Monitoring Hydroxycinnamic Acid Decarboxylation by Lactic Acid Bacteria Using High-Throughput UV-Vis Spectroscopy
Abstract
:1. Introduction
2. Results and Discussion
2.1. UV-Vis Spectra of HCAs, Decarboxylated HCAs and Growth Medium
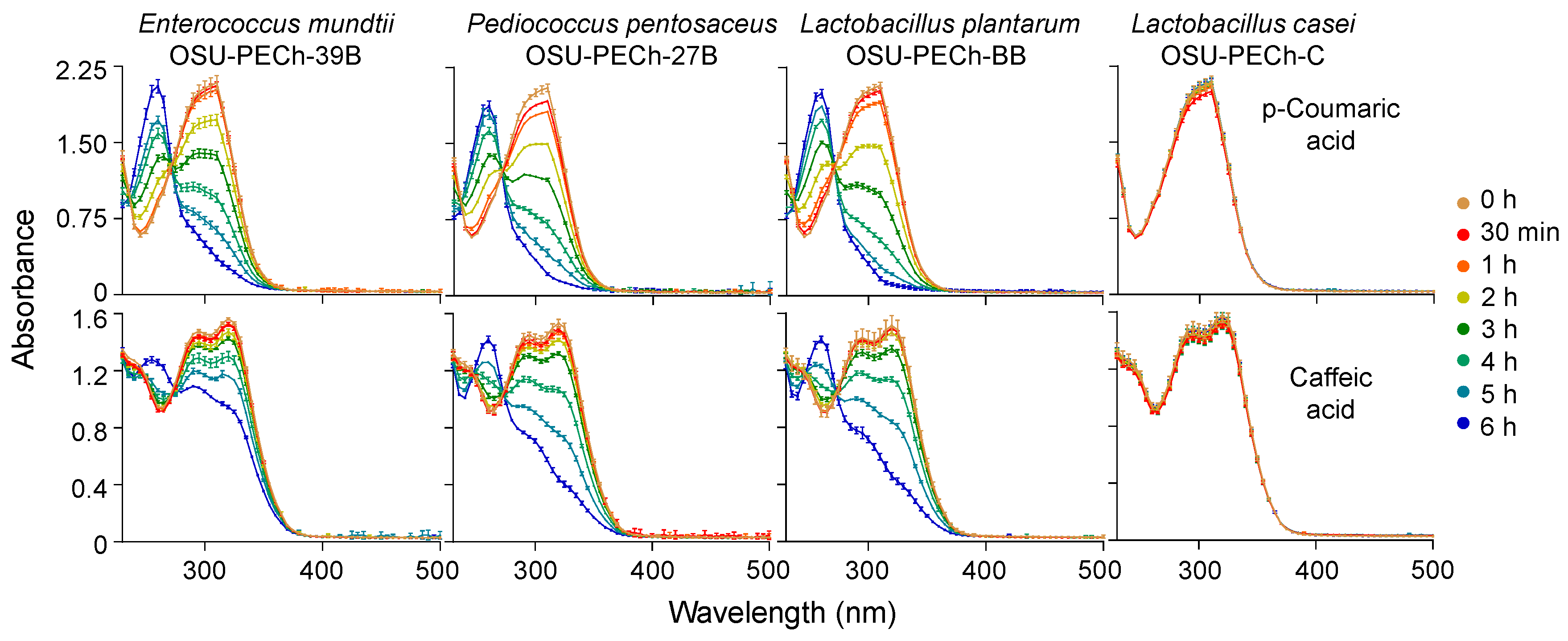
2.2. Determination of the Optimal pH for Monitoring HCA Decarboxylation
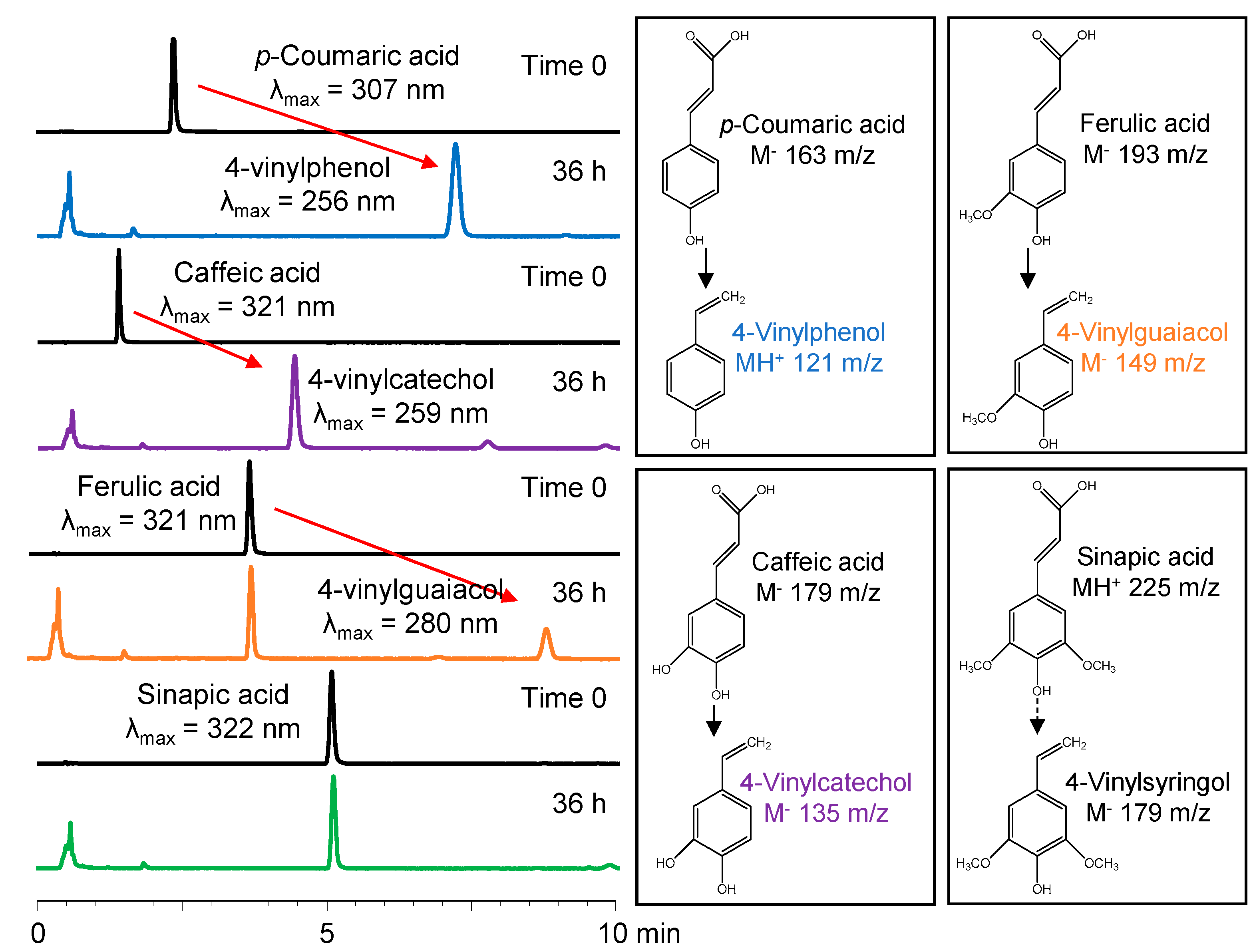
2.3. HPLC-DAD-ESI-MS Analysis of Decarboxylated Products
2.4. Screening of the OSU-PECh LAB Collection for Their Ability to Decarboxylate HCAs
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Growth Medium Selection for HCA Decarboxylation Experiments
3.3. Bacterial Strain Selection
3.4. Decarboxylation Experiments
3.5. Rapid, High-Throughput Screening of Hydroxycinnamic acid Degradation by LAB Using UV-Vis Spectroscopy and Selection of Optimal pH for Measurement
3.6. uHPLC-DAD-ESI-MS Analysis of HCAs and Degradation Products
3.7. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pereira, D.M.; Valentão, P.; Pereira, J.A.; Andrade, P.B. Phenolics: From chemistry to biology. Molecules 2009, 14, 2202–2211. [Google Scholar] [CrossRef]
- Herald, P.J.; Davidson, P.M. Antibacterial activity of selected hydroxycinnamic acids. J. Food Sci. 1983, 48, 1378–1379. [Google Scholar] [CrossRef]
- Borges, A.; Ferreira, C.; Saavedra, M.J.; Simões, M. Antibacterial activity and mode of action of ferulic and gallic acids against pathogenic bacteria. Microb. Drug Resist. 2013, 19, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Lou, Z.; Wang, H.; Rao, S.; Sun, J.; Ma, C.; Li, J. p-Coumaric acid kills bacteria through dual damage mechanisms. Food Control 2012, 25, 550–554. [Google Scholar] [CrossRef]
- Sánchez-Maldonado, A.F.; Schieber, A.; Gänzle, M.G. Structure-function relationships of the antibacterial activity of phenolic acids and their metabolism by lactic acid bacteria. J. Appl. Microbiol. 2011, 111, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Couto, J.A.; Campos, F.M.; Figueiredo, A.R.; Hogg, T.A. Ability of lactic acid bacteria to produce volatile phenols. Am. J. Enol. Vitic. 2006, 57, 166–171. [Google Scholar]
