Ion-Pair Interaction and Hydrogen Bonds as Main Features of Protein Thermostability in Mutated T1 Recombinant Lipase Originating from Geobacillus zalihae
Abstract
:1. Introduction
2. Results
2.1. Expression and Purification of Mutated Lipases
2.2. Optimum Temperature and Thermostability Study of Mutated HT1 Lipases
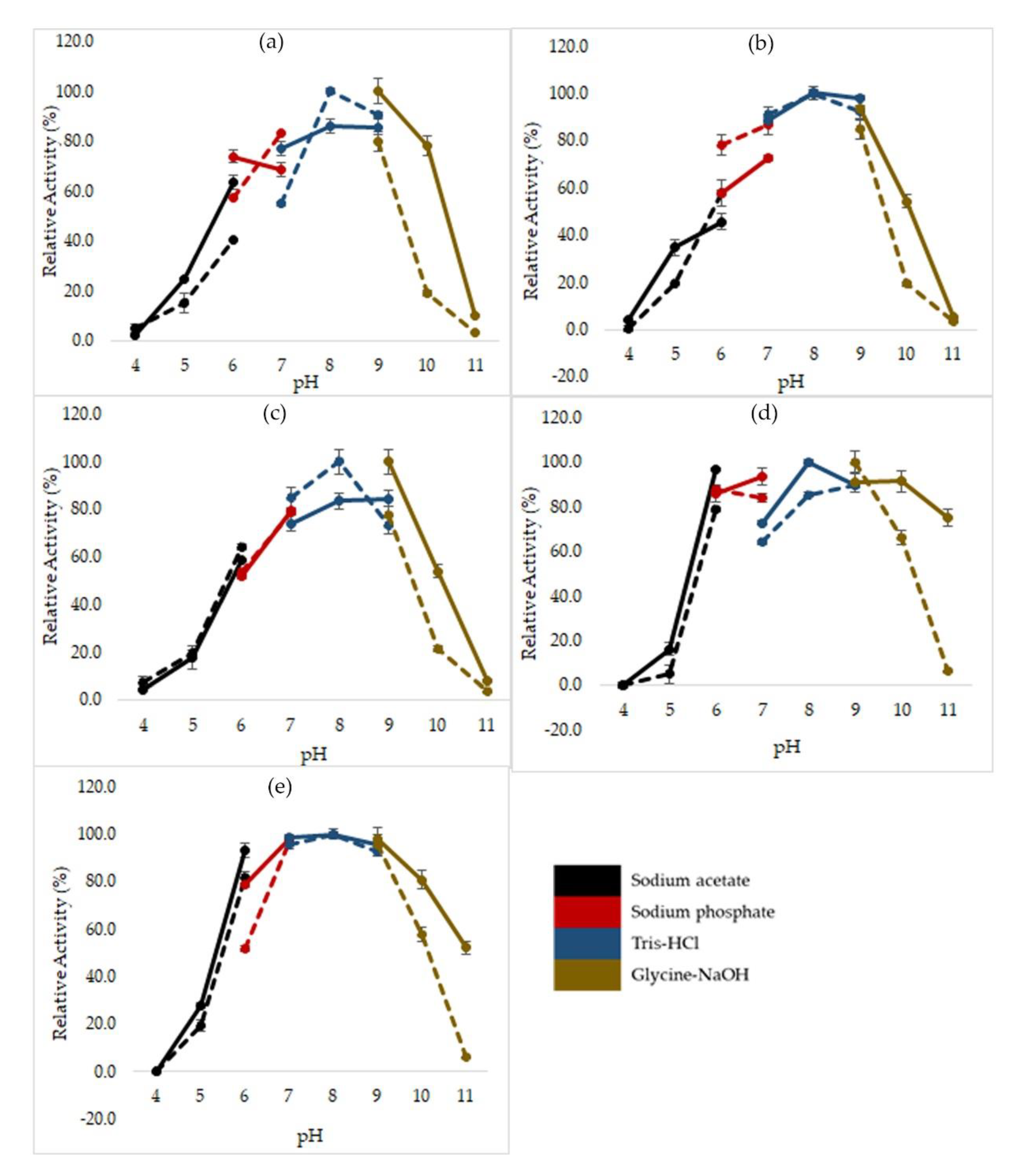
2.3. Effects of pH on Mutated HT1 Lipase Activity and Stability
2.4. Effects of Metal Ions on the Activity of Mutated HT1 Lipases
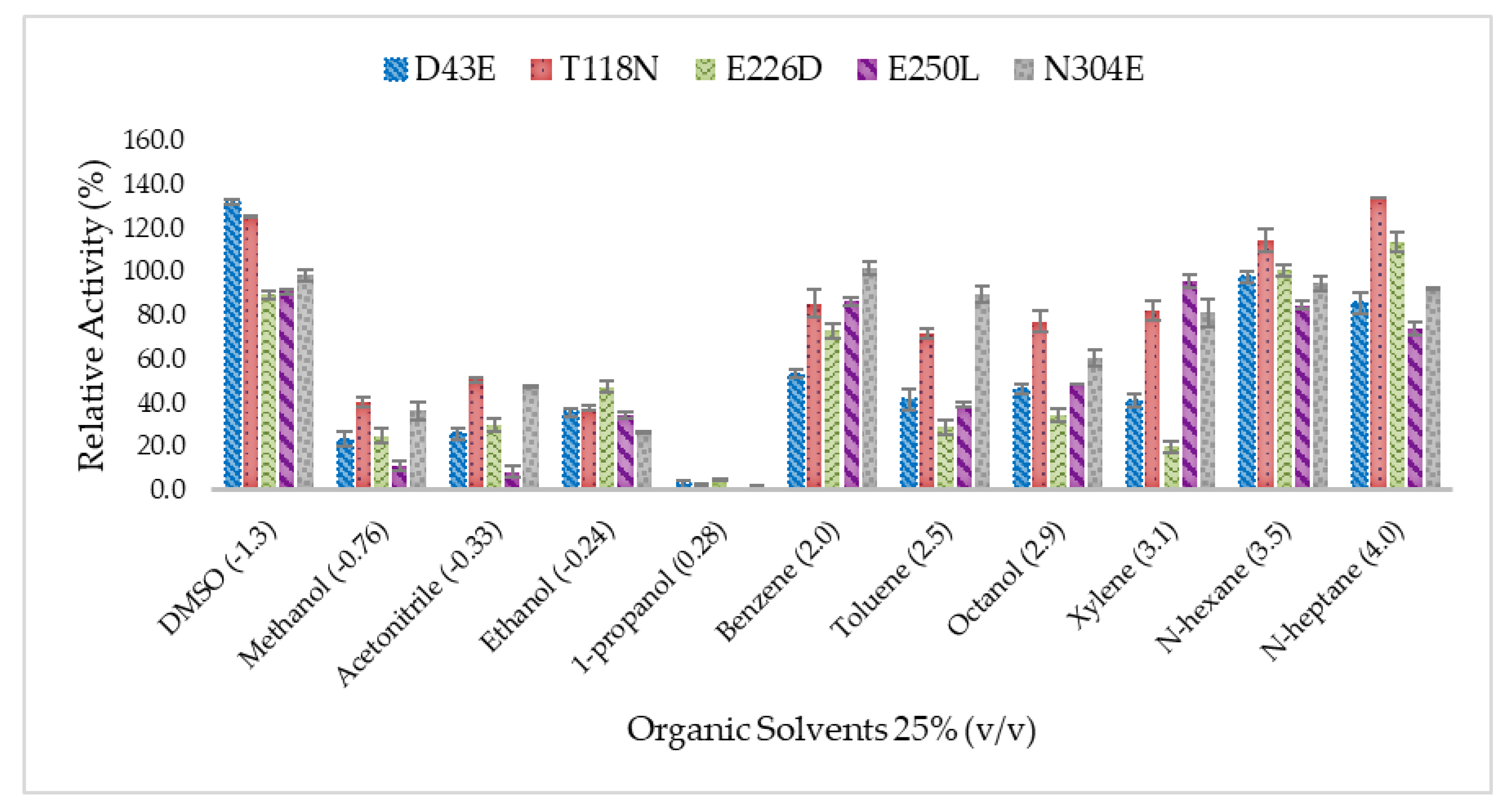
2.5. Effect of Organic Solvents on Lipase Activity of Mutated HT1 Lipases
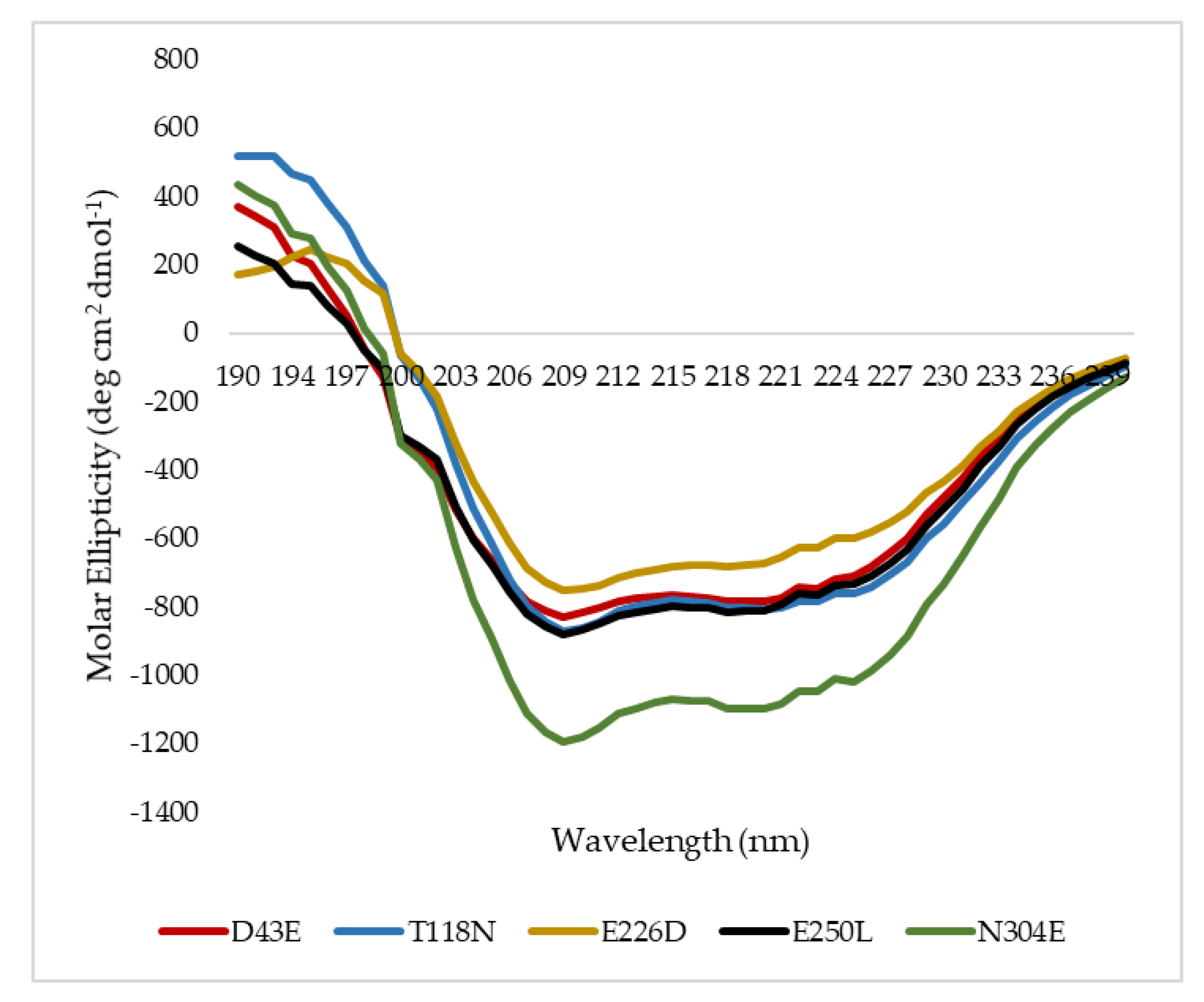
2.6. Secondary Structure Analysis of Mutant HT1 Lipases
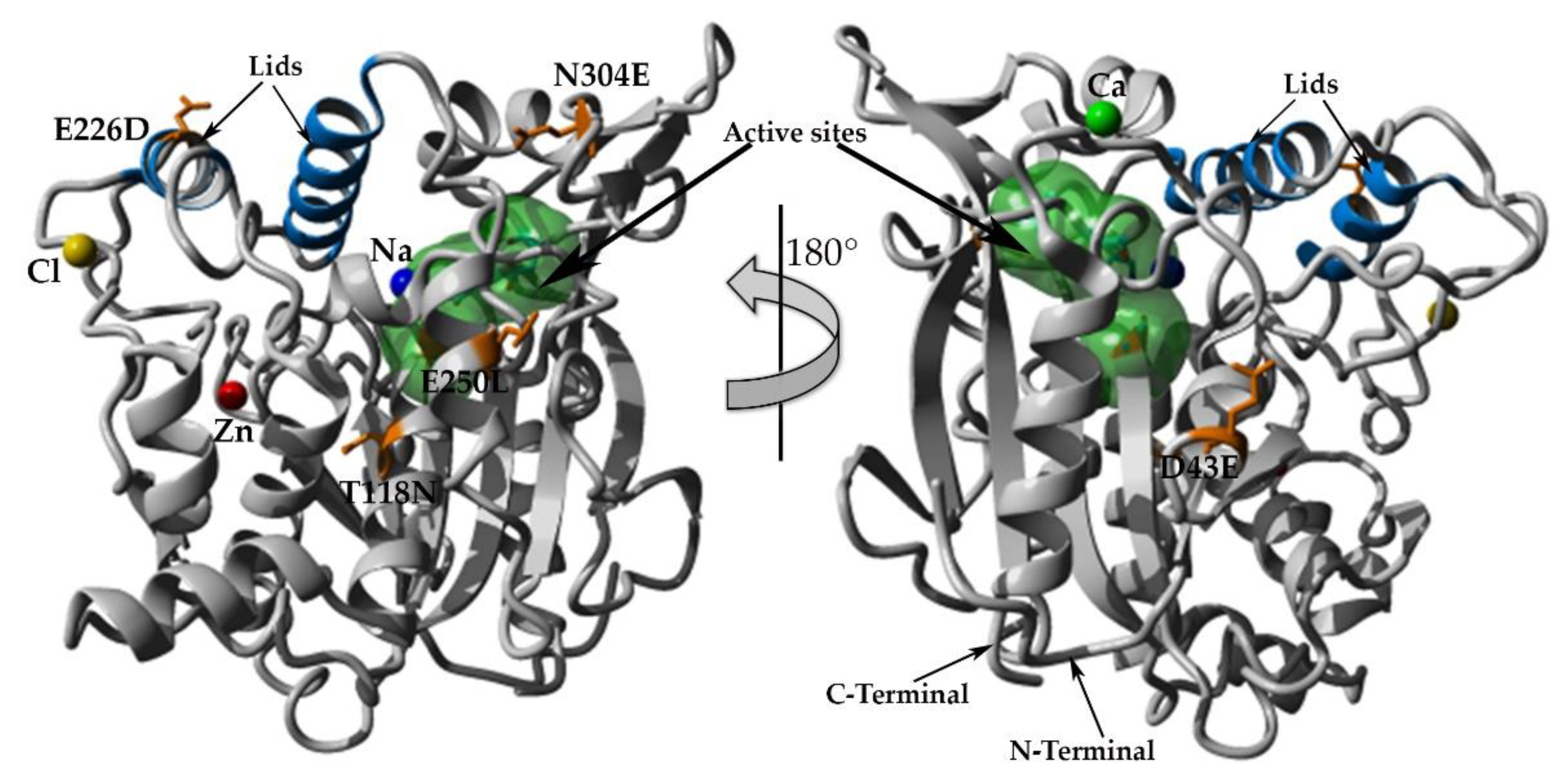
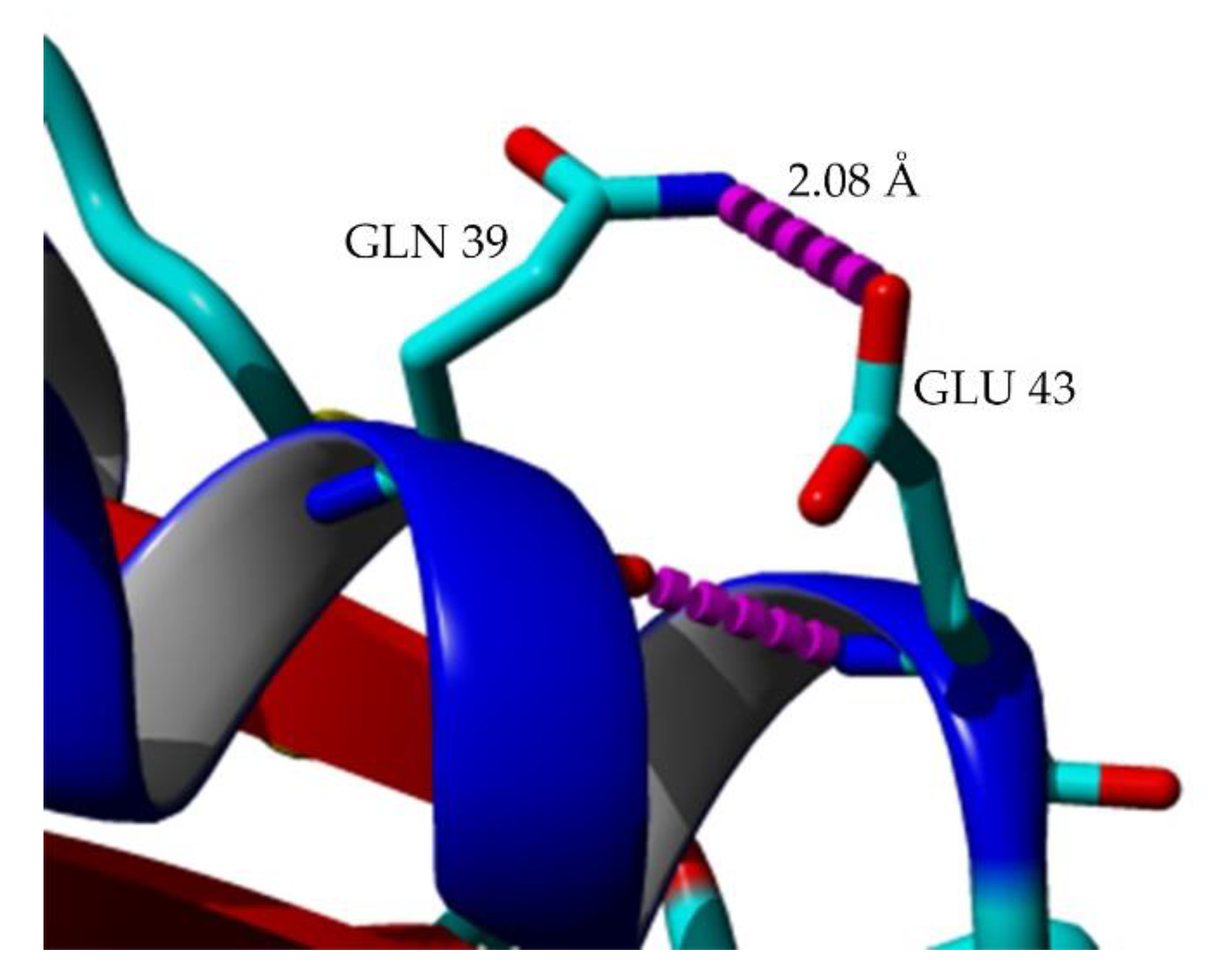
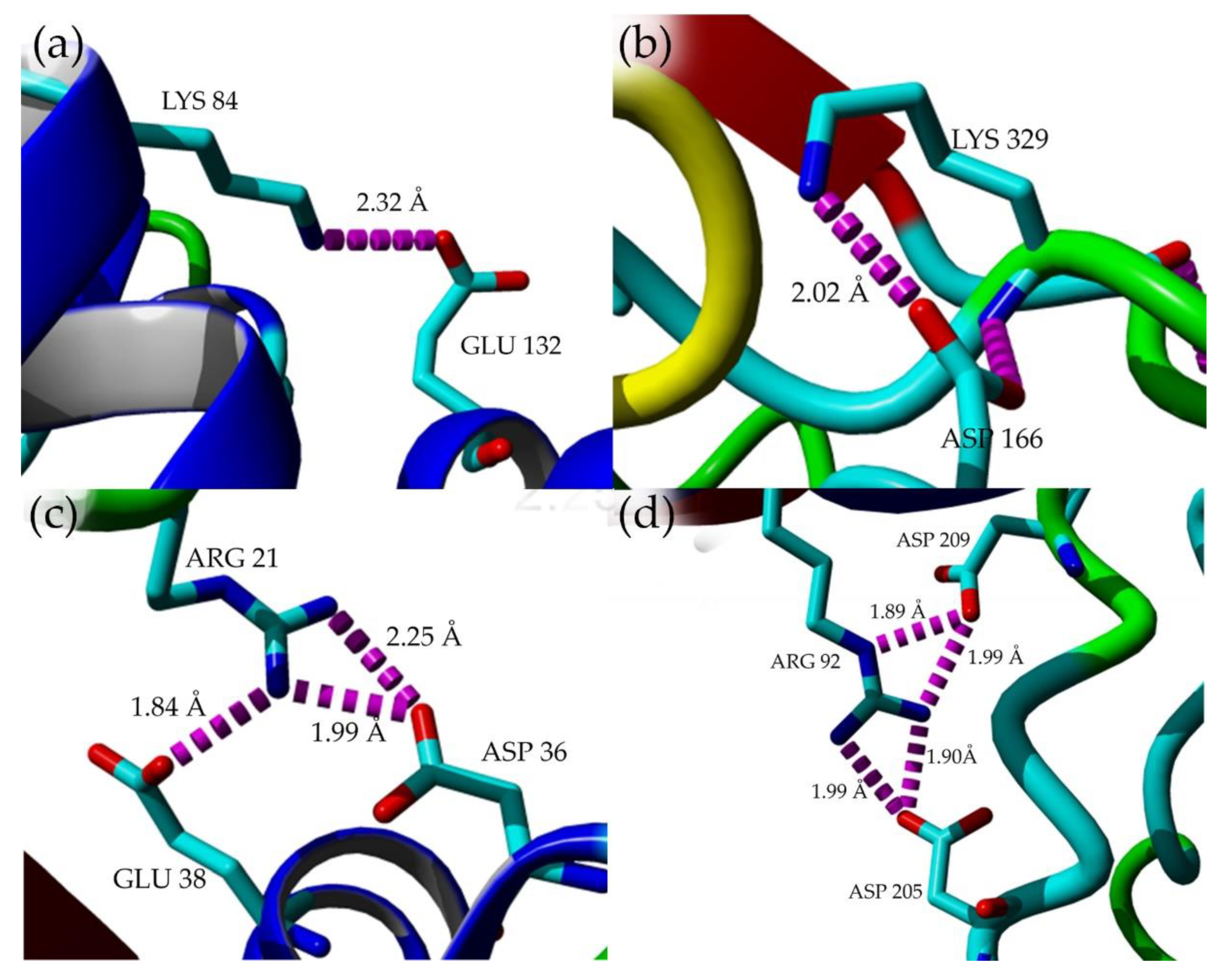
2.7. Structural Investigation
3. Discussion
4. Materials and Methods
4.1. Site-Directed Mutagenesis
4.2. Expression and Purification of Lipase
4.3. Lipase Assay and Protein Concentration Determination
4.4. Optimum Temperature and Thermostability Study of Mutated Lipases
4.5. Effect of pH on Lipase Activity and Stability
4.6. Effect of Various Metal Ions on Lipase
4.7. Effect of Organic Solvent on Lipase Stability
4.8. Circular Dichroism Studies
4.9. Homology Modeling and Structural Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Omar, K.A.; Gounga, M.E.; Liu, R.; Mlyuka, E.; Wang, X. Effects of microbial lipases on hydrolyzed milk fat at different time intervals in flavour development and oxidative stability. J. Food Sci. Technol. 2016, 53, 1035–1046. [Google Scholar] [CrossRef] [Green Version]
- Duarte, J.G.; Leone-Ignacio, K.; da Silva, J.A.C.; Fernandez-Lafuente, R.; Freire, D.M.G. Rapid determination of the synthetic activity of lipases/esterases via transesterification and esterification zymography. Fuel 2016, 177, 123–129. [Google Scholar] [CrossRef]
- Kumari, A.; Gupta, R. Phenylalanine to leucine point mutation in oxyanion hole improved catalytic efficiency of Lip12 from Yarrowia lipolytica. Enzyme Microb. Tech. 2013, 53, 386–390. [Google Scholar] [CrossRef]
