontal and Systemic Implications: A Systematic Review. Dent. J. (Basel) 2019, 7, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitorino, R.; De Morais Guedes, S.; Ferreira, R.; Lobo, M.J.C.; Duarte, J.; Ferrer-Correia, A.J.; Tomer, K.B.; Domingues, P.M.; Amado, F.M.L. Two-dimensional electrophoresis study of in vitro pellicle formation and dental caries susceptibility. Eur. J. Oral Sci. 2006, 114, 147–153. [Google Scholar] [CrossRef]
- Fidalgo, T.K.S.; Freitas-Fernandes, L.B.; Angeli, R.; Muniz, A.M.S.; Gonsalves, E.; Santos, R.; Nadal, J.; Almeida, F.C.L.; Valente, A.P.; Souza, I.P.R. Salivary metabolite signatures of children with and without dental caries lesions. Metabolomics 2013, 9, 657–666. [Google Scholar] [CrossRef]
- Chaturvedi, A.K.; Udaltsova, N.; Engels, E.A.; Katzel, J.A.; Yanik, E.L.; Katki, H.A.; Lingen, M.W.; Silverberg, M.J. Oral leukoplakia and risk of progression to oral cancer: A population-based cohort study. J. Natl. Cancer Inst. 2019. [Google Scholar] [CrossRef] [Green Version]
- Zotti, F.; Nocini, R.; Capocasale, G.; Fior, A.; Peretti, M.; Albanese, M. Malignant transformation evidences of Oral Lichen Planus: When the time is of the essence. Oral Oncol. 2020, 104594. [Google Scholar] [CrossRef]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, S.; Sugimoto, M.; Kitabatake, K.; Sugano, A.; Nakamura, M.; Kaneko, M.; Ota, S.; Hiwatari, K.; Enomoto, A.; Soga, T.; et al. Identification of salivary metabolomic biomarkers for oral cancer screening. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, S.; Sugimoto, M.; Edamatsu, K.; Sugano, A.; Kitabatake, K.; Iino, M. Discrimination of oral squamous cell carcinoma from oral lichen planus by salivary metabolomics. Oral Dis. 2020, 26, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Xie, G.; Zhou, Z.; Shi, P.; Qiu, Y.; Zheng, X.; Chen, T.; Su, M.; Zhao, A.; Jia, W. Salivary metabolite signatures of oral cancer and leukoplakia. Int. J. Cancer 2011, 129, 2207–2217. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, P.; Wang, X.; Duan, Y. The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Lohavanichbutr, P.; Zhang, Y.; Wang, P.; Gu, H.; Nagana Gowda, G.A.; Djukovic, D.; Buas, M.F.; Raftery, D.; Chen, C. Salivary metabolite profiling distinguishes patients with oral cavity squamous cell carcinoma from normal controls. PLoS ONE 2018, 13. [Google Scholar] [CrossRef] [Green Version]
- Balkwill, F.; Mantovani, A. Inflammation and cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef]
- Aziz, S.; Ahmed, S.S.; Ali, A.; Khan, F.A.; Zulfiqar, G.; Iqbal, J.; Khan, A.A.; Shoaib, M. Salivary Immunosuppressive Cytokines IL-10 and IL-13 Are Significantly Elevated in Oral Squamous Cell Carcinoma Patients. Cancer Invest. 2015, 33, 318–328. [Google Scholar] [CrossRef]
- Dowling, P.; Wormald, R.; Meleady, P.; Henry, M.; Curran, A.; Clynes, M. Analysis of the saliva proteome from patients with head and neck squamous cell carcinoma reveals differences in abundance levels of proteins associated with tumour progression and metastasis. J. Proteom. 