- Shinohara, T.; Kubodera, S.; Yanagida, F. Distribution of phenolic yeasts and production of phenolic off-flavors in wine fermentation. J. Biosci. Bioeng. 2000, 90, 90–97. [Google Scholar] [CrossRef]
- Langos, D.; Granvogl, M. Studies on the simultaneous formation of aroma-active and toxicologically relevant vinyl aromatics from free phenolic acids during wheat beer brewing. J. Agric. Food Chem. 2016, 64, 2325–2332. [Google Scholar] [CrossRef]
- Wang, H.E.; Hwang, C.F.; Tzeng, Y.M.; Hwang, W.Z.; Mau, J.L. Quality of white bread made from lactic acid bacteria-enriched dough. J. Food Process. Preserv. 2012, 36, 553–559. [Google Scholar] [CrossRef]
- Benito, S.; Morata, A.; Palomero, F.; González, M.C.; Suárez-Lepe, J.A. Formation of vinylphenolic pyranoanthocyanins by Saccharomyces cerevisiae and Pichia guillermondii in red wines produced following different fermentation strategies. Food Chem. 2011, 124, 15–23. [Google Scholar] [CrossRef]
- Farr, J.E.; Giusti, M.M. Investigating the interaction of ascorbic acid with anthocyanins and pyranoanthocyanins. Molecules 2018, 23, 744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Freitas, V.; Mateus, N. Formation of pyranoanthocyanins in red wines: A new and diverse class of anthocyanin derivatives. Anal. Bioanal. Chem. 2011, 401, 1467–1477. [Google Scholar] [CrossRef] [PubMed]
- Oh, N.S.; Lee, J.Y.; Joung, J.Y.; Kim, K.S.; Shin, Y.K.; Lee, K.W.; Kim, S.H.; Oh, S.; Kim, Y. Microbiological characterization and functionality of set-type yogurt fermented with potential prebiotic substrates Cudrania tricuspidata and Morus alba L. leaf extracts. J. Dairy Sci. 2016, 99, 6014–6025. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, S.; Straniero, R.; Pacifico, S.; Aguzzi, A.; Virgili, F. French marine bark extract pycnogenol as a possible enrichment ingredient for yogurt. J. Dairy Sci. 2008, 91, 4484–4491. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.T.; Nadeem, M.; Imran, M.; Ajmal, M.; Ali, S. Antioxidant activity, fatty acids characterization and oxidative stability of Gouda cheese fortified with mango (Mangifera indica L.) kernel fat. J. Food Sci. Technol. 2018, 55, 992–1002. [Google Scholar] [CrossRef] [PubMed]