- Abdullah, A.; Gani, S.S.A.; Yap, T.Y.H.; Haiyee, Z.A.; Zaidan, U.H.; Kassim, M.A.; Halmi, M.I.E. Lipase-catalyzed synthesis of red pitaya (Hylocereus polyrhizus) seed oil esters for cosmeceutical applications: Process optimization using response surface methodology. RSC Adv. 2019, 9, 5599–5609. [Google Scholar] [CrossRef] [Green Version]
- Turner, P.; Mamo, G.; Karlsson, E.N. Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb. Cell Fact. 2007, 6, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Li, W.F.; Zhou, X.X.; Lu, P. Structural features of thermozymes. Biotechnol. Adv. 2005, 23, 271–281. [Google Scholar] [CrossRef]
- Sangeetha, R.; Arulpandi, I.; Geetha, A. Bacterial lipases as potential industrial biocatalysts: An overview. Res. J. Microbiol. 2011, 6, 1–24. [Google Scholar] [CrossRef]
- Kumar, S.; Tsai, C.J.; Nussinov, R. Factors enhancing protein thermostability. Protein Eng. 2000, 13, 179–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Suen, W.C.; Windsor, W.; Xiao, L.; Madison, V.; Zaks, A. Improving tolerance of Candida antarctica lipase B towards irreversible thermal inactivation through directed evolution. Protein Eng. 2003, 16, 599–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.P.; Li, M.; Zhou, Y.; Yang, L.R.; Xu, G. Introducing a salt bridge into the lipase of Stenotrophomonas maltophilia results in a very large increase in thermal stability. Biotechnol. Lett. 2015, 37, 403–407. [Google Scholar] [CrossRef]
- Akbulut, N.; Ozturk, M.T.; Pijning, T.; Ozturk, S.I.; Gumusel, F. Improved activity and thermostability of Bacillus pumilus lipase by directed evolution. J. Biotechnol. 2013, 164, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Rahman, R.N.Z.R.A.; Leow, T.C.; Salleh, A.B.; Basri, M. Geobacillus zalihae sp. nov., a thermophilic lipolytic bacterium isolated from palm oil mill effluent in Malaysia. BMC Microbiol. 