Abstract
Antimicrobial peptides (AMPs) show high antibacterial activity against pathogens, which makes them potential new therapeutics to prevent and cure diseases. Porcine beta defensin 2 (pBD2) is a newly discovered AMP and has shown antibacterial activity against different bacterial species including multi-resistant bacteria. In this study, the functional mechanism of pBD2 antibacterial activity against Staphylococcus aureus was investigated. After S. aureus cells were incubated with different concentrations of pBD2, the morphological changes in S. aureus and locations of pBD2 were detected by electron microscopy. The differentially expressed genes (DEGs) were also analyzed. The results showed that the bacterial membranes were broken, bulging, and perforated after treatment with pBD2; pBD2 was mainly located on the membranes, and some entered the cytoplasm. Furthermore, 31 DEGs were detected and confirmed by quantitative real-time PCR (qRT-PCR). The known functional DEGs were associated with transmembrane transport, transport of inheritable information, and other metabolic processes. Our data suggest that pBD2 might have multiple modes of action, and the main mechanism by which pBD2 kills S. aureus is the destruction of the membrane and interaction with DNA. The results imply that pBD2 is an effective bactericide for S. aureus, and deserves further study as a new therapeutic substance against S. aureus.
1. Introduction
Defensins are a family of antimicrobial peptides (AMPs) secreted by organisms. They have high biological activity against bacteria, fungi, and viruses, which makes them alternatives as novel therapeutic drugs [1,2,3]. Mammalian defensins are classified into alpha, beta, and theta defensins based on their different connection between cysteine residues, and on an evolutionary basis, beta defensins are the oldest ones [1]. In pigs, only beta defensins have been found, and all beta defensins have been discovered through genomic analysis, except for porcine beta defensin 1 (pBD1). Relatively few studies have been carried out on these defensins [4,5]. pBD1 is the first beta defensin discovered in pigs, and it has lower bactericidal activity compared with pBD2, which has strong antibacterial activity against both Gram-negative and Gram-positive bacteria including multi-resistant bacteria [6,7,8]. To date, pBD2 has perhaps the strongest antimicrobial activity known among pBDs [6,9,10]. Oral administration of pBD2 reduces the inflammatory response in weaned piglets infected with E. coli [11], and pBD2 attenuates inflammation and mucosal lesions in dextran sodium sulfate-induced colitis in mice [12]. Transgenic pigs and mice that constitutively overexpress pBD2 show enhanced resistance to bacterial infection [13,14]. Moreover, it has high salt-resistance and low hemolytic activity [7]. These results make it a good candidate as an antibiotic.
However, for the development of pBD2 as an antibiotic agent, it is important to understand its antimicrobial mechanism against pathogens. Many studies have proposed several patterns of antimicrobial mechanisms and focused on the cell membrane [15,16,17]. It is believed that the first step in killing bacteria is the interaction between the positively charged residues of defensin and the negatively charged components in the microbial membrane, resulting in the disruption of the cell membrane. After damaging the extracellular membrane, defensins further bind to the protoplast membrane, and cell death results directly from membrane destruction and decomposition, or changing permeability of the cytoplasmic membrane, or attacking internal targets such as negatively charged DNA or RNA, etc. [18,19,20,21,22]. Antibacterial mechanisms may vary depending on the different AMP or bacterial species [17,23,24]. There is poor experimental evidence to indicate which model is applicable to beta defensins and the mechanisms of pBDs are unclear and seldom reported.
Staphylococcus aureus, as an example of Gram-positive bacteria, is considered a global threat to human and animal health [25]. In a previous report, our research team cloned the pBD2 gene and constructed the engineering strain E. coli BL21(DE3)-pET-pBD2 and reported that the purified recombinant pBD2 with His-Tag had strong antimicrobial activity against S. aureus [7]. In this study, we observed morphological changes in S. aureus and detected locations of action of pBD2 after incubating with recombinant pBD2. Moreover, differentially expressed genes (DEGs) were obtained, and their functions were analyzed. Our data suggest that pBD2 might have multiple modes of action, and the main mechanism by which pBD2 kills S. aureus is the destruction of its membrane and interaction with DNA. The results may contribute to future efforts aiming to develop this AMP as a new therapeutic substance against S. aureus infections.
2. Results
2.1. Antibacterial Activity of pBD2
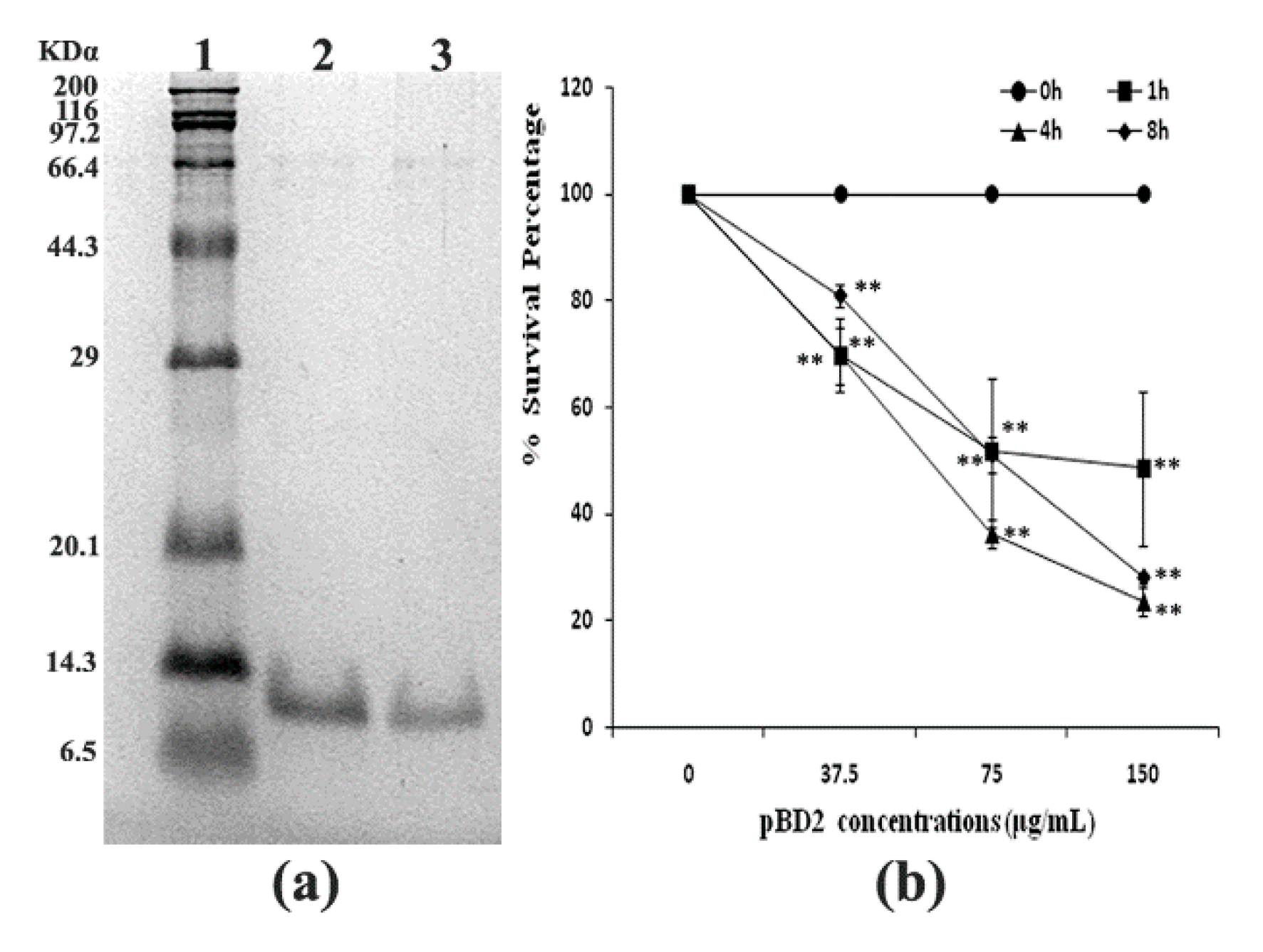
The fused pBD2 was induced and purified as described previously using the constructed engineering strain E. coli BL21(DE3)-pET-pBD2 in our lab [7]. The purified recombinant pBD2 with a His-Tag had high purity (shown in Figure 1a). The survival percentage of S. aureus (109 cfu/mL) decreased with increasing pBD2 concentrations (p < 0.01), while there was no obvious decrease with exposure time (p > 0.05), which implies that pBD2 has high antimicrobial activity against S. aureus (Figure 1b).
