RNA Granules: A View from the RNA Perspective
Abstract
:1. Importance of RNA Granules
2. Types of RNA Granules and Their Functions
2.1. P-Bodies
2.2. Stress Granules
2.3. Germ Granules
2.3.1. Germ Granules of the Early Embryo
2.3.2. Nuage
3. Formation of RNA Granules: The Role of the RNA
3.1. The Role of Long and Ribosome-Free RNAs
3.2. The Role of RNA Structure
4. Enrichment of RNAs to Granules: RNA Could Self-Recruit
4.1. Mechanisms of Enrichment
4.2. Efficiency of Enrichment
4.3. Specificity of Enrichment
4.4. Conserved RNA Properties of Diverse RNA Granules
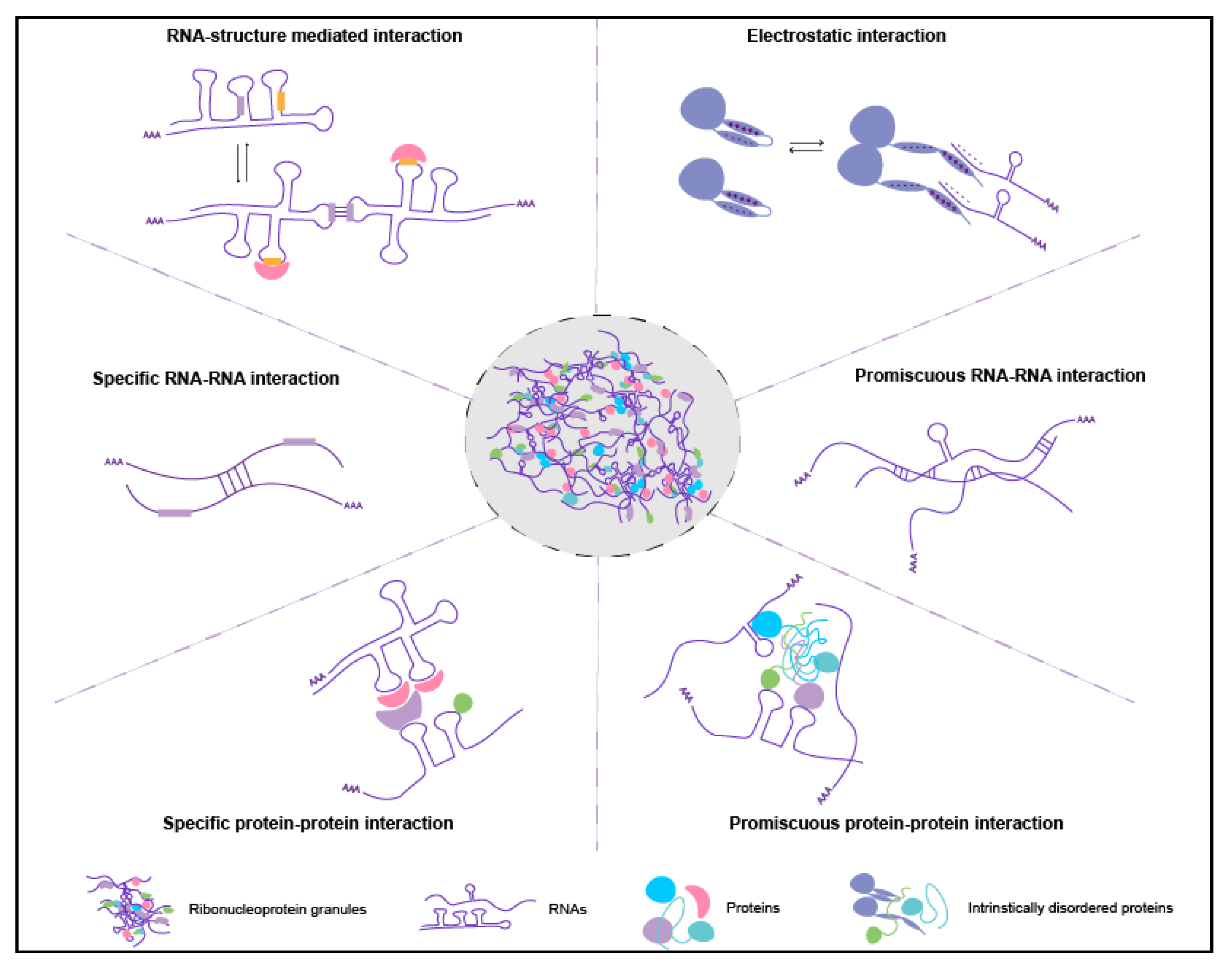
5. RNA Organization in RNA Granules: RNAs Self-Organize
5.1. Role of trans RNA-RNA Interactions
5.2. Homotypic mRNA Self-Assembly
5.3. Role of mRNA Self-Assemblies
6. Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hyman, A.A.; Weber, C.A.; Julicher, F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, S.; Parker, R. Hypo- and Hyper-Assembly Diseases of RNA-Protein Complexes. Trends Mol. Med. 2016, 22, 615–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Stefano, B.; Luo, E.C.; Haggerty, C.; Aigner, S.; Charlton, J.; Brumbaugh, J.; Ji, F.; Rabano Jimenez, I.; Clowers, K.J.; Huebner, A.J.; et al. The RNA Helicase DDX6 Controls Cellular Plasticity by Modulating P-Body Homeostasis. Cell Stem Cell 2019, 25, 622–638 e613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savas, J.N.; Makusky, A.; Ottosen, S.; Baillat, D.; Then, F.; Krainc, D.; Shiekhattar, R.; Markey, S.P.; Tanese, N. Huntington’s disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc. Natl. Acad. Sci. USA 2008, 105, 10820–10825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, P.; Kedersha, N.; Ivanov, P. Stress granules, P-bodies and cancer. Biochim. Biophys. Acta 2015, 1849, 861–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valiente-Echeverria, F.; Melnychuk, L.; Mouland, A.J. Viral modulation of stress granules. Virus Res. 2012, 169, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Trcek, T.; Lehmann, R. Germ Granules in Drosophila. Traffic 2019, 20, 650–660. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, J.P.T.; Folkmann, A.; Bernard, L.; Lee, C.Y.; Seroussi, U.; Charlesworth, A.G.; Claycomb, J.M.; Seydoux, G. P Granules Protect RNA Interference Genes from Silencing by piRNAs. Dev. Cell 2019, 50, 716–728 e716. [Google Scholar] [CrossRef]
- Sheth, U.; Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 2003, 300, 805–808. [Google Scholar] [CrossRef] [Green Version]
- Eulalio, A.; Behm-Ansmant, I.; Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007, 8, 9–22. [Google Scholar] [CrossRef]
- Standart, N.; Weil, D. P-Bodies: Cytosolic Droplets for Coordinated mRNA Storage. Trends Genet. 2018, 34, 612–626. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, V.K.; Jones, C.I.; Newbury, S.F.; Green, P.J. XRN 5′-->3′ exoribonucleases: Structure, mechanisms and functions. Biochim. Biophys. Acta 2013, 1829, 590–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira, D.; Sheth, U.; Valencia-Sanchez, M.A.; Brengues, M.; Parker, R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 2005, 11, 371–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cougot, N.; Babajko, S.; Seraphin, B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004, 165, 31–40. [Google Scholar] [CrossRef]
- Zheng, D.; Ezzeddine, N.; Chen, C.Y.; Zhu, W.; He, X.; Shyu, A.B. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 2008, 182, 89–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horvathova, I.; Voigt, F.; Kotrys, A.V.; Zhan, Y.; Artus-Revel, C.G.; Eglinger, J.; Stadler, M.B.; Giorgetti, L.; Chao, J.A. The Dynamics of mRNA Turnover Revealed by Single-Molecule Imaging in Single Cells. Mol. Cell 2017, 68, 615–625 e619. [Google Scholar] [CrossRef] [Green Version]