2008, 71, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Honarmand, M.; Farhad-Mollashahi, L.; Nakhaee, A.; Nehi, M. Salivary levels of ErbB2 and CEA in oral squamous cell carcinoma patients. Asian Pac. J. Cancer Prev. 2016, 17, 77–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jou, Y.J.; Lin, C.D.; Lai, C.H.; Tang, C.H.; Huang, S.H.; Tsai, M.H.; Chen, S.Y.; Kao, J.Y.; Lin, C.W. Salivary zinc finger protein 510 peptide as a novel biomarker for detection of oral squamous cell carcinoma in early stages. Clin. Chim. Acta 2011, 412, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Arellano, M.; Boontheung, P.; Wang, J.; Zhou, H.; Jiang, J.; Elashoff, D.; Wei, R.; Loo, J.A.; Wong, D.T. Salivary proteomics for oral cancer biomarker discovery. Clin. Cancer Res. 2008, 14, 6246–6252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.S.; Chen, Y.T.; Chiang, W.F.; Hsiao, Y.C.; Chu, L.J.; Seei, L.C.; Wu, C.S.; Tu, H.T.; Chen, H.W.; Chen, C.C.; et al. Saliva protein biomarkers To detect oral squamous cell carcinoma in a high-risk population in Taiwan. Proc. Natl. Acad. Sci. USA 2016, 113, 11549–11554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivadasan, P.; Gupta, M.K.; Sathe, G.; Sudheendra, H.V.; Sunny, S.P.; Renu, D.; Hari, P.S.; Gowda, H.; Suresh, A.; Kuriakose, M.A.; et al. Salivary proteins from dysplastic leukoplakia and oral squamous cell carcinoma and their potential for early detection. J. Proteom. 2020, 212, 103574. [Google Scholar] [CrossRef]
- Dos Santos, E.S.; Ramos, J.C.; Normando, A.G.C.; Mariano, F.V.; Paes Leme, A.F. Epigenetic alterations in salivary gland tumors. Oral Dis. 2019. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ye, J.; Zhang, Z.; Gong, Z.; Lin, Z.; Ding, M. Long non-coding RNA RBM5-AS1 promotes the aggressive behaviors of oral squamous cell carcinoma by regulation of miR-1285-3p/YAP1 axis. Biomed. Pharm. 2020, 123, 109723. [Google Scholar] [CrossRef] [PubMed]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A.; Abemayor, E.; Wong, D.T. Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 2009, 15, 5473–5477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodi, G.; Carrozzo, M.; Furness, S.; Thongprasom, K. Interventions for treating oral lichen planus: A systematic review. Br. J. Derm. 2012, 166, 938–947. [Google Scholar] [CrossRef]
- Lorenzo-Pouso, A.I.; Pérez-Sayáns, M.; Bravo, S.B.; López-Jornet, P.; García-Vence, M.; Alonso-Sampedro, M.; Carballo, J.; García-García, A.; Zalewska, A. Protein-Based Salivary Profiles as Novel Biomarkers for Oral Diseases. Dis. Markers 2018, 2018. [Google Scholar] [CrossRef]
- Yang, L.L.; Liu, X.Q.; Liu, W.; Cheng, B.; Li, M.T. Comparative analysis of whole saliva proteomes for the screening of biomarkers for oral lichen planus. Inflamm. Res. 2006, 55, 405–407. [Google Scholar] [CrossRef]
- Souza, M.M.; Florezi, G.P.; Nico, M.M.S.; de Paula, F.; Paula, F.M.; Lourenço, S.V. Salivary proteomics in lichen planus: A relationship with pathogenesis? Oral Dis. 2018, 24, 784–792. [Google Scholar] [CrossRef]
- Camisasca, D.R.; da Rós Gonçalves, L.; Soares, M.R.; Sandim, V.; Nogueira, F.C.S.; Garcia, C.H.S.; Santana, R.; de Oliveira, S.P.; Buexm, L.A.; de Faria, P.A.S.; et al. A proteomic approach to compare saliva from individuals with and without oral leukoplakia. J. Proteom. 2017, 151, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Bender, S.D. Burning Mouth Syndrome. Dent. Clin. North Am. 2018, 62, 585–596. [Google Scholar] [CrossRef]
- Ji, E.H.; Diep, C.; Liu, T.; Li, H.; Merrill, R.; Messadi, D.; Hu, S. Potential protein biomarkers for burning mouth syndrome discovered by quantitative proteomics. Mol. Pain 2017, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, D.; Zeng, C.; Liu, Y.; Huang, G.; Mei, Z. Salivary metabolomics profile of patients with recurrent aphthous ulcer as revealed by liquid chromatography–tandem mass spectrometry. J. Int. Med. Res. 2018, 46, 1052–1062. [Google Scholar] [CrossRef] [PubMed]