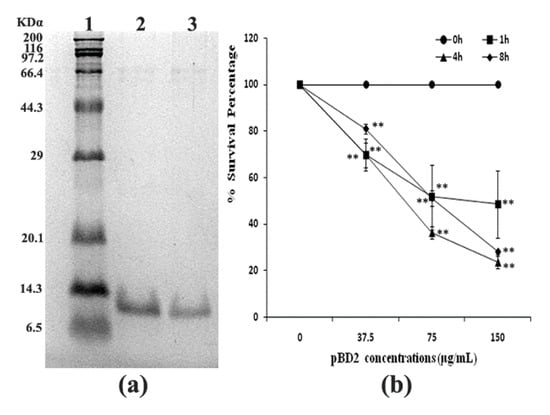
Figure 1.
Purified fused pBD2 and its antimicrobial activity. (a) SDS-PAGE analysis of purified fused pBD2. Lane 1 indicates the protein marker; Lanes 2–3 indicate purified pBD2. (b) Antimicrobial activities of recombinant pBD2 against S. aureus at different time points. The ** symbol indicates extremely significant differences compared with the control at p < 0.01 by Dunnett’s method.
2.2. Morphological Changes in Bacteria
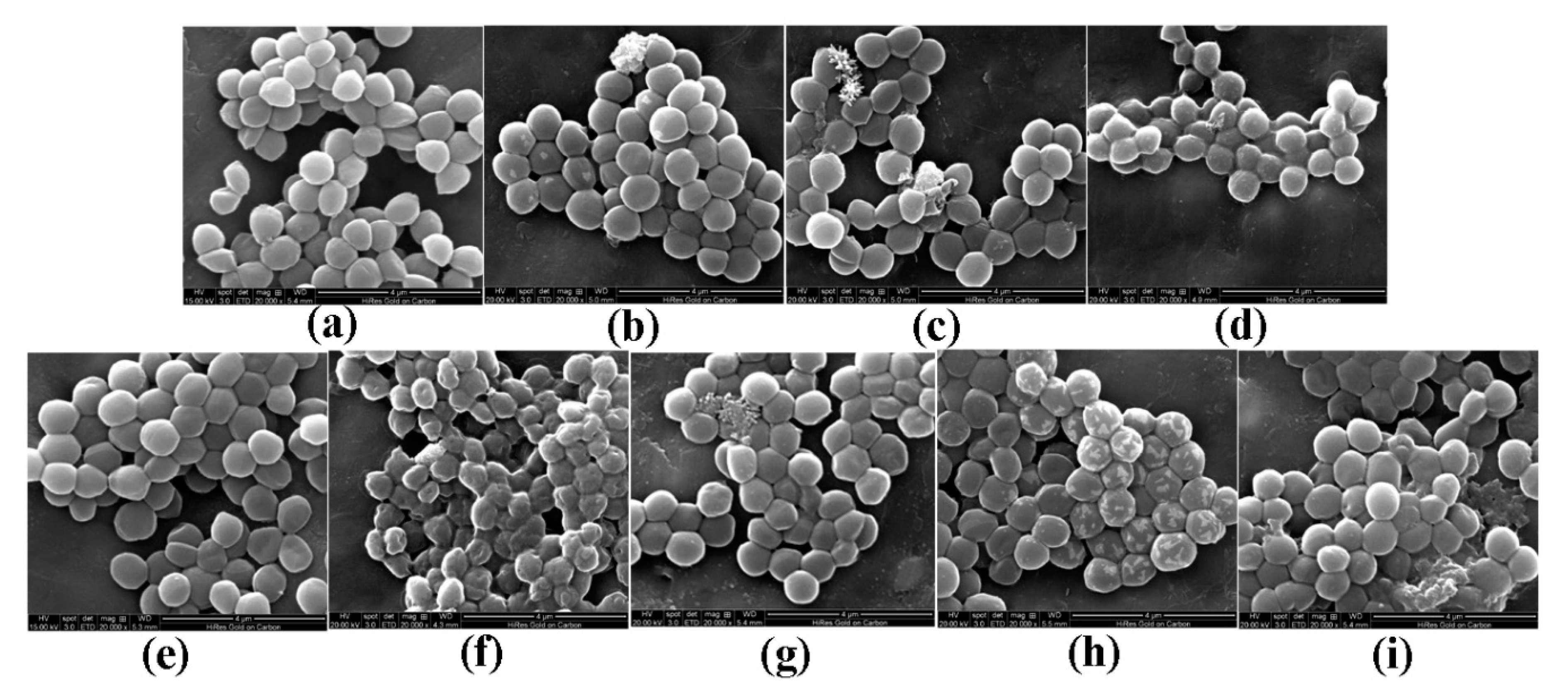
Morphological changes in S. aureus were observed by scanning electron microscopy (SEM) and are shown in Figure 2. Morphological changes in response to different pBD2 concentrations were very similar, except for those in cells incubated with 37.5 μg/mL pBD2 for 4 h. The control cells were normal without obvious damage (Figure 2a,e). After S. aureus was incubated with pBD2 for 1 h, cells were bulging, and the debris of bacteria was highly visible (Figure 2b–d). After incubation for 4 h, cells showed more bulging (Figure 2h), and more debris of bacteria was visible (Figure 2g,i). The cells were rough, extremely pitted, and perforated after incubation with 37.5 μg/mL pBD2 for 4 h, and these changes were different from those observed in other pBD2-treated cells (Figure 2f). However, some cells did not have obvious changes (Figure 2b,c,g). Furthermore, the bacterial biomass decreased with increasing pBD2 concentrations when bacteria were collected after they were treated with pBD2, and the number of cells in the total observed field of view decreased with increasing doses of pBD2, which indicates that more cells were dead with higher concentrations.
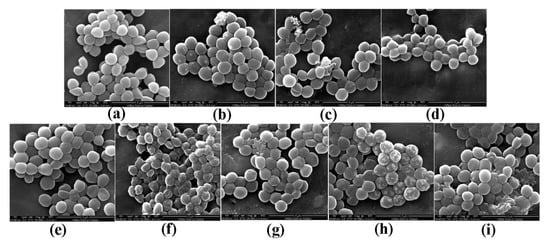
Figure 2.
Scanning electron microscopy (SEM) images of morphological changes in bacteria after they were cultured with pBD2. (a) Control cells incubated without pBD2 for 1 h; (b–d) cells were treated for 1 h with 37.5, 75, and 150 μg/mL pBD2 respectively; (e) control cells incubated without pBD2 for 4 h; (f,g) cells were treated for 4 h with 37.5 and 75 μg/mL pBD2, respectively; (h,i) cells were treated for 4 h with 150 μg/mL pBD2.
