Peptide Nucleic Acids: Applications in Biomedical Sciences
Funding
Conflicts of Interest
References
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Egholm, M.; Buchardt, O.; Christensen, L.; Behrens, C.; Freier, S.M.; Driver, D.A.; Berg, R.H.; Kim, S.K.; Norden, B.; Nielsen, P.E. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature 1993, 365, 566–568. [Google Scholar] [CrossRef]
- Demidov, V.V.; Potaman, V.N.; Frankkamenetskii, M.D.; Egholm, M.; Buchard, O.; Sonnichsen, S.H.; Nielsen, P.E. Stability of Peptide Nucleic-Acids in human serum and cellular-extracts. Biochem. Pharmacol. 1994, 48, 1310–1313. [Google Scholar] [CrossRef]
- Bendifallah, N.; Rasmussen, F.W.; Zachar, V.; Ebbesen, P.; Nielsen, P.E.; Koppelhus, U. Evaluation of cell-penetrating peptides (CPPs) as vehicles for intracellular delivery of antisense peptide nucleic acid (PNA). Bioconjug. Chem. 2006, 17, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, G.D.; Arzumanov, A.; Abes, R.; Yin, H.; Wood, M.J.A.; Lebleu, B.; Gait, M.J. Improved cell-penetrating peptide-PNA conjugates for splicing redirection in HeLa cells and exon skipping in mdx mouse muscle. Nucl. Acids Res. 2008, 36, 6418–6428. [Google Scholar] [CrossRef]
- Lebleu, B.; Moulton, H.M.; Abes, R.; Ivanova, G.D.; Abes, S.; Stein, D.A.; Iversen, P.L.; Arzumanov, A.A.; Gait, M.J. Cell penetrating peptide conjugates of steric block oligonucleotides. Adv. Drug Del. Rev. 2008, 60, 517–529. [Google Scholar] [CrossRef]
- El-Andaloussi, S.; Johansson, H.J.; Lundberg, P.; Langel, U. Induction of splice correction by cell-penetrating peptide nucleic acids. J. Gene Med. 2006, 8, 1262–1273. [Google Scholar] [CrossRef]
- Soudah, T.; Mogilevsky, M.; Karni, R.; Yavin, E. CLIP6-PNA-Peptide Conjugates: Non-Endosomal Delivery of Splice Switching Oligonucleotides. Bioconjug. Chem. 2017, 28, 3036–3042. [Google Scholar] [CrossRef]
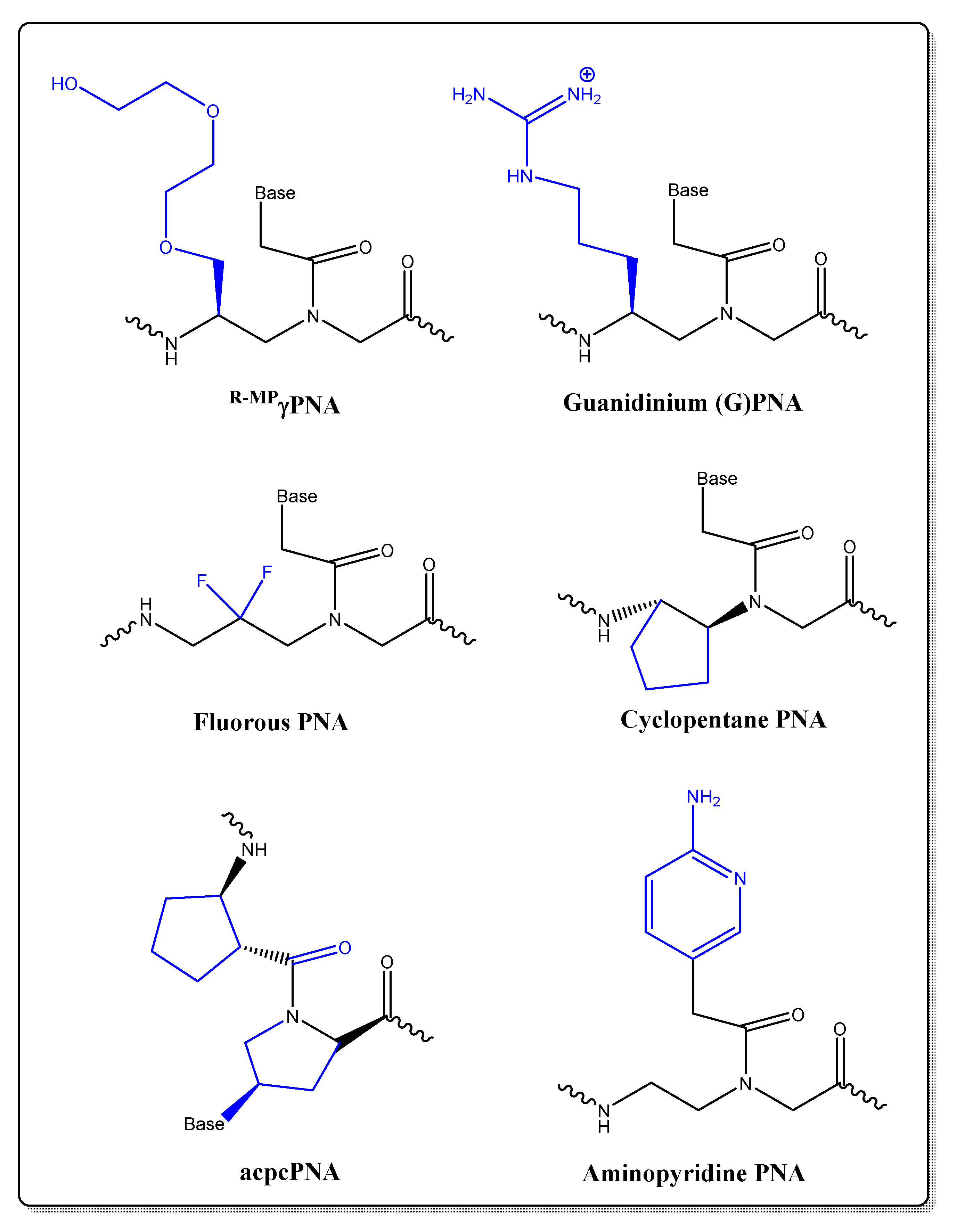
- Ellipilli, S.; Murthy, R.V.; Ganesh, K.N. Perfluoroalkylchain conjugation as a new tactic for enhancing cell permeability of peptide nucleic acids (PNAs) via reducing the nanoparticle size. Chem. Comm. 2016, 52, 521–524. [Google Scholar] [CrossRef]
- Micklitsch, C.M.; Oquare, B.Y.; Zhao, C.; Appella, D.H. Cyclopentane-peptide nucleic acids for qualitative, quantitative, and repetitive detection of nucleic acids. Anal. Chem. 2013, 85, 251–257. [Google Scholar] [CrossRef]
