Involvement of Differentially Expressed microRNAs in the PEGylated Liposome Encapsulated 188Rhenium-Mediated Suppression of Orthotopic Hypopharyngeal Tumor
Abstract
:1. Introduction
2. Results
2.1. Effects of 188Re-Liposome on HPC Derived Orthotopic Tumors Using the Repeated Dose Regime
2.2. Use of NGS Analysis to Investigate the microRNA Expressive Profile of HPC Tumor Treated with 188Re-Liposome
2.3. Validation of microRNA Identified in NGS Data Using qPCR
2.4. Investigation of Differentially Expressed microRNAs in Clinical Samples
2.5. Prediction of Genes Targeted by 188Re-Liposome-Deregulated microRNAs
2.6. Analysis of the Molecular Pathways Regulated by 188Re-Liposome-Affected microRNA
2.7. Association of 188Re-Liposome-Regulated microRNA and Patients’ Survival Rate
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Plasmid
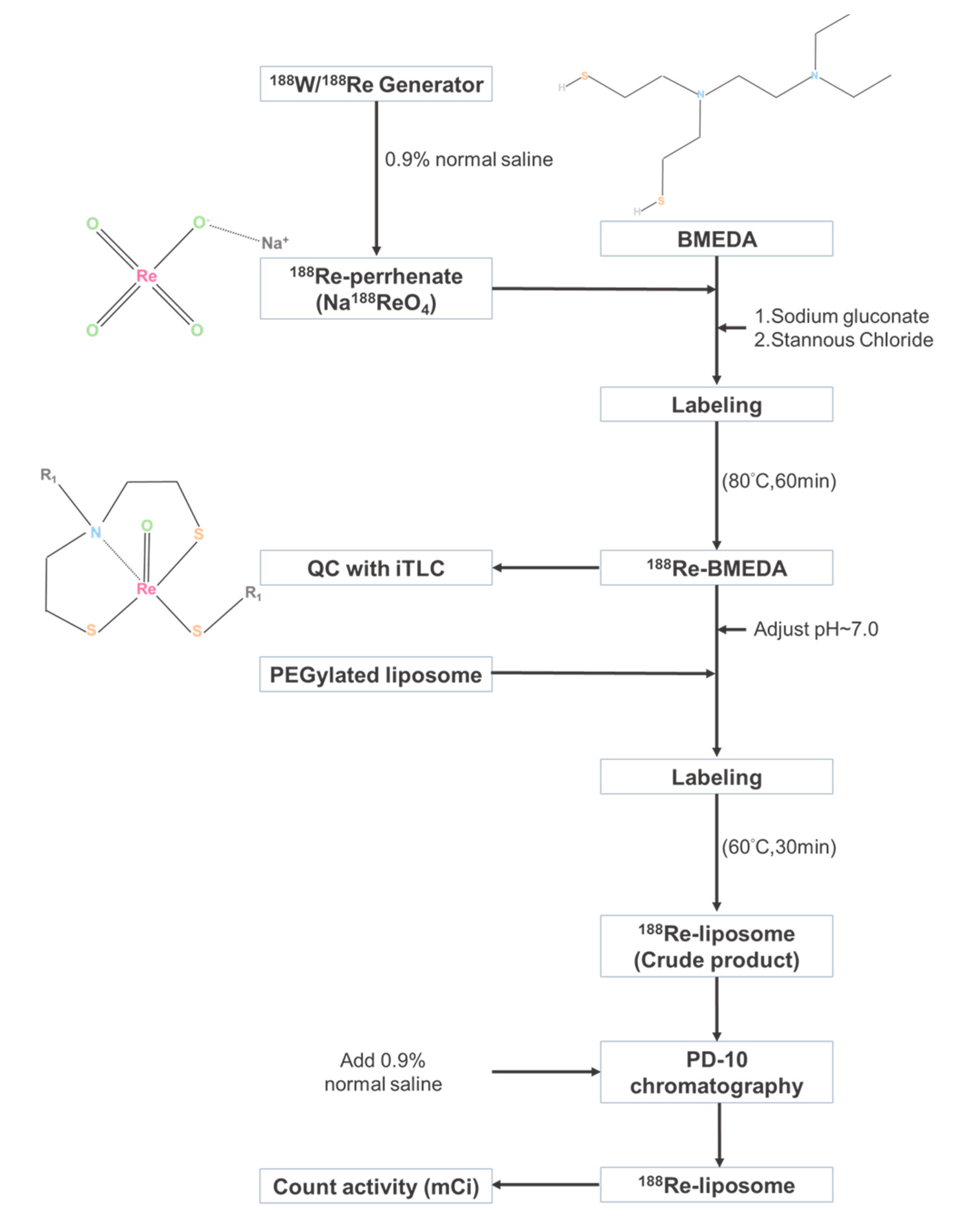
4.2. Preparation of 188Re-Liposome
4.3. Establishment of HPC Orthotopic Tumor Model for Evaluating the Therapeutic Efficacy of 188Re-Liposome
4.4. Tumor Collection and Next-Generation Sequencing (NGS)
4.5. MicroRNA Expression Analysis
4.6. Western Blot Analysis
4.7. Validation of microRNA Expression Using qPCR
4.8. Heatmap Analysis of NGS Data
4.9. Analysis of microRNA Using the Cancer Genome Atlas (TCGA)
4.10. Characterization of miRNAs
4.11. The Pathway Analysis
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Patel, R.S.; Goldstein, D.P.; Brown, D.; Irish, J.; Gullane, P.J.; Gilbert, R.W. Circumferential pharyngeal reconstruction: History, critical analysis of techniques, and current therapeutic recommendations. Head Neck 2010, 32, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.Y.; Chang, S.Y. Reconstruction of the hypopharynx after surgical treatment of squamous cell carcinoma. J. Chin. Med. Assoc. 2009, 72, 351–355. [Google Scholar] [CrossRef] [Green Version]
- Iwamoto, H. Operative treatment of cancer of the hypopharynx. Gan No Rinsho 1968, 14, 660–662. [Google Scholar] [PubMed]
- Kim, S.; Wu, H.G.; Heo, D.S.; Kim, K.H.; Sung, M.W.; Park, C.I. Advanced hypopharyngeal carcinoma treatment results according to treatment modalities. Head Neck 2001, 23, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Mura, F.; Bertino, G.; Occhini, A.; Benazzo, M. Surgical treatment of hypopharyngeal cancer: A review of the literature and proposal for a decisional flow-chart. Acta Otorhinolaryngol. Ital. 2013, 33, 299–306. [Google Scholar] [PubMed]
- Medvedeva, A.; Chernov, V.; Zeltchan, R.; Sinilkin, I.; Bragina, O.; Chijevskaya, S.; Choynzonov, E.; Goldberg, A. Nuclear medicine imaging of locally advanced laryngeal and hypopharyngeal cancer. In AIP Conference Proceedings; AIP Publishing LLC: Melville, NY, USA, 2017; Volume 1882. [Google Scholar]
- Jadvar, H.; Chen, X.; Cai, W.; Mahmood, U. Radiotheranostics in Cancer Diagnosis and Management. Radiology 2018, 286, 388–400. [Google Scholar] [CrossRef]