2.3. Localization of pBD2 Peptides
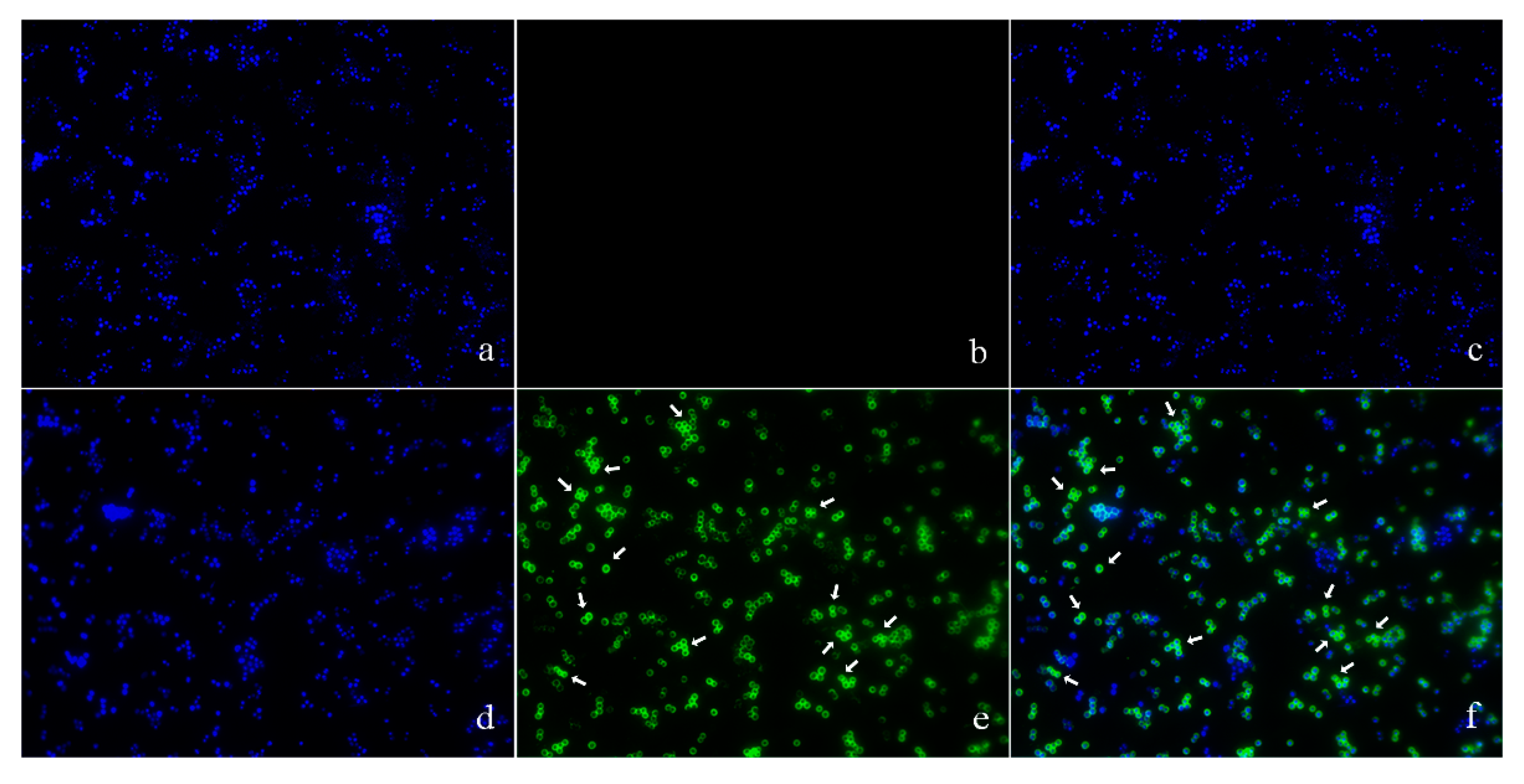
The location of pBD2 was determined by immuno-fluorescence microscopy (IFM) and is shown in Figure 3. The green fluorescence from staining with FITC (fluorescein isothiocyanate) labeled secondary antibodies was mainly concentrated on the surface of S. aureus, and distinct rings were formed, which indicates that pBD2 is mainly located on the bacterial membrane, and some entered the cytoplasm (Figure 3e,f). In addition, about 65.7% of cells were labeled with green fluorescent.
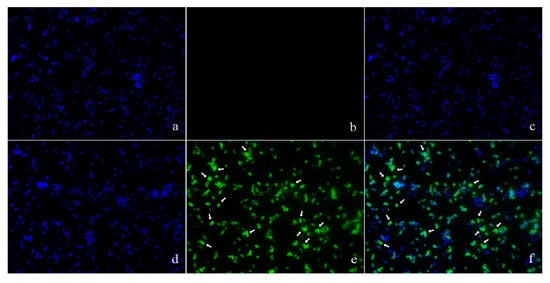
Figure 3.
The locations of pBD2 on bacteria were detected by IFM (×1000). (a–c) Control bacteria (without the first antibody) dyed by DAPI (4′,6-diamidino-2-phenylindole), FITC, and DAPI/FITC, respectively; (d–f) bacteria dyed by DAPI, FITC, and DAPI/FITC, respectively. Bacteria were magnified 1000 times. The green fluorescence, which is the result of staining with FITC-labeled secondary antibodies, indicates the location of pBD2. The blue fluorescence, which is the result of staining with DAPI, indicates the location of bacterial DNA. The arrows indicate the cells in which pDB2 is likely to be located inside.
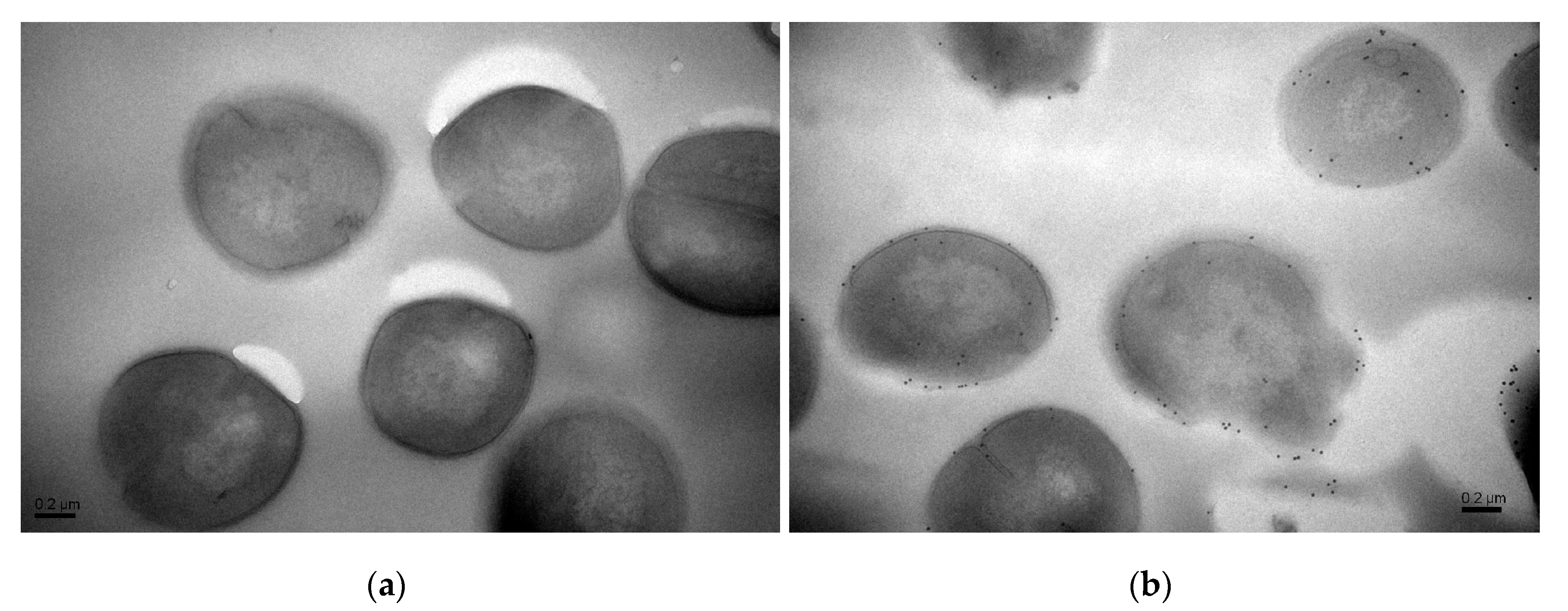
Moreover, precise localizations of pBD2 were further determined by immuno-gold transmission electron microscopy (TEM) after cells were cultured with 150 μg/mL pBD2 for 4 h (Figure 4). pBD2 marked with gold particles was mainly located on the cell membranes, and some had entered the cytoplasm, in other cells, more pBD2 was inside of the cytoplasm than on the cell membrane, without obvious membrane disruption (Figure 4b). The results from both approaches indicate that pBD2 mainly localized to the membrane and that some entered the cytoplasm.
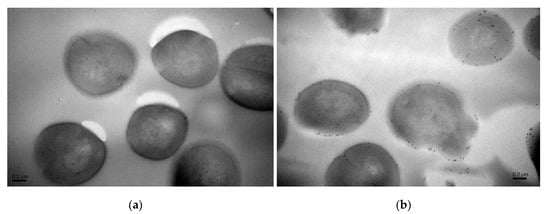
Figure 4.
The locations of pBD2 on bacteria were detected by immuno-gold TEM. (a) Control cells; (b) cells after they were treated with 150 μg/mL pBD2 for 4 h. The gold particles labeling the secondary antibody indicate the location of pBD2.
2.4. Identification of Differentially Expressed Gene Fragments by ACP (Annealing Control Primer)-Based RT-PCR
RNAs of S. aureus were extracted after cells were cultured with different pBD2 concentrations. The OD260/OD280 ratios were between 1.8 and 2.2, and RNA concentrations were above 1000 μg/mL, according to spectrophotometer analysis. The differentially expressed gene fragments were obtained by ACP-based RT-PCR, and were generally the same after different treatments for each arbitrary primer. A subset of the results is shown in Supplementary Figure S1. The fragments were cloned and sequenced.