- Bahal, R.; Sahu, B.; Rapireddy, S.; Lee, C.M.; Ly, D.H. Sequence-Unrestricted, Watson-Crick Recognition of Double Helical B-DNA by (R)-MiniPEG-γPNAs. Chembiochem 2012, 13, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.M.; Sahu, B.; Rapireddy, S.; Bahal, R.; Wheeler, S.E.; Procopio, E.M.; Kim, J.; Joyce, S.C.; Contrucci, S.; Wang, Y.; et al. Antitumor Effects of EGFR Antisense Guanidine-Based Peptide Nucleic Acids in Cancer Models. ACS Chem. Biol. 2013, 8, 345–352. [Google Scholar] [CrossRef]
- Vilaivan, T. Pyrrolidinyl PNA with α/β-Dipeptide Backbone: From Development to Applications. Acc. Chem. Res. 2015, 48, 1645–1656. [Google Scholar] [CrossRef] [PubMed]
- Zengeya, T.; Gupta, P.; Rozners, E. Triple-helical recognition of RNA using 2-aminopyridine-modified PNA at physiologically relevant conditions. Angew. Chemie Int. Ed. 2012, 51, 12593–12596. [Google Scholar] [CrossRef]
- Bahal, R.; McNeer, N.A.; Quijano, E.; Liu, Y.F.; Sulkowski, P.; Turchick, A.; Lu, Y.C.; Bhunia, D.C.; Manna, A.; Greiner, D.L.; et al. In vivo correction of anaemia in beta-thalassemic mice by gamma PNA-mediated gene editing with nanoparticle delivery. Nature Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Economos, N.G.; Oyaghire, S.; Quijano, E.; Ricciardi, A.S.; Saltzman, W.M.; Glazer, P.M. Peptide Nucleic Acids and Gene Editing: Perspectives on Structure and Repair. Molecules 2020, 25, 735. [Google Scholar] [CrossRef] [PubMed]
- Fouz, M.F.; Appella, D.H. PNA Clamping in Nucleic Acid Amplification Protocols to Detect Single Nucleotide Mutations Related to Cancer. Molecules 2020, 25, 786. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-T.; Kim, J.W.; Kim, S.K.; Joe, G.H.; Hong, I.S. Simultaneous Genotyping of Multiple Somatic Mutations by Using a Clamping PNA and PNA Detection Probes. Chembiochem 2015, 16, 209–213. [Google Scholar] [CrossRef]
- Takagi, K.; Hayashi, T.; Sawada, S.; Okazaki, M.; Hori, S.; Ogata, K.; Kato, N.; Ebara, Y.; Kaihatsu, K. SNP Discrimination by Tolane-Modified Peptide Nucleic Acids: Application for the Detection of Drug Resistance in Pathogens. Molecules 2020, 25, 769. [Google Scholar] [CrossRef]
- Hoevelmann, F.; Gaspar, I.; Chamiolo, J.; Kasper, M.; Steffen, J.; Ephrussi, A.; Seitz, O. LNA-enhanced DNA FIT-probes for multicolour RNA imaging. Chem. Sci. 2016, 7, 128–135. [Google Scholar] [CrossRef]