2007, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leow, T.C.; Rahman, R.N.Z.R.A.; Basri, M. A thermoalkaliphilic lipase of Geobacillus sp. T1. Extremophiles 2007, 11, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Aris, S.N.A.M.; Leow, T.C.; Ali, M.S.M.; Mahiran, B.; Salleh, A.B.; Rahman, R.N.Z.A. Crystallographic analysis of ground and space thermostable t1 lipase crystal obtained via counter diffusion method approach. BioMed Res. Int. 2014, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ishak, S.N.H.; Aris, S.N.A.M.; Halim, K.B.A.; Ali, M.S.M.; Leow, T.C.; Kamarudin, N.H.A.; Masomian, M.; Rahman, R.N.Z.R.A. Molecular dynamic simulation of space and earth-grown crystal structures of thermostable T1 lipase Geobacillus zalihae revealed a better structure. Molecules 2017, 22, 1574. [Google Scholar] [CrossRef] [Green Version]
- Ishak, S.N.H.; Masomian, M.; Kamarudin, N.H.A.; Ali, M.S.M.; Leow, T.C.; Rahman, R.N.Z.R.A. Changes of thermostability, organic solvent, and pH stability in Geobacillus zalihae HT1 and its mutant by calcium ion. Int. J. Mol. Sci. 2019, 20, 2561. [Google Scholar] [CrossRef] [Green Version]
- Kwon, D.Y.; Rhee, J.S. A simple and rapid colorimetric method for determination of free fatty acids for lipase assay. J. Am. Oil Chem. Soc. 1986, 63, 89–92. [Google Scholar] [CrossRef]
- Pace, C.N.; Alston, R.W.; Shaw, K.L. Charge-charge interactions influence the denatured state ensemble and contribute to protein stability. Protein Sci. 2000, 9, 1395–1398. [Google Scholar] [CrossRef]
- Xiao, S.; Patsalo, V.; Shan, B.; Bi, Y.; Green, D.F.; Raleigh, D.P. Rational modification of protein stability by targeting surface sites leads to complicated results. Proc. Natl. Acad. Sci. USA 2013, 110, 11337–11342. [Google Scholar] [CrossRef] [Green Version]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Matsumura, H.; Yamamoto, T.; Leow, T.C.; Mori, T.; Salleh, A.B.; Basri, M.; Inoue, T.; Kai, Y.; Rahman, R.N.Z.A. Novel cation-p interaction revealed by crystal structure of thermoalkalophilic lipase. Proteins 2008, 70, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Liu, B.; Hong, Y.; Sheng, D.; Shen, Y.; Ni, J. Residue Tyr224 is critical for the thermostability of Geobacillus sp. RD-2 lipase. Biotechnol. Lett. 2010, 32, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Chopra, N.; Kumar, A.; Kaur, J. Structural and functional insights into thermostable and organic solvent stable variant Pro247-Ser of Bacillus lipase. Int. J. Biol. Macromol. 