2.5. Analysis of DEGs
The sequences were analyzed using the ENSEMBL and NCBI databases, and 31 different differentially expressed gene fragments were obtained. Besides two intergenic fragments and two unknown genes (hypothetical proteins), the other 27 fragments corresponded to 31 possible functional genes and two intergenic sequences (listed in Table 1). As there are no available genome databases of S. aureus ATCC 25923, five sequences were found to correspond to different genes or intergenic sequences in different S. aureus strains (underlined in the sequence column in Table 1). The 31 possible known DEGs revealed that 10 genes were related to ion or protein transmembrane transport in the membrane, and all 10 were integral components of the membrane; 11 genes were related to the transfer of genetic information including DNA repairs, DNA replication, DNA transcription, transcriptional regulation, and translation in the cytoplasm; and 10 genes were related to glucose, lipid, and protein metabolism in the membrane or cytoplasm (listed in Table 2). In total, 13 genes were in the membranes and 12 genes were in the cytoplasm; the locations of the other five genes are uncertain, but their molecular functions suggest that they are in the cytoplasm. There were four genes next to two differentially expressed intergenic fragments and were involved in biofilm and spore formation. Taken together, pBD2 has multiple actions that affect cell transmembrane transport, the transport of inheritable information, and some metabolic processes, but its main effects are on transmembrane transport.

Table 1.
The differentially expressed gene fragments determined by the GeneFishing DEGs kit.

Table 2.
The positions, functions, and abbreviations of functional genes.
2.6. qRT-PCR Confirmation for Selected Genes
qRT-PCR was used to confirm the DEGs. The relative expression of each gene was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene expression. The results of qRT-PCR are consistent with those of the ACP-based RT-PCR (Figure 5 and Table 1).
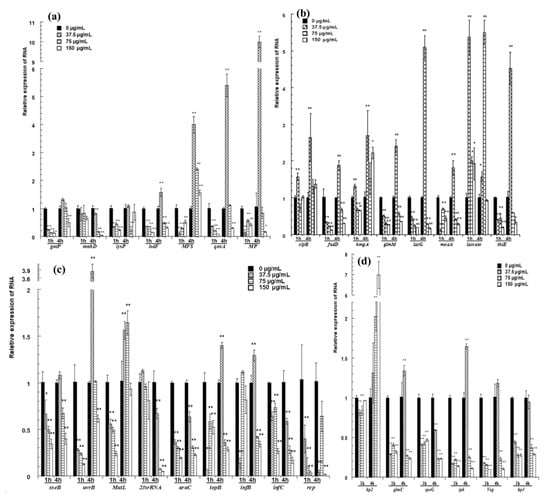
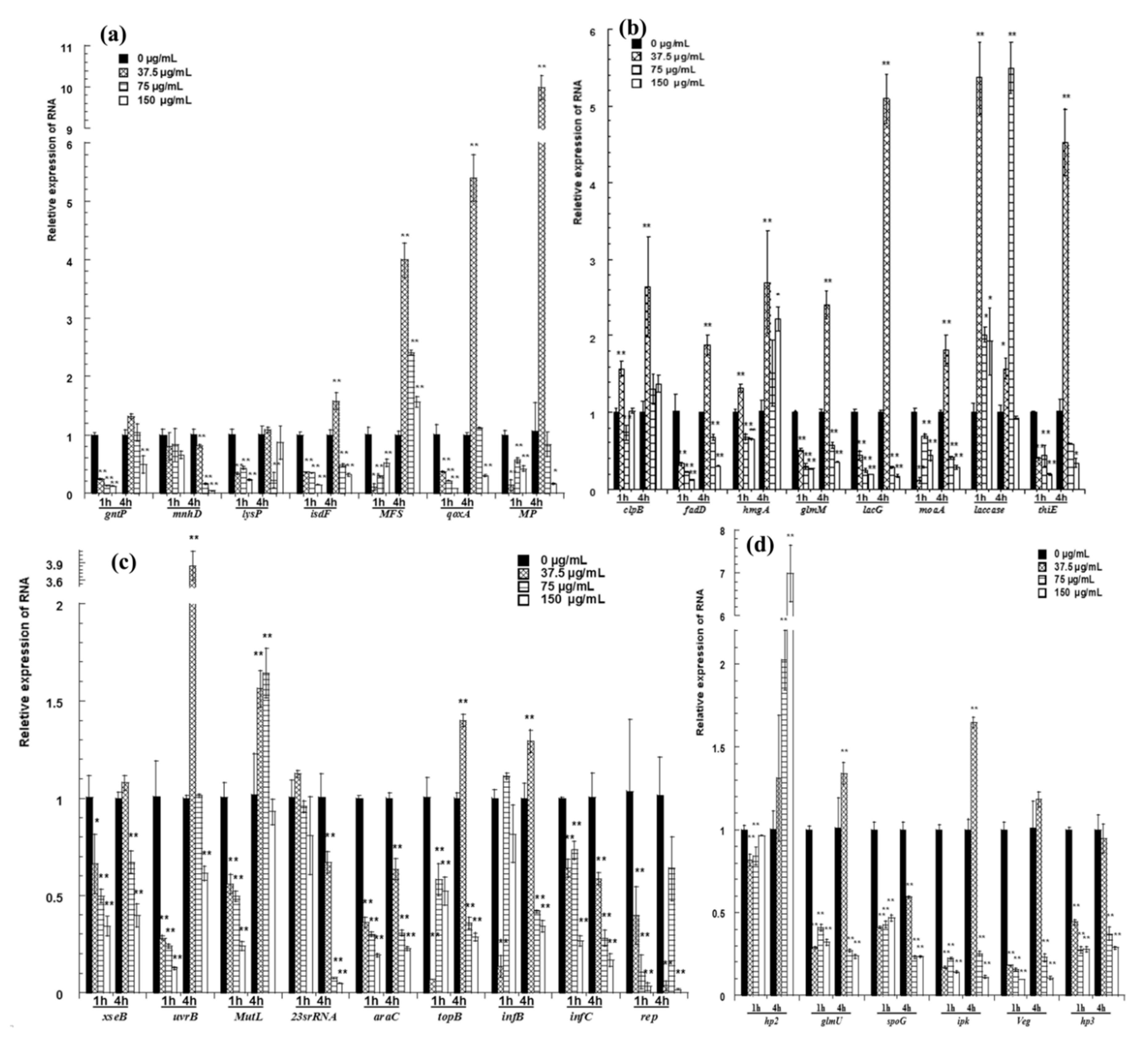
Figure 5.
qRT-PCR confirmation for selected genes. (a) The relative expression of genes for transport; (b) the expression of genes related to metabolism; (c) the relative expression of genes related to transport of inheritable information; (d) the relative expression of hypothetical proteins and the genes next to two differentially expressed intergenic fragments. The changes in gene expression were calculated by the 2−△△CT method with GAPDH as a reference gene. The * and ** symbols at the top of the column indicate significant differences from the control at p < 0.05 and p < 0.01, respectively.
The expression of genes for transport is shown in Figure 5a. After cells were treated with pBD2 for 1 h, all of these genes had lower expression levels than those of the control and showed significant differences, except for mnhD (p < 0.01). After cells were treated for 4 h, MFS was upregulated, and the expression levels of other genes were lower, except for those of isdF, qoxA and MP, which were upregulated in response to 37.5 μg/mL pBD2.
The expression of genes related to metabolism is shown in Figure 5b. Laccase was generally significantly upregulated. pBD2 treatment decreased the expression of fadD, glmM, lacG, moaA, and thiE, except for the treatment at 37.5 μg/mL pBD2 for 4 h (p < 0.01 or p < 0.05). The expression levels of clpB and hmgA were higher with treatment for 4 h and at 37.5 μg/mL pBD2 for 1 h.
The genes related to the transport of inheritable information were generally downregulated and are shown in Figure 5c. The genes related to DNA repair were generally downregulated, except for at the lower concentration for 4 h. The expression levels of genes related to DNA transcription/translation were 0.01–0.6-fold lower than those of the control, except for those of topB and infB, which had higher expression at 37.5 μg/mL pBD2 for 4 h (p < 0.01).
The genes next to two differentially expressed intergenic fragments were selected and detected (Figure 5d). glmU, spoG, ipk, and veg were downregulated, except for glmU and ipk at 37.5 μg/mL pBD2 for 4 h (p <0.01). The expression of hp3, located beside the possible intergenic sequence, also changed significantly (p < 0.01). Whether the sequences regulated the expression of nearby genes was not confirmed, although their nearby genes were changed. hp2 expression was obviously changed, and its function needs further study.