- Amann, A.; de Lacy Costello, B.; Miekisch, W.; Schubert, J.; Buszewski, B.; Pleil, J.; Ratcliffe, N.; Risby, T. The human volatilome: Volatile organic compounds (VOCs) in exhaled breath, skin emanations, urine, feces and saliva. J. Breath Res. 2014, 8, 034001. [Google Scholar] [CrossRef] [PubMed]
- Milanowski, M.; Pomastowski, P.; Ligor, T.; Buszewski, B. Saliva–Volatile Biomarkers and Profiles. Crit. Rev. Anal. Chem. 2017, 47, 251–266. [Google Scholar] [CrossRef]
- de Lacy Costello, B.; Amann, A.; Al-Kateb, H.; Flynn, C.; Filipiak, W.; Khalid, T.; Osborne, D.; Ratcliffe, N.M. A review of the volatiles from the healthy human body. J. Breath Res. 2014, 8, 014001. [Google Scholar] [CrossRef]
- Al-Kateb, H.; de Lacy Costello, B.; Ratcliffe, N. An investigation of volatile organic compounds from the saliva of healthy individuals using headspace-trap/GC-MS. J. Breath Res. 2013, 7, 036004. [Google Scholar] [CrossRef] [Green Version]
- Soini, H.A.; Klouckova, I.; Wiesler, D.; Oberzaucher, E.; Grammer, K.; Dixon, S.J.; Xu, Y.; Brereton, R.G.; Penn, D.J.; Novotny, M.V. Analysis of volatile organic compounds in human saliva by a static sorptive extraction method and gas chromatography-mass spectrometry. J. Chem. Ecol. 2010, 36, 1035–1042. [Google Scholar] [CrossRef]
- Malathi, N.; Mythili, S.; Vasanthi, H.R. Salivary diagnostics: A brief review. ISRN Dent. 2014, 2014, 158786. [Google Scholar] [CrossRef]
- Torsten, M.; Gomez-Moreno, G.; Aguilar-Salvatierra, A. Drug-related oral malodour (halitosis): A literature review. Eur. Rev. Med. Pharm. Sci. 2017, 21, 4930–4934. [Google Scholar]
- Calil, C.M.; Marcondes, F.K. Influence of anxiety on the production of oral volatile sulfur compounds. Life Sci. 2006, 79, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Milella, L. The Negative Effects of Volatile Sulphur Compounds. J. Vet. Dent. 2015, 32, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Hertel, M.; Hartwig, S.; Schutte, E.; Gillissen, B.; Preissner, R.; Schmidt-Westhausen, A.M.; Paris, S.; Kastner, I.; Preissner, S. Identification of signature volatiles to discriminate Candida albicans, glabrata, krusei and tropicalis using gas chromatography and mass spectrometry. Mycoses 2016, 59, 117–126. [Google Scholar] [CrossRef]
- Taware, R.; Taunk, K.; Pereira, J.A.M.; Shirolkar, A.; Soneji, D.; Camara, J.S.; Nagarajaram, H.A.; Rapole, S. Volatilomic insight of head and neck cancer via the effects observed on saliva metabolites. Sci. Rep. 2018, 8, 17725. [Google Scholar] [CrossRef]
- Pfaffe, T.; Cooper-White, J.; Beyerlein, P.; Kostner, K.; Punyadeera, C. Diagnostic potential of saliva: Current state and future applications. Clin. Chem. 2011, 57, 675–687. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, A.M.L.; Sorensen, C.E.; Proctor, G.B.; Carpenter, G.H.; Ekstrom, J. Salivary secretion in health and disease. J. Oral Rehabil. 2018, 45, 730–746. [Google Scholar] [CrossRef]
- Krishnan, K.; Chen, T.; Paster, B.J. A practical guide to the oral microbiome and its relation to health and disease. Oral Dis. 2017, 23, 276–286. [Google Scholar] [CrossRef] [Green Version]
- Kakoei, S.; Barkhori, F.; Mirzazadeh, A.; Mohammadi, M.; Gholamhoseinian, A. Influence of menstrual cycle and salivary ß-estradiol on volatile sulfur compound. J. Oral Health Oral Epidemiol. 2012, 1, 5. [Google Scholar]