2018, 108, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Syal, P.; Verma, V.V.; Gupta, R. Targeted mutations and MD simulations of a methanol-stable lipaseYLIP9 from Yarrowia lipolytica MSR80 to develop a biodiesel enzyme. Int. J. Biol. Macromol. 2017, 104, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Madan, B.; Mishra, P. Directed evolution of Bacillus licheniformis lipase for improvement of thermostability. Biochem. Eng. J. 2014, 91, 276–282. [Google Scholar] [CrossRef]
- Lan, D.; Wang, Q.; Xu, J.; Zhou, P.; Yang, B.; Wang, Y. Residue Asn277 affects the stability and substrate specificity of the SMG1 lipase from Malassezia globose. Int. J. Mol. Sci. 2015, 16, 7273–7288. [Google Scholar] [CrossRef] [Green Version]
- Choi, W.C.; Kim, M.H.; Ro, H.S.; Ryu, S.R.; Oh, T.K.; Lee, J.K. Zinc in lipase L1 from Geobacillus stearothermophilus L1 and structural implications on thermal stability. FEBS Lett. 2005, 579, 3461–3466. [Google Scholar] [CrossRef] [Green Version]
- Gudiukaitė, R.; Gegeckas, A.; Sadauskas, M.; Citavicius, D. Detection of Asp371, Phe375, and Tyr376 influence on GD-95-10 lipase using alanine scanning mutagenesis. Appl. Biochem. Biotechnol. 2015, 178, 654–669. [Google Scholar] [CrossRef]
- Shih, T.-W.; Pan, T.-M. Substitution of Asp189 residue alters the activity and thermostability of Geobacillus sp. NTU 03 lipase. Biotechnol. Lett. 2011, 33, 1841–1846. [Google Scholar] [CrossRef]
- Sharma, P.; Sharma, N.; Pathania, S.; Handa, S. Purification and characterization of lipase by Bacillus methylotrophicus PS3 under submerged fermentation and its application in detergent industry. J. Genet. Eng. Biotechnol. 2017, 15, 369–377. [Google Scholar] [CrossRef]