- Hashoul, D.; Shapira, R.; Falchenko, M.; Tepper, O.; Paviov, V.; Nissan, A.; Yavin, E. Red-emitting FIT-PNAs: “On site” detection of RNA biomarkers in fresh human cancer tissues. Biosens. Bioelectron. 2019, 137, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Cadoni, E.; Manicardi, A.; Madder, A. PNA-Based MicroRNA Detection Methodologies. Molecules 2020, 25, 1296. [Google Scholar] [CrossRef] [PubMed]
- Kabza, A.M.; Sczepanski, J.T. l-DNA-Based Catalytic Hairpin Assembly Circuit. Molecules 2020, 25, 947. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, M.L.; Zamecnik, P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Aartsma-Rus, A.; Corey, D.R. The 10th Oligonucleotide Therapy Approved: Golodirsen for Duchenne Muscular Dystrophy. Nucl. Acid Therap. 2020, 30, 67–70. [Google Scholar] [CrossRef]
- Ong, A.A.L.; Tan, J.; Bhadra, M.; Dezanet, C.; Patil, K.M.; Chong, M.S.; Kierzek, R.; Decout, J.-L.; Roca, X.; Chen, G. RNA Secondary Structure-Based Design of Antisense Peptide Nucleic Acids for Modulating Disease-Associated Aberrant Tau Pre-mRNA Alternative Splicing. Molecules 2019, 24, 3020. [Google Scholar] [CrossRef]
- Sultan, S.; Rozzi, A.; Gasparello, J.; Manicardi, A.; Corradini, R.; Papi, C.; Finotti, A.; Lampronti, I.; Reali, E.; Cabrini, G.; et al. A Peptide Nucleic Acid (PNA) Masking the miR-145–5p Binding Site of the 3′UTR of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) mRNA Enhances CFTR Expression in Calu-3 Cells. Molecules 2020, 25, 1677. [Google Scholar] [CrossRef]
- Wojciechowska, M.; Równicki, M.; Mieczkowski, A.; Miszkiewicz, J.; Trylska, J. Antibacterial Peptide Nucleic Acids―Facts and Perspectives. Molecules 2020, 25, 559. [Google Scholar] [CrossRef]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yavin, E. Peptide Nucleic Acids: Applications in Biomedical Sciences. Molecules 2020, 25, 3317. https://doi.org/10.3390/molecules25153317
Yavin E. Peptide Nucleic Acids: Applications in Biomedical Sciences. Molecules. 2020; 25(15):3317. https://doi.org/10.3390/molecules25153317
Chicago/Turabian StyleYavin, Eylon. 2020. "Peptide Nucleic Acids: Applications in Biomedical Sciences" Molecules 25, no. 15: 3317. https://doi.org/10.3390/molecules25153317
APA StyleYavin, E. (2020). Peptide Nucleic Acids: Applications in Biomedical Sciences. Molecules, 25(15), 3317. https://doi.org/10.3390/molecules25153317