- Hubstenberger, A.; Courel, M.; Benard, M.; Souquere, S.; Ernoult-Lange, M.; Chouaib, R.; Yi, Z.; Morlot, J.B.; Munier, A.; Fradet, M.; et al. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol. Cell 2017, 68, 144–157 e145. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Schmich, F.; Srivatsa, S.; Weidner, J.; Beerenwinkel, N.; Spang, A. Context-dependent deposition and regulation of mRNAs in P-bodies. Elife 2018, 7, e29815. [Google Scholar] [CrossRef]
- Aizer, A.; Kalo, A.; Kafri, P.; Shraga, A.; Ben-Yishay, R.; Jacob, A.; Kinor, N.; Shav-Tal, Y. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J. Cell Sci. 2014, 127, 4443–4456. [Google Scholar] [CrossRef] [Green Version]
- Eulalio, A.; Behm-Ansmant, I.; Schweizer, D.; Izaurralde, E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell Biol. 2007, 27, 3970–3981. [Google Scholar] [CrossRef] [Green Version]
- Wilbertz, J.H.; Voigt, F.; Horvathova, I.; Roth, G.; Zhan, Y.; Chao, J.A. Single-Molecule Imaging of mRNA Localization and Regulation during the Integrated Stress Response. Mol. Cell 2019, 73, 946–958 e947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brengues, M.; Parker, R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 2592–2602. [Google Scholar] [CrossRef] [Green Version]
- Brengues, M.; Teixeira, D.; Parker, R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 2005, 310, 486–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lui, J.; Castelli, L.M.; Pizzinga, M.; Simpson, C.E.; Hoyle, N.P.; Bailey, K.L.; Campbell, S.G.; Ashe, M.P. Granules harboring translationally active mRNAs provide a platform for P-body formation following stress. Cell Rep. 2014, 9, 944–954. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.H.; Barbee, S.A.; Blankenship, J.T. GW-Bodies and P-Bodies Constitute Two Separate Pools of Sequestered Non-Translating RNAs. PLoS ONE 2016, 11, e0150291. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Stoecklin, G.; Ayodele, M.; Yacono, P.; Lykke-Andersen, J.; Fritzler, M.J.; Scheuner, D.; Kaufman, R.J.; Golan, D.E.; Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005, 169, 871–884. [Google Scholar] [CrossRef] [Green Version]
- Adivarahan, S.; Livingston, N.; Nicholson, B.; Rahman, S.; Wu, B.; Rissland, O.S.; Zenklusen, D. Spatial Organization of Single mRNPs at Different Stages of the Gene Expression Pathway. Mol. Cell 2018, 72, 727–738 e725. [Google Scholar] [CrossRef] [Green Version]
- Kedersha, N.; Cho, M.R.; Li, W.; Yacono, P.W.; Chen, S.; Gilks, N.; Golan, D.E.; Anderson, P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000, 151, 1257–1268. [Google Scholar] [CrossRef]
- Khong, A.; Matheny, T.; Jain, S.; Mitchell, S.F.; Wheeler, J.R.; Parker, R. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol. Cell 2017, 68, 808–820 e805. [Google Scholar] [CrossRef]
- Mahboubi, H.; Stochaj, U. Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 884–895. [Google Scholar] [CrossRef]
- White, J.P.; Lloyd, R.E. Regulation of stress granules in virus systems. Trends Microbiol. 2012, 20, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Tourriere, H.; Chebli, K.; Zekri, L.; Courselaud, B.; Blanchard, J.M.; Bertrand, E.; Tazi, J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003, 160, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Anderson, P. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002, 30, 963–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, S.; Wheeler, J.R.; Walters, R.W.; Agrawal, A.; Barsic, A.; Parker, R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016, 164, 487–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheeler, J.R.; Matheny, T.; Jain, S.; Abrisch, R.; Parker, R. Distinct stages in stress granule assembly and disassembly. Elife 2016, 5, e18413. [Google Scholar] [CrossRef]
- Wang, B.; Maxwell, B.A.; Joo, J.H.; Gwon, Y.; Messing, J.; Mishra, A.; Shaw, T.I.; Ward, A.L.; Quan, H.; Sakurada, S.M.; et al. ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97. Mol. Cell 2019, 74, 742–757 e748. [Google Scholar] [CrossRef]