- Ali, M.S.M.; Yun, C.C.; Leow, A.T.C.; Rahman, R.N.Z.R.A.; Basri, M.; Salleh, A.B. Purification and characterisation of an F16L mutant of a thermostable lipase. Protein J. 2012, 31, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Kim, H.K.; Lee, J.K.; Park, S.Y.; Oh, T.K. Thermostable lipase of Bacillus stearothermophilus: High-level production, purification, and calcium-dependent thermostability. Biosci. Biotechnol. Biochem. 2000, 64, 280–286. [Google Scholar] [CrossRef]
- Khaleghinejad, S.H.; Motalleb, G.; Karkhane, A.A.; Aminzadeh, S.; Yakhchali, B. Study the effect of F17S mutation on the chimeric Bacillus thermocatenulatus lipase. J. Genet. Eng. Biotechnol. 2016, 14, 83–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simons, J.W.F.A.; van Kampen, M.D.; Ubarretxena-Belandia, I.; Cox, R.C.; Alves dos Santos, C.M.; Egmond, M.R.; Verheij, H.M. Identification of a calcium binding site in Staphylococcus hyicus lipase: Generation of calcium-independent variants. Biochemistry 1999, 38, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Tayyab, M.I.; Rashid, N.; Akhtar, M. Isolation and identification of lipase producing thermophilic Geobacillus sp. SBS-4S: Cloning and characterization of the lipase. J. Biosci. Bioeng. 2011, 111, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Yamada, C.; Sawano, K.; Iwase, N.; Matsuoka, M.; Arakawa, T.; Nishida, S.; Fushinobu, S. Isolation and characterization of a thermostable lipase from Bacillus thermoamylovorans NB501. J. Gen. Appl. Microbiol. 2016, 62, 313–319. [Google Scholar] [CrossRef] [Green Version]
- Johri, S.; Bhat, A.; Sayed, S.; Nargotra, A.; Jain, A.; Qazi, G.N. Novel thermostable lipase from Bacillus circulans IIIB153: Comparison with the mesostable homologue at sequence and structure level. World J. Microbiol. Biotechnol. 2012, 28, 193–203. [Google Scholar] [CrossRef]
- Sakinc, T.; Kleine, B.; Gatermann, S.G. Biochemical characterization of the surface-associated lipase of Staphylococcus saprophyticus. FEMS Microbiol. Lett. 2007, 274, 335–341. [Google Scholar] [CrossRef] [Green Version]