- Boots, A.W.; Smolinska, A.; van Berkel, J.J.; Fijten, R.R.; Stobberingh, E.E.; Boumans, M.L.; Moonen, E.J.; Wouters, E.F.; Dallinga, J.W.; Van Schooten, F.J. Identification of microorganisms based on headspace analysis of volatile organic compounds by gas chromatography-mass spectrometry. J. Breath Res. 2014, 8, 027106. [Google Scholar] [CrossRef]
- Hertel, M.; Preissner, R.; Gillissen, B.; Schmidt-Westhausen, A.M.; Paris, S.; Preissner, S. Detection of signature volatiles for cariogenic microorganisms. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Apatzidou, A.D.; Bakirtzoglou, E.; Vouros, I.; Karagiannis, V.; Papa, A.; Konstantinidis, A. Association between oral malodour and periodontal disease-related parameters in the general population. Acta Odontol. Scand. 2013, 71, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Cavaco, C.; Pereira, J.A.M.; Taunk, K.; Taware, R.; Rapole, S.; Nagarajaram, H.; Camara, J.S. Screening of salivary volatiles for putative breast cancer discrimination: An exploratory study involving geographically distant populations. Anal. Bioanal. Chem. 2018, 410, 4459–4468. [Google Scholar] [CrossRef] [PubMed]
- Cavaco, C.; Perestrelo, R.; Silva, C.; Aveiro, F.; Pereira, J.; Câmara, J. Establishment of the Saliva Volatomic Profile as an Exploratory and Non-Invasive Strategy to Find Potential Breast Cancer Biomarkers; Int. Labmate Ltd.: Hertfordshire, UK, 2014. [Google Scholar]
- Porto-Figueira, P.; Pereira, J.A.M.; Camara, J.S. Exploring the potential of needle trap microextraction combined with chromatographic and statistical data to discriminate different types of cancer based on urinary volatomic biosignature. Anal. Chim. Acta 2018, 1023, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Cavaco, C.; Perestrelo, R.; Pereira, J.; Câmara, J.S. Microextraction by packed Sorbent (MEPS) and solid-phase microextraction (SPME) as sample preparation procedures for the metabolomic profiling of urine. Metabolites 2014, 4, 71–97. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.; Silva, C.L.; Perestrelo, R.; Gonçalves, J.; Alves, V.; Câmara, J.S. Re-exploring the high-throughput potential of microextraction techniques, SPME and MEPS, as powerful strategies for medical diagnostic purposes. Innovative approaches, recent applications and future trends Microextraction Techniques. Anal. Bioanal. Chem. 2014, 406, 2101–2122. [Google Scholar] [CrossRef]
- Saigusa, D.; Okamura, Y.; Motoike, I.N.; Katoh, Y.; Kurosawa, Y.; Saijyo, R.; Koshiba, S.; Yasuda, J.; Motohashi, H.; Sugawara, J.; et al. Establishment of Protocols for Global Metabolomics by LC-MS for Biomarker Discovery. PLoS ONE 2016, 11, e0160555. [Google Scholar] [CrossRef]
- Biniecka, M.; Caroli, S. Analytical methods for the quantification of volatile aromatic compounds. TRAC—Trends Anal. Chem. 2011, 30, 1756–1770. [Google Scholar] [CrossRef]
- Taware, R.; Taunk, K.; Kumar, T.V.S.; Pereira, J.A.M.; Camara, J.S.; Nagarajaram, H.A.; Kundu, G.C.; Rapole, S. Extracellular volatilomic alterations induced by hypoxia in breast cancer cells. Metabolomics 2020, 16, 21. [Google Scholar] [CrossRef]
- Taunk, K.; Taware, R.; More, T.H.; Porto-Figueira, P.; Pereira, J.A.M.; Mohapatra, R.; Soneji, D.; Camara, J.S.; Nagarajaram, H.A.; Rapole, S. A non-invasive approach to explore the discriminatory potential of the urinary volatilome of invasive ductal carcinoma of the breast. RSC Adv. 2018, 8, 25040–25050. [Google Scholar] [CrossRef] [Green Version]