- Namkoong, S.; Ho, A.; Woo, Y.M.; Kwak, H.; Lee, J.H. Systematic Characterization of Stress-Induced RNA Granulation. Mol. Cell 2018, 70, 175–187 e178. [Google Scholar] [CrossRef] [Green Version]
- Mateju, D.; Eichenberger, B.; Eglinger, J.; Roth, G.; Chao, J.A. Single-molecule imaging reveals translation of mRNAs localized to stress granules. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Kedersha, N.; Panas, M.D.; Achorn, C.A.; Lyons, S.; Tisdale, S.; Hickman, T.; Thomas, M.; Lieberman, J.; McInerney, G.M.; Ivanov, P.; et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 2016, 212, 845–860. [Google Scholar] [CrossRef] [Green Version]
- Hegner, R.W. Effects of removing germ-cell determinants from the eggs of some chrysomelid beetles. Preliminary Report. Biol. Bull. Mar. Biol. Lab. Woods Hole 1908, 16, 19–26. [Google Scholar] [CrossRef]
- Voronina, E.; Seydoux, G. The C. elegans homolog of nucleoporin Nup98 is required for the integrity and function of germline P granules. Development 2010, 137, 1441–1450. [Google Scholar] [CrossRef] [Green Version]
- Ephrussi, A.; Dickinson, L.K.; Lehmann, R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 1991, 66, 37–50. [Google Scholar] [CrossRef]
- Markussen, F.H.; Michon, A.M.; Breitwieser, W.; Ephrussi, A. Translational Control of Oskar Generates Short Osk, the Isoform That Induces Pole Plasm Assembly. Development 1995, 121, 3723–3732. [Google Scholar] [PubMed]
- Smith, J.; Calidas, D.; Schmidt, H.; Lu, T.; Rasoloson, D.; Seydoux, G. Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. Elife 2016, 5, e21337. [Google Scholar] [CrossRef] [PubMed]
- Bontems, F.; Stein, A.; Marlow, F.; Lyautey, J.; Gupta, T.; Mullins, M.C.; Dosch, R. Bucky ball organizes germ plasm assembly in zebrafish. Curr. Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef] [Green Version]
- Pitt, J.N.; Schisa, J.A.; Priess, J.R. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev. Biol. 2000, 219, 315–333. [Google Scholar] [CrossRef] [Green Version]
- Updike, D.L.; Hachey, S.J.; Kreher, J.; Strome, S. P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 2011, 192, 939–948. [Google Scholar] [CrossRef] [Green Version]
- Sheth, U.; Pitt, J.; Dennis, S.; Priess, J.R. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development 2010, 137, 1305–1314. [Google Scholar] [CrossRef] [Green Version]
- Trcek, T.; Grosch, M.; York, A.; Shroff, H.; Lionnet, T.; Lehmann, R. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat. Commun. 2015, 6, 7962. [Google Scholar] [CrossRef] [Green Version]
- Kistler, K.E.; Trcek, T.; Hurd, T.R.; Chen, R.; Liang, F.X.; Sall, J.; Kato, M.; Lehmann, R. Phase transitioned nuclear Oskar promotes cell division of Drosophila primordial germ cells. Elife 2018, 7, e37949. [Google Scholar] [CrossRef]
- Arkov, A.L.; Wang, J.Y.; Ramos, A.; Lehmann, R. The role of Tudor domains in germline development and polar granule architecture. Development 2006, 133, 4053–4062. [Google Scholar] [CrossRef] [Green Version]
- Thomson, T.; Lasko, P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis 2004, 40, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Mahowald, A.P. Fine Structure of Pole Cells and Polar Granules in Drosophila Melanogaster. J. Exp. Zool. 1962, 151, 201–215. [Google Scholar] [CrossRef]
- Voronina, E.; Seydoux, G.; Sassone-Corsi, P.; Nagamori, I. RNA Granules in Germ Cells. Cold Spring Harb. Perspect. Biol. 2011, 3, a002774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurd, T.R.; Herrmann, B.; Sauerwald, J.; Sanny, J.; Grosch, M.; Lehmann, R. Long Oskar Controls Mitochondrial Inheritance in Drosophila melanogaster. Dev. Cell 2016, 39, 560–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frise, E.; Hammonds, A.S.; Celniker, S.E. Systematic image-driven analysis of the spatial Drosophila embryonic expression landscape. Mol. Syst. Biol. 2010, 6, 345. [Google Scholar] [CrossRef]
- Lehmann, R. Germ Plasm Biogenesis-An Oskar-Centric Perspective. Curr. Top. Dev. Biol. 2016, 116, 679–707. [Google Scholar]