- Argyrou, M.; Valassi, A.; Andreou, M.; Lyra, M. Rhenium-188 production in hospitals, by w-188/re-188 generator, for easy use in radionuclide therapy. Int. J. Mol. Imaging 2013, 2013, 290750. [Google Scholar] [CrossRef] [Green Version]
- Liepe, K.; Hliscs, R.; Kropp, J.; Gruning, T.; Runge, R.; Koch, R.; Knapp, F.F., Jr.; Franke, W.G. Rhenium-188-HEDP in the palliative treatment of bone metastases. Cancer Biother. Radiopharm. 2000, 15, 261–265. [Google Scholar] [CrossRef]
- Zhang, H.; Tian, M.; Li, S.; Liu, J.; Tanada, S.; Endo, K. Rhenium-188-HEDP therapy for the palliation of pain due to osseous metastases in lung cancer patients. Cancer Biother. Radiopharm. 2003, 18, 719–726. [Google Scholar] [CrossRef]
- Guerra Liberal, F.D.C.; Tavares, A.A.S.; Tavares, J. Palliative treatment of metastatic bone pain with radiopharmaceuticals: A perspective beyond Strontium-89 and Samarium-153. Appl. Radiat. Isot. 2016, 110, 87–99. [Google Scholar] [CrossRef] [Green Version]
- Maxon, H.R., III; Schroder, L.E.; Washburn, L.C.; Thomas, S.R.; Samaratunga, R.C.; Biniakiewicz, D.; Moulton, J.S.; Cummings, D.; Ehrhardt, G.J.; Morris, V. Rhenium-188(Sn)HEDP for treatment of osseous metastases. J. Nucl. Med. 1998, 39, 659–663. [Google Scholar] [PubMed]
- Uccelli, L.; Martini, P.; Pasquali, M.; Boschi, A. Monoclonal Antibodies Radiolabeling with Rhenium-188 for Radioimmunotherapy. BioMed Res. Int. 2017, 2017, 5923609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edelman, M.J.; Clamon, G.; Kahn, D.; Magram, M.; Lister-James, J.; Line, B.R. Targeted radiopharmaceutical therapy for advanced lung cancer: Phase I trial of rhenium Re188 P2045, a somatostatin analog. J. Thorac. Oncol. 2009, 4, 1550–1554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Bal, C.; Srivastava, D.N.; Thulkar, S.P.; Sharma, S.; Acharya, S.K.; Duttagupta, S. Management of multiple intrahepatic recurrences after radiofrequency ablation of hepatocellular carcinoma with rhenium-188-HDD-lipiodol. Eur. J. Gastroenterol. Hepatol. 2006, 18, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Lambert, B.; Bacher, K.; De Keukeleire, K.; Smeets, P.; Colle, I.; Jeong, J.M.; Thierens, H.; Troisi, R.; De Vos, F.; Van de Wiele, C. 188Re-HDD/lipiodol for treatment of hepatocellular carcinoma: A feasibility study in patients with advanced cirrhosis. J. Nucl. Med. 2005, 46, 1326–1332. [Google Scholar] [PubMed]
- Nowicki, M.L.; Cwikla, J.B.; Sankowski, A.J.; Shcherbinin, S.; Grimmes, J.; Celler, A.; Buscombe, J.R.; Bator, A.; Pech, M.; Mikolajczak, R.; et al. Initial study of radiological and clinical efficacy radioembolization using 188Re-human serum albumin (HSA) microspheres in patients with progressive, unresectable primary or secondary liver cancers. Med. Sci. Monit. 2014, 20, 1353–1362. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.M.; Lee, Y.J.; Kim, E.H.; Chang, Y.S.; Kim, Y.J.; Son, M.; Lee, D.S.; Chung, J.K.; Lee, M.C. Preparation of (188) Re-labeled paper for treating skin cancer. Appl. Radiat. Isot. 2003, 58, 551–555. [Google Scholar] [CrossRef]