2.7. Gel Retardation Assay of pBD2 Binding to DNA
The ability of pBD2 to bind DNA was assessed by a gel retardation assay. pBD2 at different concentrations was fused with DNA for 1.5 h at room temperature, and then DNA was separated by agarose gel (Supplementary Figure S2). pBD2 bound to DNA and induced gel retardation at all concentrations tested. The DNA was significantly reduced with increasing pBD2 concentrations; the pBD2-bound DNA was trapped, and its migration through pores was retarded. The results suggest that pBD2 has DNA-binding activity.
3. Discussion
pBD2 has high antibacterial activity, which makes it an ideal candidate as an antibiotic [7,8]. Our previous reports showed that pBD2 with His-Tag had high antibacterial activity against S. aureus including multi-resistant bacteria [7]. The bacterial concentration for antimicrobial tests was usually approximately 104–106 cfu/mL. In this study, 109 cfu/mL S. aureus was used because a large number of bacterial cells were needed in subsequent experiments. It was surprising that purified recombinant pBD2 had the same strong bactericidal ability, and antibacterial activity increased with pBD2 concentrations (p < 0.01), which is consistent with the results of experiments using the 105–106 cfu/mL bacteria reported previously by our lab [7].
The morphological changes in S. aureus were very similar after incubation with different pBD2 concentrations (Figure 2). The morphological changes indicated some extent of pBD2-induced damage to cell membranes. However, some cells did not have obvious damage, which may indicate that pBD2 has a clustering effect and kills S. aureus cells one by one, and about 65.7% of cells were labeled with green fluorescent by IFM, confirming the results. Our results are consistent with the effects of the antimicrobial peptides gramicidin S and PGLa [26]. In addition, the bacterial biomass collected after treatment with higher pBD2 concentrations was lower compared with those at lower concentrations, and the debris in the total observed field of view was more visible at higher pBD2 concentrations, which indicates that cells were dead.
According to IFM, and further confirmed by immune-gold TEM, pBD2 mainly localized to the membrane and also entered the cytoplasm of S. aureus. In addition, cells in which more pBD2 entered the cytoplasm than localized to the cell membrane did not show obvious membrane disruption (Figure 4b). The main reason may be that lipid–peptide interaction in live bacteria is a dynamic process that causes transient disruption of the cell membrane without destroying its structural integrity [27]. Some studies have reported that AMPs mainly acted on the membrane and led to cell death such as T9W, tachyplesin I, cathelicidin, and bovine lactoferricin derivatives [28,29,30,31,32]. Some results indicated that AMPs had different locations for killing different species of microbes such as Bac7 and SpHyastatin. Bac7 mainly localized to the cell surface of P. aeruginosa and killed bacteria, while it targeted protein synthesis when acting on other Gram-negative bacteria [24,33]. SpHyastatin acted specifically on the surface of S. aureus, whereas it could enter the cytoplasm of P. fluorescens [34]. Furthermore, some results showed that antimicrobial peptides had different locations, even if they had similar structures such as buforin II, Indolicidin, and their analogs [20,35]. Buforin II with a P11A mutation localized to the membrane, while buforin II entered the cells [35]. Indolicidin (IN), and its analogs IN-1 and IN-2 were detected on the membranes of E. coli, whereas IN-3 and IN-4 penetrated the membranes [20]. In this study, pBD2 was mainly located on the membrane, and some entered the cytoplasm, which may imply that pBD2 acts on the membrane and internal targets.
In this study, RNA was not extracted by phenol, which causes specific losses of polyadenylated-RNA [36]. Therefore, the results prove that the prokaryotic RNA obtained by ACP-based RT-PCR contained polyadenylic acid without adding poly(A). ACP-based RT-PCR technology is an easy differential genes analysis technology that uses agarose gel electrophoresis, which is widely used to identify DEGs in response to different conditions [37,38,39]. In this study, ACP-based RT-PCR was used to detect DEGs in response to pBD2 treatments. With this technique, DEGs were detected and found to be highly similar in response to different pBD2 concentration treatments for each annealing control primer, which means that ACP-based RT-PCR used in this study is credible and repeatable. Furthermore, the results of ACP-based RT-PCR were confirmed by those of qRT-PCR. In this study, 31 DEGs were obtained: the 31 possible known functional genes are involved in transport, DNA, and some metabolic processes, but they are mainly related to transport through the membrane.
Our results indicate that transmembrane transport was strongly influenced and accounted for one- third of DEGs. Among the 10 transporters, bcrB, isdF, and lysP belong to the ATP-binding cassette (ABC) superfamily protein; qoxA and MFS belong to the major facilitator superfamily. These two ubiquitous families of transporters account for nearly half of the solute transporters encoded within the genomes of microorganisms, and they transport a wide variety of substrates including ions, amino acids, peptides, proteins, lipids, sugars, and so on across extra- and intracellular membranes to import nutrients or to export waste and toxic products [40,41]. Aside from these, qoxA, mnhA, and mnhD are involved in ATP synthesis-coupled electron transfer. The expression of these genes significantly changed, indicating that transmembrane transport and energy states were strongly influenced. Using microarray analysis, Sass et al. reported that the modes of action of hBD3 against S. aureus were related to ABC transporters, and knockout of the transporter genes significantly enhanced susceptibility to hBD3 [42]. Antimicrobial peptide LL-37 has been reported to kill bacteria mainly by targeting energy metabolism [19]. Wenzelet al. reported that antimicrobial peptides had impacts on the membrane including the inhibition of the respiratory chain and a reduction in ATP levels [43]. kdpB is an important part of potassium ion (K+) transport, which is a critical determinant of growth and survival through the regulation of cytoplasmic pH and cell structure; a loss of cytoplasmic potassium leads to cell death [44,45]. It was reported that defensins could inhibit potassium channels, which was one of the antimicrobial mechanisms [46,47]. The changes in transmembrane transporters influenced transport, energy states, ion balance, and so on, which are all causes of cell death.
Eleven genes were related to the transport of genetic information. It has been reported that antimicrobial peptides interact with intracellular molecules such as DNA, which can subsequently influence DNA or RNA synthesis or protein synthesis to kill bacteria, especially proline-rich antimicrobial peptides [48,49]. N4, Oncocin and its derivatives and so on were reported to bind specifically to DNA, disrupt DNA conformation and inhibit DNA, RNA or protein synthesis [22,50,51]. Tryptophan-rich antimicrobial peptides also have this mechanism [52]. Our gel retardation assay results show that pBD2 could bind to DNA. pBD2 interacted with DNA, and according to the functional analysis of DEGs, it affected all the transfer processes of genetic information. The expression levels of these genes were generally downregulated (p < 0.01), except for those of uvrB, MutL, topB, and infB, which were upregulated after treatment with 37.5 μg/mL pBD2 for 4 h, especially uvrB and MutL, which may mean that these surviving cells repaired their damage in this condition, but this also needs further research.
The genes for metabolic processes were also influenced, and generally downregulated (p < 0.01). These were related to carbohydrate, lipid, and protein metabolism and mainly related to biosynthetic processes, especially membrane biogenesis. fadD, glmM, and oal are involved in the biogenesis of the membrane. Moreover, glmU and spoG, ipk and veg (next to two differentially expressed intergenic fragments) mainly influence membrane biogenesis and cell proliferation. It was reported that human beta defensin 3 (hBD3) killed S. aureus by inhibiting cell wall biosynthesis [53]. Dosunmuet al. reported that peptide TP359 decreased the expression of outer membrane biogenesis genes in Pseudomonas aeruginosa, which reduced the organism’s structural stability and led to cell death [54]. Synthetic cationic peptide 1037 also significantly reduced the biofilm formation of Pseudomonas aeruginosa and led to cell death [55]. Antimicrobial peptidomimetics (SAMPs) and Hyicin 4244 acted on staphylococcal biofilms [56,57]. Antimicrobial peptides C18G (Anaspec) blocked cell division [58].
Five fragments corresponded to more than one gene, and the expression levels of these genes changed markedly according to qRT-PCR, which was selected for confirmation. InfC, rep, and isdF corresponded to the same DEG, and they all changed significantly. However, based on the consistency of qRT-PCR and ACP-based RT-PCR results, the DEG is likely to be isdF, and not infC or rep. araC and lysP also corresponded to the same DEG; the results of qRT-PCR for both genes were consistent with those of ACP-based RT-PCR, and it was difficult to distinguish between them. The other three sequences were also difficult to distinguish and need further analysis. In addition, genes next to differentially expressed intergenic fragments were detected; although it is not directly evident that the sequences regulated the expression of their nearby genes, the expression of their nearby genes changed.
From the analysis above, pBD2 has multiple and variable modes of action. It acted on the cell membranes, affected DNA and influenced biosynthetic processes, among other actions. Overall, the main action of pBD2 is the disruption of the cell membrane and interaction with DNA.