- Ghori, M.I.; Iqbal, M.J.; Hameed, A. Characterization of a novel lipase from Bacillus sp. isolated from tannery wastes. Braz. J. Microbiol. 2011, 42, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Kawata, T.; Ogino, H. Amino acid residues involved in organic solvent-stability of the LST-03 lipase. Biochem. Biophys. Res. Commun. 2010, 400, 384–388. [Google Scholar] [CrossRef]
- Shokri, M.M.; Ahmadian, S.; Akbari, N.; Khajeh, K. Hydrophobic substitution of surface residues affects lipase stability in organic solvents. Mol. Biotechnol. 2014, 56, 360–368. [Google Scholar] [CrossRef]
- Park, H.J.; Joo, J.C.; Park, K.; Yoo, Y.J. Stabilization of Candida antartica Lipase B in hydrophilic organic solvent by rational design of hydrogen bond. Biotechnol. Bioprocess Eng. 2012, 17, 722–728. [Google Scholar] [CrossRef]
- Dror, A.; Shemesh, E.; Dayan, N.; Fishman, A. Protein engineering by random mutagenesis and structure-guided consensus of Geobacillus stearothermophilus lipase T6 for enhanced stability in methanol. Appl. Environ. Microbiol. 2014, 80, 1515–1527. [Google Scholar] [CrossRef] [Green Version]
- Choo, D. Analysis of the structure-stability relationship of cold-adapted lipase PsLip1 from homology modeling. Genomics Inform. 2011, 9, 79–84. [Google Scholar] [CrossRef] [Green Version]
- Charbonneau, D.M.; Beauregard, M. Role of key salt bridges in thermostability of G. Thermodenitrificans EstGtA2: Distinctive patterns within the new bacterial lipolytic enzyme subfamily XIII.2. PLoS ONE 2013, 8, e76675. [Google Scholar] [CrossRef] [PubMed]
- Hussain, C.H.A.C. Molecular Modification for Effective Purification, Scaling Up Production and Separation of T1 Lipase. Ph.D. Thesis, Universiti Putra Malaysia, Serdang, Selangor, Malaysia, 2017. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Greenfield, N.J. Analysis of the kinetics of folding of proteins and peptides using circular dichroism. Nat. Protoc. 2007, 1, 2891–2899. [Google Scholar] [CrossRef] [Green Version]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |

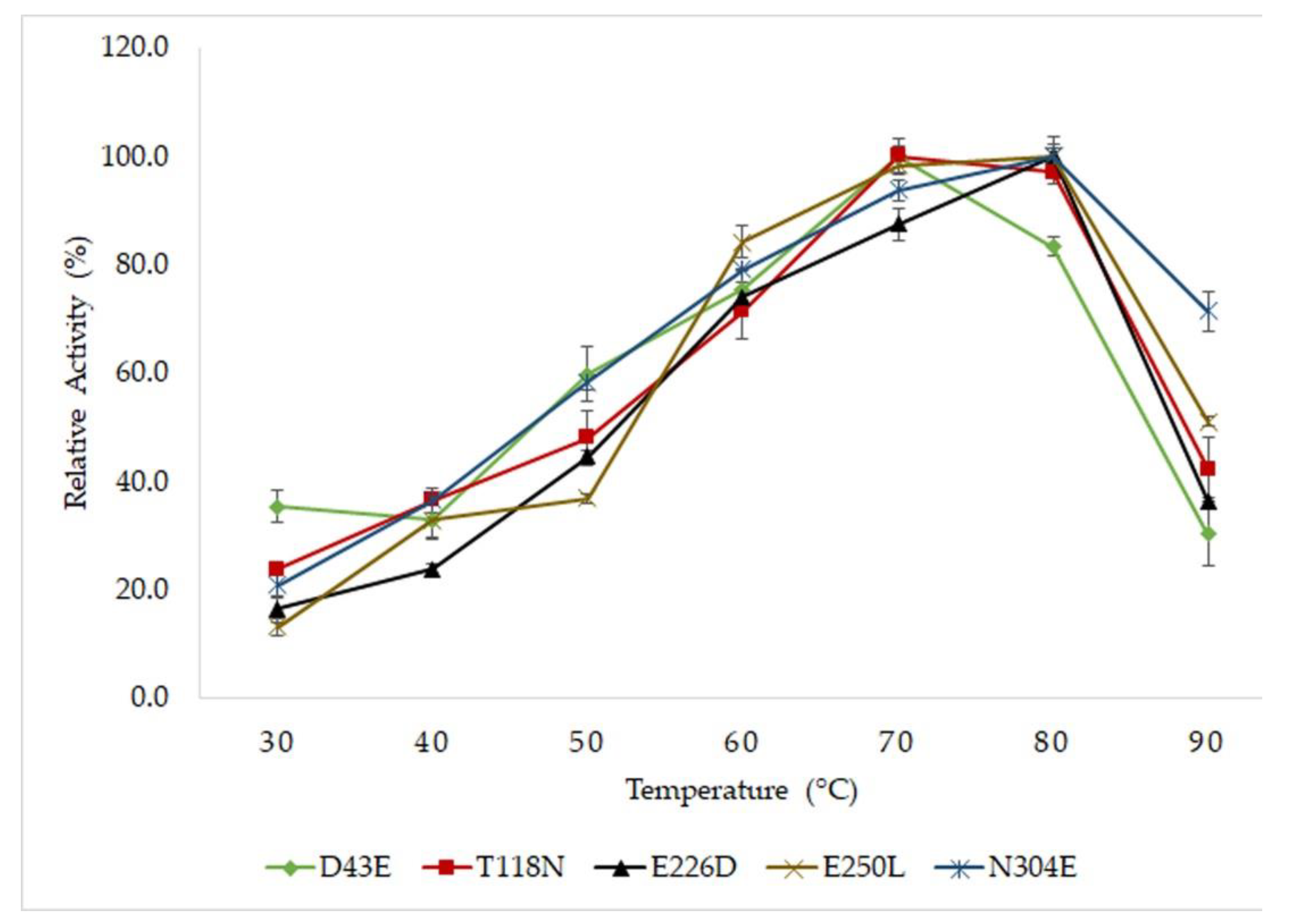
| Lipases | T½ (Min) | Thermal Denaturation(Tm) (°C) | ||