- Dahanukar, A.; Wharton, R.P. The Nanos gradient in Drosophila embryos is generated by translational regulation. Gene Dev. 1996, 10, 2610–2621. [Google Scholar] [CrossRef] [Green Version]
- Gavis, E.R.; Lunsford, L.; Bergsten, S.E.; Lehmann, R. A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development 1996, 122, 2791–2800. [Google Scholar]
- Smibert, C.A.; Wilson, J.E.; Kerr, K.; Macdonald, P.M. smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996, 10, 2600–2609. [Google Scholar] [CrossRef] [Green Version]
- Zaessinger, S.; Busseau, I.; Simonelig, M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 2006, 133, 4573–4583. [Google Scholar] [CrossRef] [Green Version]
- Asaoka-Taguchi, M.; Yamada, M.; Nakamura, A.; Hanyu, K.; Kobayashi, S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1999, 1, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, R.; Nusslein-Volhard, C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development 1991, 112, 679–691. [Google Scholar]
- Putnam, A.; Cassani, M.; Smith, J.; Seydoux, G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol. 2019, 26, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Julicher, F.; Hyman, A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef] [PubMed]
- Hanazawa, M.; Yonetani, M.; Sugimoto, A. PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. J. Cell Biol. 2011, 192, 929–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.S.; Putnam, A.; Lu, T.; He, S.; Ouyang, J.P.T.; Seydoux, G. Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. Elife 2020, 9, e52896. [Google Scholar] [CrossRef]
- Lee, C.S.; Lu, T.; Seydoux, G. Nanos promotes epigenetic reprograming of the germline by down-regulation of the THAP transcription factor LIN-15B. Elife 2017, 6, e30201. [Google Scholar] [CrossRef]
- Gallo, C.M.; Wang, J.T.; Motegi, F.; Seydoux, G. Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science 2010, 330, 1685–1689. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Fejes Toth, K.; Aravin, A.A. piRNA Biogenesis in Drosophila melanogaster. Trends Genet. 2017, 33, 882–894. [Google Scholar] [CrossRef] [Green Version]
- Kasper, D.M.; Gardner, K.E.; Reinke, V. Homeland security in the C. elegans germ line: Insights into the biogenesis and function of piRNAs. Epigenetics 2014, 9, 62–74. [Google Scholar] [CrossRef] [Green Version]
- Wan, G.; Fields, B.D.; Spracklin, G.; Shukla, A.; Phillips, C.M.; Kennedy, S. Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 2018, 557, 679–683. [Google Scholar] [CrossRef] [PubMed]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Choi, J.M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174, 688–699 e616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brangwynne, C.P. Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 2013, 203, 875–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, P.R.; Milin, A.N.; Moosa, M.M.; Onuchic, P.L.; Deniz, A.A. Reentrant Phase Transition Drives Dynamic Substructure Formation in Ribonucleoprotein Droplets. Angew. Chem. Int. Ed. Engl. 2017, 56, 11354–11359. [Google Scholar] [CrossRef]
- Maharana, S.; Wang, J.; Papadopoulos, D.K.; Richter, D.; Pozniakovsky, A.; Poser, I.; Bickle, M.; Rizk, S.; Guillen-Boixet, J.; Franzmann, T.M.; et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 2018, 360, 918–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ries, R.J.; Zaccara, S.; Klein, P.; Olarerin-George, A.; Namkoong, S.; Pickering, B.F.; Patil, D.P.; Kwak, H.; Lee, J.H.; Jaffrey, S.R. m(6)A enhances the phase separation potential of mRNA. Nature 2019, 571, 424–428. [Google Scholar] [CrossRef]