4. Materials and Methods
4.1. Preparation of Porcine Beta Defensin 2
Based on the constructed engineering strain E. coli BL(DE3)-pET-pBD2 constructed in our laboratory, recombinant pBD2 was induced and purified by the His-Tag affinity column according to the procedure reported previously [7]. The concentrations were determined by the bicinchoninic-acid (BCA) method.
4.2. Bacteria with pBD2 Treatment
The killing kinetics of pBD2 were determined by the turbidimetric method [59]. Overnight cultured S. aureus ATCC 25923 was transferred into fresh medium, cultured at 37 °C, and 220 rpm for 2–4 h until the OD600 value was about 1 (bacteria were in the logarithmic phase, and the density was about 109 cfu/mL), and then S. aureus ATCC 25923 cells were incubated with different pBD2 concentrations in a polypropylene 96-well microtiter plate for different times. The growth of S. aureus was measured by optical density (OD) at dual wavelengths (630 nm and 405 nm) by a microplate reader (Stat Fax 2100, Awarenss Technology Inc., Palm, FL, USA). The survival percentages were calculated and defined as the ratio of cell density in the presence of pBD2 to the cell density without pBD2, according to a previous report [59]. In addition, after S. aureus cells were incubated with pBD2 at 37 °C for 1 h or 4 h in tubes, the cells were harvested, washed, and then prepared for scanning electron microscopy (SEM), immuno-fluorescence microscopy (IFM), immuno-gold transmission electron microscopy (TEM), or RNA extraction.
4.3. Morphological Changes in S. aureus
Morphological changes in S. aureus were observed according to previous reports with some modifications [26,28]. In brief, after pBD2 treatment, S. aureus cells were fixed with 2.5% glutaraldehyde overnight at 4 °C, subsequently dehydrated with gradient concentrations of ethanol, and then placed in an ethanol and tert-butyl alcohol mixed solution (1:1). The treated samples were dropped on tin foil, and dried in the air. Sample were gold-coated by an ion spray instrument (MSP-2S, IXRF, Austin, TX, America) and then observed by environmental scanning electron microscopy (FEI Quanta 250, Hillsboro, OR, USA).
4.4. Localization of pBD2 Peptides
The locations of pBD2 were detected by IFM according to a previous report with some modification [60]. In brief, S. aureus cells were fixed with 4% paraformaldehyde for 2–4 h at 4 °C on anti-off slides (Wuhan Boster Biological Engineering Co., Ltd, Wuhan, China), and treated with 0.5% Triton-100 for 15 min. After incubation with 10% normal goat serum and then further incubated with the pBD2 polyclonal antibody (prepared by our group from rabbit [61], diluted to 1:500) at 4 °C overnight, these cells were finally incubated with FITC (fluorescein isothiocyanate)-labeled secondary antibodies for 1 h at 37 °C in the box. After that, cells were dyed by DAPI (4′,6-diamidino-2-phenylindole) solution for 5 min in the dark, blocked by a blocking agent containing anti-fluorescent quencher, and sealed with cover slips. Controls for IFM were performed by incubation with phosphate buffer (PB) instead of primary antibodies. The cells were observed by immunofluorescence microscopy (DM6000B, Leica, Wetzlar, Germany).
Immuno-gold TEM was used to further determine the localization of pBD2 and performed by the method reported previously [60]. The cells were embedded in 2% low-melting- point agarose and fixed with 3% paraformaldehyde (pH 7.4), dehydrated with gradient concentrations of methanol and finally infiltrated in pure Lowicryl K4M. The samples were cut into 0.1 μm thick sheets by a frozen slicer (CM1900, Leica, Wetzlar, Germany). Sections were incubated with 0.05% Triton X-100, 0.05% Tween, and 1% bovine serum albumin (BSA) in 0.01 M phosphate-buffered saline (PBS, pH 7.4) for 5 min at 20 °C. Then, they were incubated with the pBD2 antibody (1:500) for 2 h and protein-A gold (1:50) for 1 h. Finally, specimens were stained with 3% uranium acetate for 4 min and lead citrate for 1 min and observed by electron microscope (Tecnai F12 S-Twin, FEI, USA). The results from the control cells were obtained by incubating cells with PBS buffer without primary antibodies.
4.5. RNA Extraction
The total RNAs of S. aureus were extracted according to the procedure reported previously [36]. The extracted total RNA was treated with DNase for 30 min and purified again. The quality of RNAs was detected by 1% agarose gel electrophoresis, and the concentrations were determined by UV absorption using a spectrophotometer (Nanodrop 2000/2000C, Thermo Scientific, Waltham, MA, USA).
4.6. Identification and Analysis of DEGs
DEGs were identified according to the manufacturer’s instructions for the GeneFishingTM DEG Kit using 20 different arbitrary primers in combination with an oligo-dT anchor primer (Seegene, Seoul, South Korea). The differentially expressed gene fragments were obtained by ACP-based RT-PCR; the amplified PCR products were separated on 1% agarose gel, and DEGs were extracted from the gel using a DNA Purification Kit (TIANGEN, BeiJing, China) and directly cloned and sequenced. Sequences were compared with the databases in GenBank and Ensembl using the BLAST programs to find their functions.
4.7. Quantitative Real-Time PCR
qRT-PCR was performed with SYBR green dye, according to the manufacturer’s protocol (TaKaRa, Dalian, China) using a qRT-PCR platform (LightCycler 96, Roche, Base, Switzerland). All samples were analyzed in triplicate, and values were calculated by the 2−△△CT methods with GAPDH as a reference gene.
4.8. Gel Retardation Assay
A gel retardation assay was performed as described previously with some modifications [62]. In brief, 10 μL of different concentrations of pBD2 (0, 37.5, 75 and 150 μg/mL) were incubated with 10 μL of 129.2 ng/μL DNA for 1.5 h at room temperature, Then, 5 μL of the reaction solution was mixed with 2.5 μL of 6X gel loading buffer, and electrophoresed on 1% agarose gel for 30 min at 100 V.
4.9. Statistical Analysis
Analysis of variance (ANOVA) was performed with the software SPSS 17.0 (IBM, Armonk, NY, USA). LSD or Dunnett’s method was used to compare treatment means. Significance and extremely significance were defined as p < 0.05 and p < 0.01, respectively.
5. Conclusions
After S. aureus was cultured with different pBD2 concentrations, the morphology of S. aureus changed markedly, and the debris was highly visible. According to IFM and immune-gold TEM, pBD2 mainly localized to the cell membrane, and some entered the cytoplasm. The detected DEGs were related to transmembrane transport, DNA, and some metabolic processes. In addition, they were generally predominantly downregulated. In sum, the main mechanism by which pBD2 kills S. aureus is the destruction of cell membranes and interaction with DNA. Our study contributes to understanding the mechanism of pBD2 and lays the foundation for further research on its application in practice.
Supplementary Materials
The following are available online. Figure S1: Electrophoresis analysis of the DEGs by ACP-based RT-PCR. Figure S2: Gel retardation assay of pBD2 binding to DNA.
Author Contributions
K.Z. and C.G. performed the scanning electron microscopy and immuno-fluorescence microscopy experiments; H.Z. and R.C. performed the DEG analysis, qRT-PCR, and gel retardation assay. C.L. designed the experiments and supervised the whole study. C.L. and K.Z. analyzed the data and wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
The research was funded by the National Natural Science Foundation of China, grant number 31101694, and the University Science and Technology Talented Person Support Plan in Henan Province, grant number 14HASTIT021.