- Taware, R.; Taunk, K.; Pereira, J.; Dhakne, R.; Kannan, N.; Soneji, D.; Câmara, J.; Nagarajaram, H.; Rapole, S. Investigation of urinary volatomic alterations in head and neck cancer: A non-invasive approach towards diagnosis and prognosis. Metabolomics 2017, 13, 111. [Google Scholar] [CrossRef]
- Trefz, P.; Kischkel, S.; Hein, D.; James, E.S.; Schubert, J.K.; Miekisch, W. Needle trap micro-extraction for VOC analysis: Effects of packing materials and desorption parameters. J. Chromatogr. A 2012, 1219, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Porto-Figueira, P.; Pereira, J.; Miekisch, W.; Camara, J.S. Exploring the potential of NTME/GC-MS, in the establishment of urinary volatomic profiles. Lung cancer patients as case study. Sci. Rep. 2018, 8, 13113. [Google Scholar] [CrossRef] [PubMed]
- Bruheim, I.; Liu, X.; Pawliszyn, J. Thin-film microextraction. Anal. Chem. 2003, 75, 1002–1010. [Google Scholar] [CrossRef]
- Shigeyama, H.; Wang, T.; Ichinose, M.; Ansai, T.; Lee, S.W. Identification of volatile metabolites in human saliva from patients with oral squamous cell carcinoma via zeolite-based thin-film microextraction coupled with GC–MS. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2019, 1104, 49–58. [Google Scholar] [CrossRef]
- Ide, A.H.; Nogueira, J.M.F. Hollow fiber microextraction: A new hybrid microextraction technique for trace analysis. Anal. Bioanal. Chem. 2018, 410, 2911–2920. [Google Scholar] [CrossRef]
- Nogueira, J.M. Novel sorption-based methodologies for static microextraction analysis: A review on SBSE and related techniques. Anal. Chim. Acta 2012, 757, 1–10. [Google Scholar] [CrossRef]
- Penn, D.J.; Oberzaucher, E.; Grammer, K.; Fischer, G.; Soini, H.A.; Wiesler, D.; Novotny, M.V.; Dixon, S.J.; Xu, Y.; Brereton, R.G. Individual and gender fingerprints in human body odour. J. R. Soc. Interface 2007, 4, 331–340. [Google Scholar] [CrossRef]
- Kumar, S.; Huang, J.; Abbassi-Ghadi, N.; Španěl, P.; Smith, D.; Hanna, G.B. Selected ion flow tube mass spectrometry analysis of exhaled breath for volatile organic compound profiling of esophago-gastric cancer. Anal. Chem. 2013, 85, 6121–6128. [Google Scholar] [CrossRef]
- Sukul, P.; Trefz, P.; Schubert, J.K.; Miekisch, W. Immediate effects of breath holding maneuvers onto composition of exhaled breath. J. Breath Res. 2014, 8. [Google Scholar] [CrossRef]
- Yuan, B.; Koss, A.R.; Warneke, C.; Coggon, M.; Sekimoto, K.; De Gouw, J.A. Proton-Transfer-Reaction Mass Spectrometry: Applications in Atmospheric Sciences. Chem. Rev. 2017, 117, 13187–13229. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Bean, H.D.; Wargo, M.J.; Leclair, L.W.; Hill, J.E. Detecting bacterial lung infections: In vivo evaluation of in vitro volatile fingerprints. J. Breath Res. 2013, 7. [Google Scholar] [CrossRef] [PubMed]
- Sukul, P.; Schubert, J.K.; Oertel, P.; Kamysek, S.; Taunk, K.; Trefz, P.; Miekisch, W. FEV manoeuvre induced changes in breath VOC compositions: An unconventional view on lung function tests. Sci. Rep. 2016, 6, 28029. [Google Scholar] [CrossRef]
- Sukul, P.; Schubert, J.K.; Trefz, P.; Miekisch, W. Natural menstrual rhythm and oral contraception diversely affect exhaled breath compositions. Sci. Rep. 2018, 8, 10838. [Google Scholar] [CrossRef] [PubMed]