- Langdon, E.M.; Qiu, Y.; Ghanbari Niaki, A.; McLaughlin, G.A.; Weidmann, C.A.; Gerbich, T.M.; Smith, J.A.; Crutchley, J.M.; Termini, C.M.; Weeks, K.M.; et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science 2018, 360, 922–927. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Elbaum-Garfinkle, S.; Langdon, E.M.; Taylor, N.; Occhipinti, P.; Bridges, A.A.; Brangwynne, C.P.; Gladfelter, A.S. RNA Controls PolyQ Protein Phase Transitions. Mol. Cell 2015, 60, 220–230. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, J.C.; Wang, X.; Podell, E.R.; Cech, T.R. RNA seeds higher-order assembly of FUS protein. Cell Rep. 2013, 5, 918–925. [Google Scholar] [CrossRef] [Green Version]
- Fuller, G.G.; Han, T.; Freeberg, M.A.; Moresco, J.J.; Ghanbari Niaki, A.; Roach, N.P.; Yates, J.R., 3rd; Myong, S.; Kim, J.K. RNA promotes phase separation of glycolysis enzymes into yeast G bodies in hypoxia. Elife 2020, 9, e48480. [Google Scholar] [CrossRef] [PubMed]
- Rhine, K.; Vidaurre, V.; Myong, S. RNA Droplets. Annu. Rev. Biophys. 2020, 49, 247–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, N.; Buchan, J.R. RPS28B mRNA acts as a scaffold promoting cis-translational interaction of proteins driving P-body assembly. Nucleic Acids Res. 2020, 48, 6265–6279. [Google Scholar] [CrossRef] [PubMed]
- Trcek, T.; Douglas, T.E.; Grosch, M.; Yin, Y.; Eagle, W.V.I.; Gavis, E.R.; Shroff, H.; Rothenberg, E.; Lehmann, R. Sequence-Independent Self-Assembly of Germ Granule mRNAs into Homotypic Clusters. Mol. Cell 2020, 78, 941–950.e1. [Google Scholar] [CrossRef]
- Ma, W.; Zhen, G.; Xie, W.; Mayr, C. Unstructured mRNAs form multivalent RNA-RNA interactions to generate TIS granule networks. Biorxiv 2020. [Google Scholar] [CrossRef]
- Rangan, P.; DeGennaro, M.; Jaime-Bustamante, K.; Coux, R.X.; Martinho, R.G.; Lehmann, R. Temporal and spatial control of germ-plasm RNAs. Curr. Biol. 2009, 19, 72–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Chan, C.Y.; Lawrence, C.E. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA 2005, 11, 1157–1166. [Google Scholar] [CrossRef] [Green Version]
- Van Treeck, B.; Protter, D.S.W.; Matheny, T.; Khong, A.; Link, C.D.; Parker, R. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. USA 2018, 115, 2734–2739. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg, H.; Felsenfeld, G. Studies of the temperature-dependent conformation and phase separation of polyriboadenylic acid solutions at neutral pH. J. Mol. Biol. 1967, 30, 17–37. [Google Scholar] [CrossRef]
- Jain, A.; Vale, R.D. RNA phase transitions in repeat expansion disorders. Nature 2017, 546, 243–247. [Google Scholar] [CrossRef] [Green Version]
- Tauber, D.; Tauber, G.; Khong, A.; Van Treeck, B.; Pelletier, J.; Parker, R. Modulation of RNA Condensation by the DEAD-Box Protein eIF4A. Cell 2020, 180, 411–426. [Google Scholar] [CrossRef]
- Holmstrom, E.D.; Liu, Z.; Nettels, D.; Best, R.B.; Schuler, B. Disordered RNA chaperones can enhance nucleic acid folding via local charge screening. Nat. Commun. 2019, 10, 2453. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Mathieu, C.; Kolaitis, R.M.; Zhang, P.; Messing, J.; Yurtsever, U.; Yang, Z.; Wu, J.; Li, Y.; Pan, Q.; et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345 e328. [Google Scholar] [CrossRef] [PubMed]
- Sanchez de Groot, N.; Armaos, A.; Grana-Montes, R.; Alriquet, M.; Calloni, G.; Vabulas, R.M.; Tartaglia, G.G. RNA structure drives interaction with proteins. Nat. Commun. 2019, 10, 3246. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; He, C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013, 29, 108–115. [Google Scholar] [CrossRef] [Green Version]
- Spitale, R.C.; Flynn, R.A.; Zhang, Q.C.; Crisalli, P.; Lee, B.; Jung, J.W.; Kuchelmeister, H.Y.; Batista, P.J.; Torre, E.A.; Kool, E.T.; et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 2015, 519, 486–490. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Zhuang, X. m6A-binding YTHDF proteins promote stress granule formation by modulating phase separation of stress granule proteins. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Anders, M.; Chelysheva, I.; Goebel, I.; Trenkner, T.; Zhou, J.; Mao, Y.; Verzini, S.; Qian, S.B.; Ignatova, Z. Dynamic m(6)A methylation facilitates mRNA triaging to stress granules. Life Sci. Alliance 2018, 1, e201800113. [Google Scholar] [CrossRef] [Green Version]