Acknowledgments
We would like to acknowledge Yun-qin Li at Zhejiang University for her help performing the immuno-gold TEM.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Peschel, A.; Sahl, H.G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006, 4, 529–536. [Google Scholar] [CrossRef]
- Lehrer, R.; Ganz, T. Defensins of vertbrate animal. Curr. Opin. Immunol. 2002, 14, 96–102. [Google Scholar] [CrossRef]
- Silva, J.P.; Appelberg, R.; Gama, F.M. Antimicrobial peptides as novel anti-tuberculosis therapeutics. Biotechnol. Adv. 2016, 34, 924–940. [Google Scholar] [CrossRef]
- Sang, Y.; Patil, A.A.; Zhang, G.; Ross, C.R.; Blecha, F. Bioinformatic and expression analysis of novel porcine beta-defensins. Mamm. Genome 2006, 17, 332–339. [Google Scholar] [CrossRef]
- Choi, M.K.; Le, M.T.; Nguyen, D.T.; Choi, H.; Kim, W.; Kim, J.H.; Chun, J.; Hyeon, J.; Seo, K.; Park, C. Genome-level identification, gene expression, and comparative analysis of porcine β-defensin genes. BMC Genet. 2012, 13, 98. [Google Scholar] [CrossRef]
- Li, C.L.; Xu, T.T.; Chen, R.B.; Huang, X.X.; Zhao, Y.C.; Bao, Y.Y.; Zhao, W.D.; Zheng, Z.Y. Cloning, expression and characterization of antimicrobial porcine beta defensin 1 in Escherichia coli. Protein Expr. Purif. 2013, 88, 47–53. [Google Scholar] [CrossRef]
- Li, C.L.; Zhao, Y.C.; Song, X.Y.; Huang, X.X.; Zhao, W.D. Molecular cloning, expression and characterization of the porcine beta defensin 2 in E. coli. Protein Pept. Lett. 2012, 20, 715–723. [Google Scholar] [CrossRef]
- Veldhuizen, E.J.; Rijnders, M.; Claassen, E.A.; van Dijk, A.; Haagsman, H.P. Porcine beta-defensin 2 displays broad antimicrobial activity against pathogenic intestinal bacteria. Mol. Immunol. 2008, 45, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Xie, H.; Su, G.; Chen, D.; Yu, B.; Mao, X.; Huang, Z.; Yu, J.; Luo, J.; Zheng, P.; et al. Beta-defensin 129 attenuates bacterial endotoxin-induced inflammation and intestinal epithelial cell apoptosis. Front. Immunol. 2019, 10, 2333. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Xie, K.; Chen, D.; Yu, B.; Huang, Z.; Luo, Y.; Mao, X.; Zheng, P.; Yu, J.; Luo, J.; et al. Differential expression, molecular cloning, and characterization of porcine beta defensin 114. J. Anim. Sci. Biotechnol. 2019, 10, 60. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Xu, L.; Shi, B.; Deng, H.; Lai, X.; Liu, J.; Sun, Z. Oral administration of synthetic porcine beta-defensin-2 improves growth performance and cecal microbial flora and down-regulates the expression of intestinal toll-like receptor-4 and inflammatory cytokines in weaned piglets challenged with enterotoxigenic Escherichia coli. Anim. Sci. J. 2016, 87, 1258–1266. [Google Scholar] [CrossRef]
- Han, F.; Zhang, H.; Xia, X.; Xiong, H.; Song, D.; Zong, X.; Wang, Y. Porcine beta-defensin 2 attenuates inflammation and mucosal lesions in dextran sodium sulfate-induced colitis. J. Immunol. 2015, 194, 1882–1893. [Google Scholar] [CrossRef]
- Yang, X.; Cheng, Y.T.; Tan, M.F.; Zhang, H.W.; Liu, W.Q.; Zou, G.; Zhang, L.S.; Zhang, C.Y.; Deng, S.M.; Yu, L.; et al. Overexpression of porcine beta-defensin 2 enhances resistance to actinobacillus pleuropneumoniae infection in pigs. Infect. Immun. 2015, 83, 2836–2843. [Google Scholar] [CrossRef]
- Huang, C.; Yang, X.; Huang, J.; Liu, X.; Jin, H.; Huang, Q.; Li, L.; Zhou, R. Porcine beta-defensin 2 provides protection against bacterial infection by a direct bactericidal activity and alleviates inflammation via interference with the TLR4/NF-kappaB pathway. Front. Immunol. 2019, 10, 1673. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Hong, M. Structure and mechanism of β-hairpin antimicrobial peptides in lipid bilayers from solid-state NMR spectroscopy. Mol. BioSyst. 2009, 5, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, W.; Hong, M. The membrane-bound structure and topology of a human α-defensin indicate a dimer pore mechanism for membrane disruption. Biochemistry 2010, 49, 9770–9782. [Google Scholar] [CrossRef] [PubMed]
- Sani, M.A.; Separovic, F. How membrane-active peptides get into lipid membranes. Acc. Chem. Res. 2016, 49, 1130–1138. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Haney, E.; Vogel, H. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Dong, S.L.; Xu, F.; Wang, X.Q.; Withers, T.R.; Yu, H.D.; Wang, X. Effect of intracellular expression of antimicrobial peptide LL-37 on growth of escherichia coli strain TOP10 under aerobic and anaerobic conditions. Antimicrob. Agents Chemother. 2013, 57, 4707–4716. [Google Scholar] [CrossRef]
- Nan, Y.H.; Park, K.H.; Park, Y.; Jeon, Y.J.; Kim, Y.; Park, I.S.; Hahm, K.S.; Shin, S.Y. Investigating the effects of positive charge and hydrophobicity on the cell selectivity, mechanism of action and anti-inflammatory activity of a Trp-rich antimicrobial peptide indolicidin. FEMS Microbiol. Lett. 2009, 292, 134–140. [Google Scholar] [CrossRef]
- Peters, B.M.; Shirtliff, M.E.; Jabra-Rizk, M.A. Antimicrobial peptides: Primeval molecules or future drugs? PLoS Pathog. 2010, 6, e1001067. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Teng, D.; Mao, R.; Yang, N.; Hao, Y.; Wang, J. Combined systems approaches reveal a multistage mode of action of a marine antimicrobial peptide against pathogenic Escherichia coli and its protective effect against bacterial peritonitis and endotoxemia. Antimicrob. Agents Chemother. 2017, 61, e01056-16. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, C.; Hu, D.; Ulmschneider, M.; Ulmschneider, J. Spontaneous formation of structurally diverse membrane channel architectures from a single antimicrobial peptide. Nat. Commun. 2016, 7, 13535. [Google Scholar] [CrossRef] [PubMed]
- Runti, G.; Benincasa, M.; Giuffrida, G.; Devescovi, G.; Venturi, V.; Gennaro, R.; Scocchi, M. The mechanism of killing by the proline-rich peptide Bac7(1-35) against clinical strains of Pseudomonas aeruginosa differs from that against other Gram-negative bacteria. Antimicrob. Agents Chemother. 2017, 61, e01660-16. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Hartmann, M.; Berditsch, M.; Hawecker, J.; Ardakani, M.F.; Gerthsen, D.; Ulrich, A.S. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob. Agents Chemother. 2010, 54, 3132–3142. [Google Scholar] [CrossRef]
- Gee, M.L.; Burton, M.; Grevis-James, A.; Hossain, M.A.; McArthur, S.; Palombo, E.A.; Wade, J.D.; Clayton, A.H. Imaging the action of antimicrobial peptides on living bacterial cells. Sci. Rep. 2013, 3, 1557. [Google Scholar] [CrossRef]
- Zhu, X.; Shan, A.; Ma, Z.; Xu, W.; Wang, J.; Chou, S.; Cheng, B. Bactericidal efficiency and modes of action of the novel antimicrobial peptide T9W against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2015, 59, 3008–3017. [Google Scholar] [CrossRef]
- Benincasa, M.; Scocchi, M.; Pacor, S.; Tossi, A.; Nobili, D.; Basaglia, G.; Busetti, M.; Gennaro, R. Fungicidal activity of five cathelicidin peptides against clinically isolated yeasts. J. Antimicrob. Chemother. 2006, 58, 950–959. [Google Scholar] [CrossRef]
- Hong, J.; Guan, W.; Jin, G.; Zhao, H.; Jiang, X.; Dai, J. Mechanism of tachyplesin I injury to bacterial membranes and intracellular enzymes, determined by laser confocal scanning microscopy and flow cytometry. Microbiol. Res. 2015, 170, 69–77. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, P.; Wang, Z.; Liu, H.; Zhang, Y.; Jiang, S.; Han, W.; Xia, Q.; Zhao, P. Antibacterial mechanism of gloverin 2 from silkworm, Bombyx mori. Int. J. Mol. Sci. 2018, 19, 2275. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Li, Y.; Cao, S.; Wang, H.; Jiang, C.; Pang, S.; Hussain, M.A.; Hou, J. Antibacterial activity and mechanism of action of bovine lactoferricin derivatives with symmetrical amino acid sequences. Int. J. Mol. Sci. 2018, 19, 2951. [Google Scholar] [CrossRef] [PubMed]