- Karaaslan, M.; Ozden, M.; Vardin, H.; Turkoglu, H. Phenolic fortification of yogurt using grape and callus extracts. Lwt Food Sci. Technol. 2011, 44, 1065–1072. [Google Scholar] [CrossRef]
- Coïsson, J.D.; Travaglia, F.; Piana, G.; Capasso, M.; Arlorio, M. Euterpe oleracea juice as a functional pigment for yogurt. Food Res. Int. 2005, 38, 893–897. [Google Scholar] [CrossRef]
- Curiel, J.A.; Rodríguez, H.; Landete, J.M.; de las Rivas, B.; Muñoz, R. Ability of Lactobacillus brevis strains to degrade food phenolic acids. Food Chem. 2010, 120, 225–229. [Google Scholar] [CrossRef] [Green Version]
- Thurston, P.A.; Tubb, R.S. Screening yeast strains for their ability to produce phenolic off-flavours: A simple method for determining phenols in wort and beer. J. Inst. Brew. 1981, 87, 177–179. [Google Scholar] [CrossRef]
- Cavin, J.F.; Andioc, V.; Etievant, X.; Diviès, C. Ability of wine lactic acid bacteria to metabolize phenol carboxylic acids. Am. J. Enol. Vitic. 1993, 44, 76–80. [Google Scholar]
- Robbins, R.J. Phenolic acids in foods: An overview of analytical methodology. J. Agric. Food Chem. 2003, 51, 2866–2887. [Google Scholar] [CrossRef]
- Ong, B.Y.; Nagel, C.W. High-pressure liquid chromatographyic analysis of hydroxycinnamic acid-tartaric acid esters and their glucose esters in vitis vinifera. J. Chromatogr. A 1978, 157, 345–355. [Google Scholar] [CrossRef]
- Erdemgil, F.Z.; Şanli, S.; Şanli, N.; Özkan, G.; Barbosa, J.; Guiteras, J.; Beltrán, J.L. Determination of pKa values of some hydroxylated benzoic acids in methanol-water binary mixtures by LC methodology and potentiometry. Talanta 2007, 72, 489–496. [Google Scholar] [CrossRef]
- Sanli, N.; Fonrodona, G.; Barrón, D.; Özkan, G.; Barbosa, J. Prediction of chromatographic retention, pKa values and optimization of the separation of polyphenolic acids in strawberries. J. Chromatogr. A 2002, 975, 299–309. [Google Scholar] [CrossRef]
- De Las Rivas, B.; Rodríguez, H.; Curiel, J.A.; Landete, J.M.; Muñoz, R. Molecular screening of wine lactic acid bacteria degrading hydroxycinnamic acids. J. Agric. Food Chem. 2009, 57, 490–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fras, P.; Campos, F.M.; Hogg, T.; Couto, J.A. Production of volatile phenols by Lactobacillus plantarum in wine conditions. Biotechnol. Lett. 2014, 36, 281–285. [Google Scholar] [CrossRef]