- Wang, S.J.; Huang, W.S.; Chuang, C.M.; Chang, C.H.; Lee, T.W.; Ting, G.; Chen, M.H.; Chang, P.M.; Chao, T.C.; Teng, H.W.; et al. A phase 0 study of the pharmacokinetics, biodistribution, and dosimetry of (188)Re-liposome in patients with metastatic tumors. Ejnmmi. Res. 2019, 9, 46. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.J.; Chang, C.H.; Chang, T.J.; Yu, C.Y.; Chen, L.C.; Jan, M.L.; Luo, T.Y.; Lee, T.W.; Ting, G. Biodistribution, pharmacokinetics and microSPECT/CT imaging of 188Re-bMEDA-liposome in a C26 murine colon carcinoma solid tumor animal model. Anticancer Res. 2007, 27, 2217–2225. [Google Scholar]
- Chang, C.H.; Liu, S.Y.; Chi, C.W.; Yu, H.L.; Chang, T.J.; Tsai, T.H.; Lee, T.W.; Chen, Y.J. External beam radiotherapy synergizes (1)(8)(8)Re-liposome against human esophageal cancer xenograft and modulates (1)(8)(8)Re-liposome pharmacokinetics. Int. J. Nanomed. 2015, 10, 3641–3649. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.Y.; Lee, T.W.; Chang, C.H.; Chen, L.C.; Hsu, W.H.; Chang, C.W.; Lo, J.M. Evaluation of 188Re-labeled PEGylated nanoliposome as a radionuclide therapeutic agent in an orthotopic glioma-bearing rat model. Int. J. Nanomed. 2015, 10, 463–473. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.T.; Chang, C.H.; Yu, H.L.; Liu, R.S.; Wang, H.E.; Chiu, S.J.; Chen, F.D.; Lee, T.W.; Lee, Y.J. Evaluation of the therapeutic and diagnostic effects of PEGylated liposome-embedded 188Re on human non-small cell lung cancer using an orthotopic small-animal model. J. Nucl. Med. 2014, 55, 1864–1870. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.T.; Chang, C.Y.; Chang, C.H.; Wang, H.E.; Chiou, S.H.; Liu, R.S.; Lee, T.W.; Lee, Y.J. Involvement of let-7 microRNA for the therapeutic effects of Rhenium-188-embedded liposomal nanoparticles on orthotopic human head and neck cancer model. Oncotarget 2016, 7, 65782–65796. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.A.; Lan, K.L.; Chang, C.H.; Lin, L.T.; He, C.L.; Chen, P.H.; Lee, T.W.; Lee, Y.J.; Chuang, C.M. Intraperitoneal (188)Re-Liposome delivery switches ovarian cancer metabolism from glycolysis to oxidative phosphorylation and effectively controls ovarian tumour growth in mice. Radiother. Oncol. 2016, 119, 282–290. [Google Scholar] [CrossRef]
- Chang, C.M.; Lan, K.L.; Huang, W.S.; Lee, Y.J.; Lee, T.W.; Chang, C.H.; Chuang, C.M. 188Re-Liposome Can Induce Mitochondrial Autophagy and Reverse Drug Resistance for Ovarian Cancer: From Bench Evidence to Preliminary Clinical Proof-of-Concept. Int. J. Mol. Sci. 2017, 18, 903. [Google Scholar] [CrossRef] [Green Version]
- Rutman, A.M.; Kuo, M.D. Radiogenomics: Creating a link between molecular diagnostics and diagnostic imaging. Eur. J. Radiol. 2009, 70, 232–241. [Google Scholar] [CrossRef]