|---|---|---|---|---|
| 60 °C | 70 °C | 80 °C | ||
| D43E | 900 | 135 | 33 | 76.0 ± 1.2 |
| T118N | 360 | 75 | 10 | 69.3 ± 0.3 |
| E226D | 1680 | 165 | 47 | 77.4 ± 2.6 |
| E250L | 540 | 115 | 17 | 71.7 ± 0.1 |
| N304E | 600 | 120 | 30 | 74.3 ± 0.2 |
| Lipase | Concentration (mM) | Na+ | Ca2+ | Mg2+ | Fe3+ | Ni2+ | Cu2+ | Zn2+ |
|---|---|---|---|---|---|---|---|---|
| D43E | 1 | 114.3 ± 3.5 | 123.2 ± 2.9 | 109.5 ± 4.8 | 70.6 ± 0.5 | 102.9 ± 2.2 | 22.5 ± 0.5 | 5.5 ± 0.1 |
| 5 | 84.4 ± 0.4 | 156.4 ± 3.0 | 111.3 ± 1.8 | 9.4 ± 0.1 | 19.2 ± 3.6 | 6.3 ± 0.4 | 4.7 ± 0.1 | |
| T118N | 1 | 100.6 ± 4.6 | 154.0 ± 5.1 | 115.9 ± 0.3 | 129.9 ± 4.8 | 32.8 ± 1.1 | 66.6 ± 3.0 | 5.7 ± 1.0 |
| 5 | 91.7 ± 3.5 | 172.8 ± 2.3 | 111.4 ± 1.9 | 70.8 ± 2.5 | 10.5 ± 0.4 | 28.9 ± 1.2 | 5.3 ± 0.4 | |
| E226D | 1 | 112.7 ± 4.5 | 110.1 ± 1.8 | 74.6 ± 4.9 | 72.3 ± 1.4 | 44.7 ± 4.3 | 18.6 ± 2.6 | 8.8 ± 1.5 |
| 5 | 100.2 ± 5.1 | 119.7 ± 3.1 | 77.8 ± 4.0 | 5.6 ± 0.4 | 12.5 ± 1.9 | 3.5 ± 1.0 | 2.2 ± 0.6 | |
| E250L | 1 | 103.3 ± 5.3 | 150.0 ± 4.6 | 97.5 ± 2.7 | 20.0 ± 2.2 | 11.7 ± 2.7 | 64.6 ± 1.8 | 2.7 ± 1.3 |
| 5 | 106.6 ± 4.2 | 156.0 ± 4.2 | 101.4 ± 5.9 | 2.0 ± 0.9 | 0.8 ± 2.4 | 7.0 ± 0.5 | 11.2 ± 0.8 | |
| N304E | 1 | 91.8 ± 4.0 | 101.1 ± 4.9 | 88.5 ± 4.7 | 54.1 ± 1.2 | 46.3 ± 4.3 | 27.4 ± 3.5 | 47.7 ± 6.7 |
| 5 | 96.9 ± 3.7 | 106.8 ± 3.3 | 61.0 ± 3.9 | 8.1 ± 2.1 | 40.7 ± 6.1 | 7.9 ± 4.0 | 9.8 ± 5.4 |
| D43E | T118N | E226D | E250L | N304E | |
|---|---|---|---|---|---|
| α-Helix | 20.80 | 29.10 | 24.40 | 21.60 | 24.40 |
| β-Sheet | 23.60 | 9.80 | 17.90 | 19.80 | 14.20 |
| Turn | 18.10 | 24.30 | 23.40 | 21.10 | 23.30 |
| Random | 37.50 | 36.80 | 34.30 | 37.50 | 38.10 |
| Lipases | Hydrogen Bond | Ion-Pair Interaction | Hydrophobic Interaction (Sidechain) | Aromatic Interaction |
|---|---|---|---|---|
| D43E | 338 | 30 | 674 | 41 |
| T118N | 304 | 7 | 653 | 37 |
| E226D | 331 | 28 | 662 | 39 |
| E250L | 337 | 25 | 680 | 41 |
| N304E | 342 | 22 | 681 | 42 |
| Thermostability (T1/2) (Min) | Tm (°C) | Relative Activity in DMSO (%) | Relative Activity in Ca2+ (%) | |||
|---|---|---|---|---|---|---|