- Kan, L.; Grozhik, A.V.; Vedanayagam, J.; Patil, D.P.; Pang, N.; Lim, K.S.; Huang, Y.C.; Joseph, B.; Lin, C.J.; Despic, V.; et al. The m(6)A pathway facilitates sex determination in Drosophila. Nat. Commun. 2017, 8, 15737. [Google Scholar] [CrossRef]
- Forrest, K.M.; Clark, I.E.; Jain, R.A.; Gavis, E.R. Temporal complexity within a translational control element in the nanos mRNA. Development 2004, 131, 5849–5857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padron, A.; Iwasaki, S.; Ingolia, N.T. Proximity RNA Labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules. Mol. Cell 2019, 75, 875–887 e875. [Google Scholar] [CrossRef]
- Pitchiaya, S.; Mourao, M.D.A.; Jalihal, A.P.; Xiao, L.; Jiang, X.; Chinnaiyan, A.M.; Schnell, S.; Walter, N.G. Dynamic Recruitment of Single RNAs to Processing Bodies Depends on RNA Functionality. Mol. Cell 2019, 74, 521–533 e526. [Google Scholar] [CrossRef] [PubMed]
- Gavis, E.R.; Lehmann, R. Translational regulation of nanos by RNA localization. Nature 1994, 369, 315–318. [Google Scholar] [CrossRef]
- Moon, S.L.; Morisaki, T.; Khong, A.; Lyon, K.; Parker, R.; Stasevich, T.J. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol. 2019, 21, 162–168. [Google Scholar] [CrossRef]
- Andrei, M.A.; Ingelfinger, D.; Heintzmann, R.; Achsel, T.; Rivera-Pomar, R.; Luhrmann, R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 2005, 11, 717–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadyrova, L.Y.; Habara, Y.; Lee, T.H.; and Wharton, R.P. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 2007, 134, 1519–1527. [Google Scholar] [CrossRef] [Green Version]
- Clark, I.E.; Wyckoff, D.; Gavis, E.R. Synthesis of the posterior determinant Nanos is spatially restricted by a novel cotranslational regulatory mechanism. Curr. Biol. 2000, 10, 1311–1314. [Google Scholar] [CrossRef] [Green Version]
- Bergsten, S.E.; Gavis, E.R. Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development 1999, 126, 659–669. [Google Scholar] [PubMed]
- Gavis, E.R.; Curtis, D.; Lehmann, R. Identification of cis-acting sequences that control nanos RNA localization. Dev. Biol. 1996, 176, 36–50. [Google Scholar] [CrossRef] [Green Version]
- Matheny, T.; Rao, B.S.; Parker, R. Transcriptome-Wide Comparison of Stress Granules and P-Bodies Reveals that Translation Plays a Major Role in RNA Partitioning. Mol. Cell Biol. 2019, 39. [Google Scholar] [CrossRef]
- Thomsen, S.; Anders, S.; Janga, S.C.; Huber, W.; Alonso, C.R. Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol. 2010, 11, R93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baugh, L.R.; Hill, A.A.; Slonim, D.K.; Brown, E.L.; Hunter, C.P. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 2003, 130, 889–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Gao, M.; Huynh, N.; Tindell, S.J.; Vo, H.D.; McDonald, W.H.; Arkov, A.L. In vivo mapping of a dynamic ribonucleoprotein granule interactome in early Drosophila embryos. FEBS Open Bio. 2016, 6, 1248–1256. [Google Scholar] [CrossRef] [PubMed]
- Jeske, M.; Bordi, M.; Glatt, S.; Muller, S.; Rybin, V.; Muller, C.W.; Ephrussi, A. The Crystal Structure of the Drosophila Germline Inducer Oskar Identifies Two Domains with Distinct Vasa Helicase- and RNA-Binding Activities. Cell Rep. 2015, 12, 587–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, N.; Yu, Z.; Hu, M.; Wang, M.; Lehmann, R.; Xu, R.M. Structure of Drosophila Oskar reveals a novel RNA binding protein. Proc. Natl. Acad. Sci. USA 2015, 112, 11541–11546. [Google Scholar] [CrossRef] [Green Version]
- Niepielko, M.G.; Eagle, W.V.I.; Gavis, E.R. Stochastic Seeding Coupled with mRNA Self-Recruitment Generates Heterogeneous Drosophila Germ Granules. Curr. Biol. 2018, 28, 1872–1881 e1873. [Google Scholar] [CrossRef] [Green Version]