- Kang, J.; Rancati, T.; Lee, S.; Oh, J.H.; Kerns, S.L.; Scott, J.G.; Schwartz, R.; Kim, S.; Rosenstein, B.S. Machine Learning and Radiogenomics: Lessons Learned and Future Directions. Front. Oncol. 2018, 8, 228. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Mermel, C.H.; Robinson, J.T.; Garraway, L.A.; Golub, T.R.; Meyerson, M.; Gabriel, S.B.; Lander, E.S.; Getz, G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014, 505, 495–501. [Google Scholar] [CrossRef] [Green Version]
- Schulze, K.; Nault, J.C.; Villanueva, A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J. Hepatol. 2016, 65, 1031–1042. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Roh, S.W.; Abell, G.C.; Kim, K.H.; Nam, Y.D.; Bae, J.W. Comparing microarrays and next-generation sequencing technologies for microbial ecology research. Trends Biotechnol. 2010, 28, 291–299. [Google Scholar] [CrossRef]
- Doostparast Torshizi, A.; Wang, K. Next-generation sequencing in drug development: Target identification and genetically stratified clinical trials. Drug Discov. Today 2018, 23, 1776–1783. [Google Scholar] [CrossRef]
- Liu, F.; Zhao, X.; Qian, Y.; Zhang, J.; Zhang, Y.; Yin, R. MiR-206 inhibits Head and neck squamous cell carcinoma cell progression by targeting HDAC6 via PTEN/AKT/mTOR pathway. Biomed. Pharm. 2017, 96, 229–237. [Google Scholar] [CrossRef]
- Koshizuka, K.; Hanazawa, T.; Fukumoto, I.; Kikkawa, N.; Matsushita, R.; Mataki, H.; Mizuno, K.; Okamoto, Y.; Seki, N. Dual-receptor (EGFR and c-MET) inhibition by tumor-suppressive miR-1 and miR-206 in head and neck squamous cell carcinoma. J. Hum. Genet. 2017, 62, 113–121. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Trachtenberg, A.J.; Kuo, W.P.; Cheng, Y.S. Genomewide Study of Salivary MicroRNAs for Detection of Oral Cancer. J. Dent. Res. 2014, 93, 86S–93S. [Google Scholar] [CrossRef]
- Shin, K.H.; Pucar, A.; Kim, R.H.; Bae, S.D.; Chen, W.; Kang, M.K.; Park, N.H. Identification of senescence-inducing microRNAs in normal human keratinocytes. Int. J. Oncol. 2011, 39, 1205–1211. [Google Scholar] [CrossRef] [Green Version]
- Lou, C.; Xiao, M.; Cheng, S.; Lu, X.; Jia, S.; Ren, Y.; Li, Z. MiR-485-3p and miR-485-5p suppress breast cancer cell metastasis by inhibiting PGC-1alpha expression. Cell Death Dis. 2016, 7, e2159. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.Q.; Zhang, M.Y.; Deng, M.L.; Weng, N.Q.; Wang, H.Y.; Wu, S.X. Low serum level of miR-485-3p predicts poor survival in patients with glioblastoma. PLoS ONE 2017, 12, e0184969. [Google Scholar] [CrossRef]
- Zhang, Y.; Sui, R.; Chen, Y.; Liang, H.; Shi, J.; Piao, H. Downregulation of miR-485-3p promotes glioblastoma cell proliferation and migration via targeting RNF135. Exp. Med. 2019, 18, 475–482. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Chen, C.; Yan, X.; Wang, P. The role of miR-382-5p in glioma cell proliferation, migration and invasion. Onco. Targets 2019, 12, 4993–5002. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Qi, B.; Guo, L.; Chen, L.Y.; Wei, X.F.; Liu, Y.Z.; Zhao, B.S. miR-382 functions as a tumor suppressor against esophageal squamous cell carcinoma. World J. Gastroenterol. 2017, 23, 4243–4251. [Google Scholar] [CrossRef]