- Trefz, P.; Schmidt, S.C.; Sukul, P.; Schubert, J.K.; Miekisch, W.; Fischer, D.C. Non-Invasive Assessment of Metabolic Adaptation in Paediatric Patients Suffering from Type 1 Diabetes Mellitus. J. Clin. Med. 2019, 8, 1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, A.D.; Baietto, M. Applications and advances in electronic-nose technologies. Sensors 2009, 9, 5099–5148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lv, Y.; Wang, H.; Tao, X.; Zhang, X. Classification identification of abalone flavoring liquids based on metal sensor array. Carpathian J. Food Sci. Technol. 2016, 8, 107–112. [Google Scholar]
- Wilson, A.D. Diverse applications of electronic-nose technologies in agriculture and forestry. Sensors 2013, 13, 2295–2348. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.S.; Prada, P.A.; Curran, A.M.; Furton, K.G. Applicability of emanating volatile organic compounds from various forensic specimens for individual differentiation. Forensic Sci. Int. 2013, 226, 173–182. [Google Scholar] [CrossRef]
- Monedeiro, F.; Milanowski, M.; Ratiu, I.A.; Zmyslowski, H.; Ligor, T.; Buszewski, B. VOC Profiles of Saliva in Assessment of Halitosis and Submandibular Abscesses Using HS-SPME-GC/MS Technique. Molecules 2019, 24, 2977. [Google Scholar] [CrossRef] [Green Version]
- Ramdzan, A.N.; Mornane, P.J.; McCullough, M.J.; Mazurek, W.; Kolev, S.D. Determination of acetaldehyde in saliva by gas-diffusion flow injection analysis. Anal. Chim. Acta 2013, 786, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Alagendran, S.; Archunan, G.; Rameshkumar, K.; Kadalmani, B.; Rodriguez, J.A.; Fernandez, G.; Guzman, R. 2-Nonenal-Ovulatory Specific Volatiles in Human Saliva throughout Menstrual Cycle by Gas Chromatography and Mass Spectrometry Analysis. Am. J. Biochem. Biotechnol. 2010, 6, 187–194. [Google Scholar] [CrossRef]
- Bijlsma, S.; Bobeldijk, I.; Verheij, E.R.; Ramaker, R.; Kochhar, S.; Macdonald, I.A.; Van Ommen, B.; Smilde, A.K. Large-scale human metabolomics studies: A strategy for data (pre-) processing and validation. Anal. Chem. 2006, 78, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wang, X. Saliva metabolomics opens door to biomarker discovery, disease diagnosis, and treatment. Appl. Biochem. Biotechnol. 2012, 168, 1718–1727. [Google Scholar] [CrossRef]
- Kettaneh, N.; Berglund, A.; Wold, S. PCA and PLS with very large data sets. Comput. Stat. Data Anal. 2005, 48, 69–85. [Google Scholar] [CrossRef]
- McGeer, P.L.; Lee, M.; Kennedy, K.; McGeer, E.G. Saliva Diagnosis as a Disease Predictor. J. Clin. Med. 2020, 9, 377. [Google Scholar] [CrossRef] [Green Version]
- Rabe, A.; Gesell Salazar, M.; Michalik, S.; Fuchs, S.; Welk, A.; Kocher, T.; Volker, U. Metaproteomics analysis of microbial diversity of human saliva and tongue dorsum in young healthy individuals. J. Oral Microbiol. 2019, 11, 1654786. [Google Scholar] [CrossRef]
- Schulz, A.; Lang, R.; Behr, J.; Hertel, S.; Reich, M.; Kummerer, K.; Hannig, M.; Hannig, C.; Hofmann, T. Targeted metabolomics of pellicle and saliva in children with different caries activity. Sci. Rep. 2020, 10, 697. [Google Scholar] [CrossRef] [Green Version]
- Tothova, L.; Kamodyova, N.; Cervenka, T.; Celec, P. Salivary markers of oxidative stress in oral diseases. Front. Cell. Infect. Microbiol. 2015, 5, 73. [Google Scholar] [CrossRef] [Green Version]
- Nanayakkara, S.; Zhou, X.; Spallek, H. Impact of big data on oral health outcomes. Oral Dis. 2019, 25, 1245–1252. [Google Scholar] [CrossRef]
- Malley, J.D.; Dasgupta, A.; Moore, J.H. The limits of p-values for biological data mining. Biodata Min. 2013, 6. [Google Scholar] [CrossRef] [Green Version]