| 60 °C | 70 °C | 80 °C | 1 mM | 5 mM | ||
| 540 | 85 | 16 | 70.9 ± 0.1 | 105.8 ± 4.2 | 122.0 ± 1.7 | 123.0 ± 4.3 |
| Primer | Nucleotide Sequence (5′–3′) |
|---|---|
| D43E | Forward 5′3′: caatggctgaacgagaacggttatcgaac Reverse 5′3′: gttcgataaccgttctcgttcagccattg |
| T118N | Forward 5′3′: caaggggggcagaacgcccgcatgcttg Reverse 5′3′: caagcatgcgggcgttctgccccccttg |
| E226D | Forward 5′3′: gaccattattttgatcggctcaagcgctc Reverse 5′3′: gagcgcttgagccgatcaaaataatggtc |
| E250L | Forward 5′3′: gatttatccgtttccggagctttgaagttgaatcaatggtccac Reverse 5′3′: gtggaccattgattcaacttcaaagctccggaaacggataaatc |
| N304E | Forward 5′3′: ggttcgtaccgcgagccgacgctcggc Reverse 5′3′: gccgagcgtcggctcgcggtacgaacc |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishak, S.N.H.; Kamarudin, N.H.A.; Ali, M.S.M.; Leow, A.T.C.; Rahman, R.N.Z.R.A. Ion-Pair Interaction and Hydrogen Bonds as Main Features of Protein Thermostability in Mutated T1 Recombinant Lipase Originating from Geobacillus zalihae. Molecules 2020, 25, 3430. https://doi.org/10.3390/molecules25153430
Ishak SNH, Kamarudin NHA, Ali MSM, Leow ATC, Rahman RNZRA. Ion-Pair Interaction and Hydrogen Bonds as Main Features of Protein Thermostability in Mutated T1 Recombinant Lipase Originating from Geobacillus zalihae. Molecules. 2020; 25(15):3430. https://doi.org/10.3390/molecules25153430
Chicago/Turabian StyleIshak, Siti Nor Hasmah, Nor Hafizah Ahmad Kamarudin, Mohd Shukuri Mohamad Ali, Adam Thean Chor Leow, and Raja Noor Zaliha Raja Abd. Rahman. 2020. "Ion-Pair Interaction and Hydrogen Bonds as Main Features of Protein Thermostability in Mutated T1 Recombinant Lipase Originating from Geobacillus zalihae" Molecules 25, no. 15: 3430. https://doi.org/10.3390/molecules25153430
APA StyleIshak, S. N. H., Kamarudin, N. H. A., Ali, M. S. M., Leow, A. T. C., & Rahman, R. N. Z. R. A. (2020). Ion-Pair Interaction and Hydrogen Bonds as Main Features of Protein Thermostability in Mutated T1 Recombinant Lipase Originating from Geobacillus zalihae. Molecules, 25(15), 3430. https://doi.org/10.3390/molecules25153430