- Seefeldt, A.C.; Graf, M.; Perebaskine, N.; Nguyen, F.; Arenz, S.; Mardirossian, M.; Scocchi, M.; Wilson, D.N.; Innis, C.A. Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 2016, 44, 2429–2438. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Zhu, K.; Peng, H.; Chen, B.; Liu, J.; Chen, F.; Ma, X.; Wang, S.; Qiao, K.; Wang, K. The new antimicrobial peptide SpHyastatin from the Mud Crab Scylla paramamosain with multiple antimicrobial mechanisms and high effect on bacterial infection. Front. Microbiol. 2016, 7, 1140. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; LaBouyer, M.A.; Darling, L.E.; Elmore, D.E. Bacterial spheroplasts as a model for visualizing membrane translocation of antimicrobial peptides. Antimicrob. Agents Chemother. 2016, 60, 6350–6352. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishna, Y.; Langley, D.; Sarkar, N. Detection of high levels of polyadenylate-containing RNA in bacteria by the use of a single-step RNA isolation procedure. Nucleic Acids Res. 1981, 9, 3545–3554. [Google Scholar] [CrossRef][Green Version]
- Hwang, K.C.; Lee, H.Y.; Cui, X.S.; Kim, J.H.; Kim, N.H. Identification of maternal mRNAs in porcine parthenotes at the 2-cell stage: A comparison with the blastocyst stage. Mol. Reprod. Dev. 2005, 70, 314–323. [Google Scholar] [CrossRef]
- Han, S.H.; Odathurai Saminathan, S.; Kim, S.J. Insulin stimulates gene expression of ferritin light chain in osteoblast cells. J. Cell. Biochem. 2010, 111, 1493–1500. [Google Scholar] [CrossRef]
- Yang, Z.; Peng, Z.; Wei, S.; Yu, Y.; Cai, P. Identification of differentially expressed genes in three-pistil mutation in wheat using annealing control primer system. Gene 2011, 485, 81–84. [Google Scholar] [CrossRef]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H., Jr. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998, 62, 1–34. [Google Scholar] [CrossRef]
- Dean, M.; Hamon, Y.; Chimini, G. The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid Res. 2001, 42, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Sass, V.; Pag, U.; Tossi, A.; Bierbaum, G.; Sahl, H.-G. Mode of action of human β-defensin 3 against Staphylococcus aureus and transcriptional analysis of responses to defensin challenge. Int. J. Med. Microbiol. 2008, 298, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, M.; Chiriac, A.I.; Otto, A.; Zweytick, D.; May, C.; Schumacher, C.; Gust, R.; Albada, H.B.; Penkova, M.; Kramer, U.; et al. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc. Natl. Acad. Sci. USA 2014, 111, E1409–E1418. [Google Scholar] [CrossRef] [PubMed]
- Roosild, T.P.; Castronovo, S.; Healy, J.; Miller, S.; Pliotas, C.; Rasmussen, T.; Bartlett, W.; Conway, S.J.; Booth, I.R. Mechanism of ligand-gated potassium efflux in bacterial pathogens. Proc. Natl. Acad. Sci. USA 2010, 107, 19784–19789. [Google Scholar] [CrossRef]
- Lee, H.; Hwang, J.S.; Lee, J.; Kim, J.I.; Lee, D.G. Scolopendin 2, a cationic antimicrobial peptide from centipede, and its membrane-active mechanism. Biochim. Biophys. Acta 2015, 1848, 634–642. [Google Scholar] [CrossRef]
- Yang, W.; Feng, J.; Xiang, F.; Xie, Z.; Zhang, G.; Sabatier, J.M.; Cao, Z.; Li, W.; Chen, Z.; Wu, Y. Endogenous animal toxin-like human beta-defensin 2 inhibits own K(+) channels through interaction with channel extracellular pore region. Cell. Mol. Life Sci. 2015, 72, 845–853. [Google Scholar] [CrossRef]
- Xie, Z.; Feng, J.; Yang, W.; Xiang, F.; Yang, F.; Zhao, Y.; Cao, Z.; Li, W.; Chen, Z.; Wu, Y. Human alpha-defensins are immune-related Kv1.3 channel inhibitors: New support for their roles in adaptive immunity. FASEB J. 2015, 29, 4324–4333. [Google Scholar] [CrossRef]
- Florin, T.; Maracci, C.; Graf, M.; Karki, P.; Klepacki, D.; Berninghausen, O.; Beckmann, R.; Vázquez-Laslop, N.; Wilson, D.; Rodnina, M.; et al. An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome. Nat. Struct. Mol. Biol. 2017, 24, 752–757. [Google Scholar] [CrossRef]
- Graf, M.; Mardirossian, M.; Nguyen, F.; Seefeldt, A.C.; Guichard, G.; Scocchi, M.; Innis, C.A.; Wilson, D.N. Proline-rich antimicrobial peptides targeting protein synthesis. Nat. Prod. Rep. 2017, 34, 702–711. [Google Scholar] [CrossRef]
- Gagnon, M.; Roy, R.; Lomakin, I.; Florin, T.; Mankin, A.; Steitz, T. Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Res. 2016, 44, 2439–2450. [Google Scholar] [CrossRef]
- Roy, R.; Lomakin, I.; Gagnon, M.; Steitz, T. The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat. Struct. Mol. Biol. 2015, 22, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Petersen, A.P.; Lau, C.K.; Jing, W.; Storey, D.G.; Vogel, H.J. Mechanism of action of puroindoline derived tryptophan-rich antimicrobial peptides. Biochim. Biophys. Acta 2013, 1828, 1802–1813. [Google Scholar] [CrossRef] [PubMed]
- Sass, V.; Schneider, T.; Wilmes, M.; Korner, C.; Tossi, A.; Novikova, N.; Shamova, O.; Sahl, H.G. Human beta-defensin 3 inhibits cell wall biosynthesis in Staphylococci. Infect. Immun. 2010, 78, 2793–2800. [Google Scholar] [CrossRef] [PubMed]
- Dosunmu, E.F.; Chaudhari, A.A.; Bawage, S.; Bakeer, M.K.; Owen, D.R.; Singh, S.R.; Dennis, V.A.; Pillai, S.R. Novel cationic peptide TP359 down-regulates the expression of outer membrane biogenesis genes in Pseudomonas aeruginosa: A potential TP359 anti-microbial mechanism. BMC Microbiol. 2016, 16, 192. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente-Núñez, C.; Korolik, V.; Bains, M.; Nguyen, U.; Breidenstein, E.B.; Horsman, S.; Lewenza, S.; Burrows, L.; Hancock, R.E. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 2012, 56, 2696–2704. [Google Scholar] [CrossRef]
- Flemming, K.; Klingenberg, C.; Cavanagh, J.P.; Sletteng, M.; Stensen, W.; Svendsen, J.S.; Flaegstad, T. High in vitro antimicrobial activity of synthetic antimicrobial peptidomimetics against staphylococcal biofilms. J. Antimicrob. Chemother. 2009, 63, 136–145. [Google Scholar] [CrossRef]
- Duarte, A.F.S.; Ceotto-Vigoder, H.; Barrias, E.S.; Souto-Padron, T.; Nes, I.F.; Bastos, M. Hyicin 4244, the first sactibiotic described in staphylococci, exhibits an anti-staphylococcal biofilm activity. Int. J. Antimicrob. Agents 2018, 51, 349–356. [Google Scholar] [CrossRef]
- Yadavalli, S.S.; Carey, J.N.; Leibman, R.S.; Chen, A.I.; Stern, A.M.; Roggiani, M.; Lippa, A.M.; Goulian, M. Antimicrobial peptides trigger a division block in Escherichia coli through stimulation of a signalling system. Nat. Commun. 2016, 7, 12340. [Google Scholar] [CrossRef]
- Huang, L.; Leong, S.S.; Jiang, R. Soluble fusion expression and characterization of bioactive human beta-defensin 26 and 27. Appl. Microbiol. Biotechnol. 2009, 84, 301–308. [Google Scholar] [CrossRef]
- Schneider, V.A.; Coorens, M.; Ordonez, S.R.; Tjeerdsma-van Bokhoven, J.L.; Posthuma, G.; van Dijk, A.; Haagsman, H.P.; Veldhuizen, E.J. Imaging the antimicrobial mechanism(s) of cathelicidin-2. Sci. Rep. 2016, 6, 32948. [Google Scholar] [CrossRef]
- Bao, Y.Y.; Li, L.; Zhang, H.; Gao, C.Y.; Xiao, C.B.; Li, C.L. Preparation of polyclonal antibody against porcine beta defensin 2 and identification of its distribution in tissues of pig. Genet. Mol. Res. 2015, 14, 18863–18871. [Google Scholar] [CrossRef] [PubMed]
- Velivelli, S.L.S.; Islam, K.T.; Hobson, E.; Shah, D.M. Modes of action of a bi-domain plant defensin MtDef5 against a bacterial pathogen Xanthomonas campestris. Front. Microbiol. 2018, 9, 934. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).