- Takeda, I.; Stretch, C.; Barnaby, P.; Bhatnager, K.; Rankin, K.; Fu, H.; Weljie, A.; Jha, N.; Slupsky, C. Understanding the human salivary metabolome. NMR Biomed. 2009, 22, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Ballabio, D.; Consonni, V. Classification tools in chemistry. Part 1: Linear models. PLS-DA. Anal. Methods 2013, 5, 3790. [Google Scholar] [CrossRef]
- Pereira, J.L.; Duarte, D.; Carneiro, T.J.; Ferreira, S.; Cunha, B.; Soares, D.; Costa, A.L.; Gil, A.M. Saliva NMR metabolomics: Analytical issues in pediatric oral health research. Oral Dis. 2019, 25, 1545–1554. [Google Scholar] [CrossRef]
- Casas-Ferreira, A.M.; Nogal-Sánchez, M.d.; Rodríguez-Gonzalo, E.; Moreno-Cordero, B.; Pérez-Pavón, J.L. Determination of leucine and isoleucine/allo-isoleucine by electrospray ionization-tandem mass spectrometry and partial least square regression: Application to saliva samples. Talanta 2020. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; van Velzen, E.J.J.; van Duijnhoven, J.P.M.; van Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Szymańska, E.; Saccenti, E.; Smilde, A.; Westerhuis, J. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Shayea, Q.K. Artificial neural networks in medical diagnosis. Int. J. Comput. Sci. Issues 2011, 8, 150–154. [Google Scholar]
- Amato, F.; López, A.; Peña-Méndez, E.M.; Vaňhara, P.; Hampl, A.; Havel, J. Artificial neural networks in medical diagnosis. J. Appl. Biomed. 2013, 11, 47–58. [Google Scholar] [CrossRef]
- Mohsen, A.A.; Alsurori, M.; Aldobai, B.; Mohsen, G.A. New Approach to Medical Diagnosis Using Artificial Neural Network and Decision Tree Algorithm: Application to Dental Diseases. Int. J. Inf. Eng. Electron. Bus. 2019, 11, 52–60. [Google Scholar] [CrossRef]
- Nakano, Y.; Takeshita, T.; Kamio, N.; Shiota, S.; Shibata, Y.; Suzuki, N.; Yoneda, M.; Hirofuji, T.; Yamashita, Y. Supervised machine learning-based classification of oral malodor based on the microbiota in saliva samples. Artif. Intell. Med. 2014, 60, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Barnes, V.M.; Kennedy, A.D.; Panagakos, F.; Devizio, W.; Trivedi, H.M.; Jonsson, T.; Guo, L.; Cervi, S.; Scannapieco, F.A. Global metabolomic analysis of human saliva and plasma from healthy and diabetic subjects, with and without periodontal disease. PLoS ONE 2014, 9, e105181. [Google Scholar] [CrossRef] [PubMed]

| Condition Putative Volatile Biomarker | Metabolic Context | Ref |
|---|---|---|
| Periodontal disease | ||
| pyridine and three methylpyridine isomers (picolines) | detected in patients but not in controls | [78] |
| hydrogen sulphide | oral bacteria infection | |
| methyl mercaptan | oral bacteria infection | |
| Halitosis | ||
| dimethyl disulphide | oral bacterial infection | [78] |
| dimethyl disulphide, carbon disulphide, VSCs | drug-related metabolism | [83] |
| VSCs | microbial degradation products of the sulphur-containing amino acids cysteine, cystine and methionine | [78] |
| VSCs | augmented levels detected upon anxiety challenge | [84] |
| VSCs, aliphatic amines, branched chain fatty acids, indole and phenol | oral bacteria metabolism | [77] |
| Putrescine, cadaverine, histamine, tyramine, indole, skatole, mercaptans and sulphides | microbial metabolism of proteinaceous substrates | [85] |
| 2,3-butanedione; 2,3-pentanedione; Phenol; pyrrole; indole and dimethyl disulphide | bacterial metabolism of lipids and carbohydrates | [80] |