- Zhu, W.J.; Chen, X.; Wang, Y.W.; Liu, H.T.; Ma, R.R.; Gao, P. MiR-1268b confers chemosensitivity in breast cancer by targeting ERBB2-mediated PI3K-AKT pathway. Oncotarget 2017, 8, 89631–89642. [Google Scholar] [CrossRef] [Green Version]
- Pu, Y.; Zhao, F.; Cai, W.; Meng, X.; Li, Y.; Cai, S. MiR-193a-3p and miR-193a-5p suppress the metastasis of human osteosarcoma cells by down-regulating Rab27B and SRR, respectively. Clin. Exp. Metastasis 2016, 33, 359–372. [Google Scholar] [CrossRef] [Green Version]
- Chou, N.H.; Lo, Y.H.; Wang, K.C.; Kang, C.H.; Tsai, C.Y.; Tsai, K.W. MiR-193a-5p and -3p Play a Distinct Role in Gastric Cancer: miR-193a-3p Suppresses Gastric Cancer Cell Growth by Targeting ETS1 and CCND1. Anticancer Res. 2018, 38, 3309–3318. [Google Scholar] [CrossRef]
- Yin, C.Y.; Kong, W.; Jiang, J.; Xu, H.; Zhao, W. miR-7-5p inhibits cell migration and invasion in glioblastoma through targeting SATB1. Oncol. Lett. 2019, 17, 1819–1825. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Luo, X.; Li, P.; Tan, J.; Wang, X.; Xiang, T.; Ren, G. miR-7-5p suppresses cell proliferation and induces apoptosis of breast cancer cells mainly by targeting REGgamma. Cancer Lett. 2015, 358, 27–36. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, B.; Ji, X.; Deng, Y.; Zhang, T.; Zhang, X.; Gao, H.; Sun, H.; Wu, H.; Chen, X.; et al. MicroRNA-378-5p suppresses cell proliferation and induces apoptosis in colorectal cancer cells by targeting BRAF. Cancer Cell Int. 2015, 15, 40. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Dai, S.; Zhen, T.; Shi, H.; Zhang, F.; Yang, Y.; Kang, L.; Liang, Y.; Han, A. Clinical and biological significance of miR-378a-3p and miR-378a-5p in colorectal cancer. Eur. J. Cancer. 2014, 50, 1207–1221. [Google Scholar] [CrossRef]
- Ostadrahimi, S.; Abedi Valugerdi, M.; Hassan, M.; Haddad, G.; Fayaz, S.; Parvizhamidi, M.; Mahdian, R.; Fard Esfahani, P. miR-1266-5p and miR-185-5p Promote Cell Apoptosis in Human Prostate Cancer Cell Lines. Asian Pac. J. Cancer Prev. 2018, 19, 2305–2311. [Google Scholar] [CrossRef]
- Shen, X.P.; Ling, X.; Lu, H.; Zhou, C.X.; Zhang, J.K.; Yu, Q. Low expression of microRNA-1266 promotes colorectal cancer progression via targeting FTO. Eur. Rev. Med. Pharm. Sci. 2018, 22, 8220–8226. [Google Scholar] [CrossRef]