- Al-Husini, N.; Tomares, D.T.; Pfaffenberger, Z.J.; Muthunayake, N.S.; Samad, M.A.; Zuo, T.; Bitar, O.; Aretakis, J.R.; Bharmal, M.M.; Gega, A.; et al. BR-Bodies Provide Selectively Permeable Condensates that Stimulate mRNA Decay and Prevent Release of Decay Intermediates. Mol. Cell 2020, 78, 670–682 e678. [Google Scholar] [CrossRef]
- Antar, L.N.; Dictenberg, J.B.; Plociniak, M.; Afroz, R.; Bassell, G.J. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005, 4, 350–359. [Google Scholar] [CrossRef]
- De Diego Otero, Y.; Severijnen, L.A.; van Cappellen, G.; Schrier, M.; Oostra, B.; Willemsen, R. Transport of fragile X mental retardation protein via granules in neurites of PC12 cells. Mol. Cell Biol. 2002, 22, 8332–8341. [Google Scholar] [CrossRef] [Green Version]
- Greenblatt, E.J.; Spradling, A.C. Fragile X mental retardation 1 gene enhances the translation of large autism-related proteins. Science 2018, 361, 709–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.; Mayr, C. A Membraneless Organelle Associated with the Endoplasmic Reticulum Enables 3’UTR-Mediated Protein-Protein Interactions. Cell 2018, 175, 1492–1506 e1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souquere, S.; Mollet, S.; Kress, M.; Dautry, F.; Pierron, G.; Weil, D. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci. 2009, 122, 3619–3626. [Google Scholar] [CrossRef] [Green Version]
- Little, S.C.; Sinsimer, K.S.; Lee, J.J.; Wieschaus, E.F.; Gavis, E.R. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat. Cell Biol. 2015, 17, 558–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrandon, D.; Koch, I.; Westhof, E.; Nusslein-Volhard, C. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 1997, 16, 1751–1758. [Google Scholar] [CrossRef] [Green Version]
- Jambor, H.; Brunel, C.; Ephrussi, A. Dimerization of oskar 3′ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA 2011, 17, 2049–2057. [Google Scholar] [CrossRef] [Green Version]
- Clever, J.L.; Wong, M.L.; Parslow, T.G. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 1996, 70, 5902–5908. [Google Scholar] [CrossRef] [Green Version]
- Marquet, R.; Baudin, F.; Gabus, C.; Darlix, J.L.; Mougel, M.; Ehresmann, C.; Ehresmann, B. Dimerization of human immunodeficiency virus (type 1) RNA: Stimulation by cations and possible mechanism. Nucleic Acids Res. 1991, 19, 2349–2357. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Agarwal, S.; Agarwal, D.; and Phadke, S.R. Myotonic dystrophy type 1 (DM1): A triplet repeat expansion disorder. Gene 2013, 522, 226–230. [Google Scholar] [CrossRef]
- Mateos-Aierdi, A.J.; Goicoechea, M.; Aiastui, A.; Fernandez-Torron, R.; Garcia-Puga, M.; Matheu, A.; Lopez de Munain, A. Muscle wasting in myotonic dystrophies: A model of premature aging. Front. Aging Neurosci. 2015, 7, 125. [Google Scholar] [CrossRef]
- Bou-Nader, C.; Zhang, J. Structural Insights into RNA Dimerization: Motifs, Interfaces and Functions. Molecules 2020, 25, 2881. [Google Scholar] [CrossRef]
- Ennifar, E.; Walter, P.; Ehresmann, B.; Ehresmann, C.; Dumas, P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 2001, 8, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Schulz, E.C.; Seiler, M.; Zuliani, C.; Voigt, F.; Rybin, V.; Pogenberg, V.; Mucke, N.; Wilmanns, M.; Gibson, T.J.; Barabas, O. Intermolecular base stacking mediates RNA-RNA interaction in a crystal structure of the RNA chaperone Hfq. Sci. Rep. 2017, 7, 9903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lech, C.J.; Heddi, B.; Phan, A.T. Guanine base stacking in G-quadruplex nucleic acids. Nucleic Acids Res. 2013, 41, 2034–2046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 1–16. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, M.; Duncan, S.; Yang, X.; Abdelhamid, M.A.S.; Huang, L.; Zhang, H.; Benfey, P.N.; Waller, Z.A.E.; Ding, Y. G-quadruplex structures trigger RNA phase separation. Nucleic Acids Res. 2019, 47, 11746–11754. [Google Scholar] [CrossRef]