| indole and skatole | bacterial fermentation products of tryptophan | [78] |
| phenol and p-cresol | bacterial putrefaction metabolites of phenolic amino acids | |
| Oral candidiasis | ||
| 3-methyl-2-butanone and styrene | Candida albicans infection | [86] |
| p-xylene, 2-octanone, 2-heptanone and n-butyl acetate | Candida krusei infection | |
| Oral cancer | ||
| 1,4-dichlorobenzene; 1,2-decanediol; 2,5-di-tert-butylphenol and E-3-decen-2-ol | identified in head and neck cancer cohorts | [87] |
| Dietary origin | ||
| 2-heptanone, benzaldehyde, dodecanal, 2-butyl-1-octanol, allyl isothiocyanate | examples of ketones, aldehydes, alcohols, esters and VSCs obtained from our diet | [80] |
| Oxidative stress | ||
| hexanal and nonanal | general markers for oxidative damage (endogenously produced from membrane lipid oxidation) | [80] |
| Environmental contaminants (air pollutants) | ||
| long-chain alkane derivatives (hexane, octane and undecane); aromatic compounds (as benzene, toluene, xylenes and styrene) | common air pollutants found in saliva | [78] |
| Experimental Layout/Condition | Relevant VOCs Identified | Ref |
|---|---|---|
| HS-SPME/GC-MS | ||
| Breast cancer | 3-methyl-pentanoic acid, 4-methyl-pentanoic acid, phenol and p-tert-butyl-phenol (Portuguese samples) and acetic, propanoic, benzoic acids, 1,2-decanediol, 2-decanone, and decanal (Indian samples) | [95] |
| Control subjects | twenty-one VOCs detected in saliva samples, mostly aldehydes | [122] |
| Halitosis and Submandibular Abscesses | 23 VOCs specific for halitosis and 41 for abscess | [123] |
| TFME-GC/MS | ||
| OSCC | Twelve salivary VOCs were characteristic of OSCC patients | [108] |
| HS-trap/GC-MS | ||
| Control subjects | 34 VOCs present in all samples analysed (n = 100) | [80] |
| SBSE-GC/MS | ||
| Control subjects | Excellent reproducibility for a wide range of salivary compounds, including alcohols, aldehydes, ketones, carboxylic acids, esters, amines, amides, lactones, and hydrocarbons | [81] |
| Control subjects | Comparison of individual and gender fingerprints using different biofluids (sweat, urine and saliva) | [111] |
| gas-diffusion flow injection analysis-GC/MS | ||
| acetaldehyde | [124] | |
| DCM extraction and derivatization followed by GC/MS analysis | ||
| women | 2-Nonenal-ovulatory specific salivary VOCs throughout menstrual cycle | [125] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, J.A.M.; Porto-Figueira, P.; Taware, R.; Sukul, P.; Rapole, S.; Câmara, J.S. Unravelling the Potential of Salivary Volatile Metabolites in Oral Diseases. A Review. Molecules 2020, 25, 3098. https://doi.org/10.3390/molecules25133098
Pereira JAM, Porto-Figueira P, Taware R, Sukul P, Rapole S, Câmara JS. Unravelling the Potential of Salivary Volatile Metabolites in Oral Diseases. A Review. Molecules. 2020; 25(13):3098. https://doi.org/10.3390/molecules25133098
Chicago/Turabian StylePereira, Jorge A. M., Priscilla Porto-Figueira, Ravindra Taware, Pritam Sukul, Srikanth Rapole, and José S. Câmara. 2020. "Unravelling the Potential of Salivary Volatile Metabolites in Oral Diseases. A Review" Molecules 25, no. 13: 3098. https://doi.org/10.3390/molecules25133098
APA StylePereira, J. A. M., Porto-Figueira, P., Taware, R., Sukul, P., Rapole, S., & Câmara, J. S. (2020). Unravelling the Potential of Salivary Volatile Metabolites in Oral Diseases. A Review. Molecules, 25(13), 3098. https://doi.org/10.3390/molecules25133098