- Edlin, D.A.N.; Narbad, A.; Gasson, M.J.; Dickinson, J.R.; Lloyd, D. Purification and characterization of hydroxycinnamate decarboxylase from Brettanomyces anomalus. Enzym. Microb. Technol. 1998, 22, 232–239. [Google Scholar] [CrossRef]
- Ripari, V.; Bai, Y.; Gänzle, M.G. Metabolism of phenolic acids in whole wheat and rye malt sourdoughs. Food Microbiol. 2019, 77, 43–51. [Google Scholar] [CrossRef]
- Nsogning Dongmo, S.; Sacher, B.; Kollmannsberger, H.; Becker, T. Key volatile aroma compounds of lactic acid fermented malt based beverages–impact of lactic acid bacteria strains. Food Chem. 2017, 229, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, H.; Angulo, I.; De Rivas, B.L.; Campillo, N.; Páez, J.A.; Muñoz, R.; Mancheño, J.M. p-Coumaric acid decarboxylase from Lactobacillus plantarum: Structural insights into the active site and decarboxylaction catalytic mechanism. Proteins 2010, 78, 1662–1676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha-Mendoza, D.; Kosmerl, E.; Miyagusuku-Cruzado, G.; Giusti, M.M.; Jiménez-Flores, R.; García-Cano, I. Growth of lactic acid bacteria in milk phospholipids enhances their adhesion to Caco-2 cells. J. Dairy Sci. 2020, in press. [Google Scholar] [CrossRef]
- García-Cano, I.; Rocha-Mendoza, D.; Ortega-Anaya, J.; Wang, K.; Kosmerl, E.; Jiménez-Flores, R. Lactic acid bacteria isolated from dairy products as potential producers of lipolytic, proteolytic and antibacterial proteins. Appl. Microbiol. Biotechnol. 2019, 103, 5243–5257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, 61–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavin, J.F.; Dartois, V.; Diviès, C. Gene cloning, transcriptional analysis, purification, and characterization of phenolic acid decarboxylase from Bacillus subtilis. Appl. Env. Microbiol. 1998, 64, 1466–1471. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |



| Lactic Acid Bacteria Strain | Accession Code | pCA | CA | FA | SA |
|---|---|---|---|---|---|
| Enterococcus mundtii | OSU-PECh-39B | (++) | (++) | (+) | (−) |