- Fay, M.M.; Anderson, P.J.; Ivanov, P. ALS/FTD-Associated C9ORF72 Repeat RNA Promotes Phase Transitions In Vitro and in Cells. Cell Rep. 2017, 21, 3573–3584. [Google Scholar] [CrossRef] [Green Version]
- Van Treeck, B.; Parker, R. Emerging Roles for Intermolecular RNA-RNA Interactions in RNP Assemblies. Cell 2018, 174, 791–802. [Google Scholar] [CrossRef] [Green Version]
- Khong, A.; Parker, R. mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J. Cell Biol. 2018, 217, 4124–4140. [Google Scholar] [CrossRef] [Green Version]
- Adams, M.D.; Celniker, S.E.; Holt, R.A.; Evans, C.A.; Gocayne, J.D.; Amanatides, P.G.; Scherer, S.E.; Li, P.W.; Hoskins, R.A.; Galle, R.F.; et al. The genome sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef] [Green Version]
- Nott, T.J.; Craggs, T.D.; Baldwin, A.J. Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat. Chem. 2016, 8, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Roovers, E.F.; Kaaij, L.J.T.; Redl, S.; Bronkhorst, A.W.; Wiebrands, K.; de Jesus Domingues, A.M.; Huang, H.Y.; Han, C.T.; Riemer, S.; Dosch, R.; et al. Tdrd6a Regulates the Aggregation of Buc into Functional Subcellular Compartments that Drive Germ Cell Specification. Dev. Cell 2018, 46, 285–301 e289. [Google Scholar] [CrossRef] [Green Version]
- Eno, C.; Hansen, C.L.; Pelegri, F. Aggregation, segregation, and dispersal of homotypic germ plasm RNPs in the early zebrafish embryo. Dev. Dyn. 2019, 248, 306–318. [Google Scholar] [CrossRef]
- Weil, T.T.; Parton, R.M.; Herpers, B.; Soetaert, J.; Veenendaal, T.; Xanthakis, D.; Dobbie, I.M.; Halstead, J.M.; Hayashi, R.; Rabouille, C.; et al. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat. Cell Biol. 2012, 14, 1305–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bashirullah, A.; Halsell, S.R.; Cooperstock, R.L.; Kloc, M.; Karaiskakis, A.; Fisher, W.W.; Fu, W.; Hamilton, J.K.; Etkin, L.D.; Lipshitz, H.D. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J. 1999, 18, 2610–2620. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Protter, D.S.; Rosen, M.K.; Parker, R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell 2015, 60, 208–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, S.; Weber, C.A.; Nousch, M.; Adame-Arana, O.; Hoege, C.; Hein, M.Y.; Osborne-Nishimura, E.; Mahamid, J.; Jahnel, M.; Jawerth, L.; et al. Polar Positioning of Phase-Separated Liquid Compartments in Cells Regulated by an mRNA Competition Mechanism. Cell 2016, 166, 1572–1584 e1516. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| RNA Granule | Enriched RNAs (Examples) | Depleted RNAs (Examples) | Prefer Long mRNAs? | Prefer Translationally Repressed mRNAs? |
|---|---|---|---|---|
| P-bodies | Translationally repressed mRNAs [17] | 18S and 28S rRNAs [17] | N/A | Yes |
| Stress granules | mRNAs with longer coding regions and 3′UTRs [29,37] lncRNAs [29] 18S rRNA [124] | Membrane-associated mRNAs [29] gapdh [29] 28S rRNA [124] | Yes | Yes |
| P granules (C. elegans) | Long mRNAs with low ribosome occupancy [67] Germ-cell specific mRNAs [67] | N/A | Yes | Yes |
| Polar granules (D. melanogaster) | Germ-cell specific mRNAs [49,87,125] | oskar [49,125] | N/A | Yes, but with exceptions [58,59,87,105,108,109,110,111] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, S.; Curnutte, H.A.; Trcek, T. RNA Granules: A View from the RNA Perspective. Molecules 2020, 25, 3130. https://doi.org/10.3390/molecules25143130
Tian S, Curnutte HA, Trcek T. RNA Granules: A View from the RNA Perspective. Molecules. 2020; 25(14):3130. https://doi.org/10.3390/molecules25143130
Chicago/Turabian StyleTian, Siran, Harrison A. Curnutte, and Tatjana Trcek. 2020. "RNA Granules: A View from the RNA Perspective" Molecules 25, no. 14: 3130. https://doi.org/10.3390/molecules25143130
APA StyleTian, S., Curnutte, H. A., & Trcek, T. (2020). RNA Granules: A View from the RNA Perspective. Molecules, 25(14), 3130. https://doi.org/10.3390/molecules25143130