| Lactobacillus acidophilus | OSU-PECh-LA5 | (++) | (−) | (−) | (−) |
| Lactobacillus helveticus | OSU-PECh-25 | (−) | (−) | (−) | (−) |
| OSU-PECh-26 | (−) | (−) | (−) | (−) | |
| OSU-PECh-33 | (−) | (−) | (−) | (−) | |
| OSU-PECh-40 | (−) | (−) | (−) | (−) | |
| OSU-PECh-57B | (−) | (−) | (−) | (−) | |
| OSU-PECh-60 | (−) | (−) | (−) | (−) | |
| OSU-PECh-LH1B | (++) | (−) | (−) | (−) | |
| OSU-PECh-LH4A | (−) | (−) | (−) | (−) | |
| OSU-PECh-LH7 | (++) | (++) | (−) | (−) | |
| OSU-PECh-LH15A | (−) | (−) | (−) | (−) | |
| OSU-PECh-LH19 | (−) | (++) | (−) | (−) | |
| Lactobacillus pentosus | OSU-PECh-LP6C | (++) | (++) | (−) | (−) |
| Lactobacillus plantarum | OSU-PECh-A | (++) | (++) | (−) | (−) |
| OSU-PECh-BB | (++) | (++) | (+) | (−) | |
| Lactobacillus rhamnosus | OSU-PECh-24 | (−) | (−) | (−) | (−) |
| Pediococcus acidilactici | OSU-PECh-PA3A | (++) | (++) | (−) | (−) |
| OSU-PECh-PAL | (−) | (−) | (−) | (−) | |
| Pediococcus pentosaceus | OSU-PECh-27B | (++) | (++) | (+) | (−) |
| OSU-PECh-PP6A | (++) | (++) | (−) | (−) | |
| OSU-PECh-PP13 | (++) | (++) | (−) | (−) | |
| Lactobacillus casei | OSU-PECh-C | (−) | (−) | (−) | (−) |
| Staphylococcus epidermidis | ATCC 1222 | (−) | (−) | (−) | (−) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miyagusuku-Cruzado, G.; García-Cano, I.; Rocha-Mendoza, D.; Jiménez-Flores, R.; Giusti, M.M. Monitoring Hydroxycinnamic Acid Decarboxylation by Lactic Acid Bacteria Using High-Throughput UV-Vis Spectroscopy. Molecules 2020, 25, 3142. https://doi.org/10.3390/molecules25143142
Miyagusuku-Cruzado G, García-Cano I, Rocha-Mendoza D, Jiménez-Flores R, Giusti MM. Monitoring Hydroxycinnamic Acid Decarboxylation by Lactic Acid Bacteria Using High-Throughput UV-Vis Spectroscopy. Molecules. 2020; 25(14):3142. https://doi.org/10.3390/molecules25143142
Chicago/Turabian StyleMiyagusuku-Cruzado, Gonzalo, Israel García-Cano, Diana Rocha-Mendoza, Rafael Jiménez-Flores, and M. Monica Giusti. 2020. "Monitoring Hydroxycinnamic Acid Decarboxylation by Lactic Acid Bacteria Using High-Throughput UV-Vis Spectroscopy" Molecules 25, no. 14: 3142. https://doi.org/10.3390/molecules25143142