- Hironaka-Mitsuhashi, A.; Matsuzaki, J.; Takahashi, R.U.; Yoshida, M.; Nezu, Y.; Yamamoto, Y.; Shiino, S.; Kinoshita, T.; Ushijima, T.; Hiraoka, N.; et al. A tissue microRNA signature that predicts the prognosis of breast cancer in young women. PLoS ONE 2017, 12, e0187638. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yuan, L.; Wang, G.; Cao, R.; Peng, J.; Shu, B.; Qian, G.; Wang, X.; Xiao, Y. Identification and bioinformatics analysis of miRNAs associated with human muscle invasive bladder cancer. Mol. Med. Rep. 2017, 16, 8709–8720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedersen, N.J.; Jensen, D.H.; Lelkaitis, G.; Kiss, K.; Charabi, B.W.; Ullum, H.; Specht, L.; Schmidt, A.Y.; Nielsen, F.C.; von Buchwald, C. MicroRNA-based classifiers for diagnosis of oral cavity squamous cell carcinoma in tissue and plasma. Oral. Oncol. 2018, 83, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, Y.; Lu, Q.; Fei, X.; Lu, C.; Li, C.; Chen, H. MiR-34a-5p Inhibits Proliferation, Migration, Invasion and Epithelial-mesenchymal Transition in Esophageal Squamous Cell Carcinoma by Targeting LEF1 and Inactivation of the Hippo-YAP1/TAZ Signaling Pathway. J. Cancer 2020, 11, 3072–3081. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Li, N.; Dong, Y.; Li, S.; Xu, L.; Li, X.; Li, Y.; Li, Z.; Ng, S.S.; Sung, J.J.; et al. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene 2015, 34, 4142–4152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Li, Q.; Niu, J.; Li, B.; Jiang, D.; Wan, Z.; Yang, Q.; Jiang, F.; Wei, P.; Bai, S. microRNA-342-5p and miR-608 inhibit colon cancer tumorigenesis by targeting NAA10. Oncotarget 2016, 7, 2709–2720. [Google Scholar] [CrossRef] [Green Version]
- Soriano, A.; Masanas, M.; Boloix, A.; Masia, N.; Paris-Coderch, L.; Piskareva, O.; Jimenez, C.; Henrich, K.O.; Roma, J.; Westermann, F.; et al. Functional high-throughput screening reveals miR-323a-5p and miR-342-5p as new tumor-suppressive microRNA for neuroblastoma. Cell Mol. Life Sci. 2019, 76, 2231–2243. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Yi, J.; Liu, X.; Chen, J.; Han, S.; Jin, L.; Chen, L.; Song, H. MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial-mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol. Cancer 2016, 15, 51. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Chen, X.; Hu, Y.; Ying, F.; Zou, R.; Lin, F.; Shi, Z.; Zhu, X.; Yan, X.; Li, S.; et al. LncRNA-TCONS_00026907 is involved in the progression and prognosis of cervical cancer through inhibiting miR-143-5p. Cancer Med. 2017, 6, 1409–1423. [Google Scholar] [CrossRef]
- Yang, F.; Xiao, Z.; Zhang, S. Knockdown of miR-194-5p inhibits cell proliferation, migration and invasion in breast cancer by regulating the Wnt/beta-catenin signaling pathway. Int. J. Mol. Med. 2018, 42, 3355–3363. [Google Scholar] [CrossRef] [Green Version]
- Jiang, M.J.; Chen, Y.Y.; Dai, J.J.; Gu, D.N.; Mei, Z.; Liu, F.R.; Huang, Q.; Tian, L. Dying tumor cell-derived exosomal miR-194-5p potentiates survival and repopulation of tumor repopulating cells upon radiotherapy in pancreatic cancer. Mol. Cancer 2020, 19, 68. [Google Scholar] [CrossRef] [PubMed]
- Kosela-Paterczyk, H.; Paziewska, A.; Kulecka, M.; Balabas, A.; Kluska, A.; Dabrowska, M.; Piatkowska, M.; Zeber-Lubecka, N.; Ambrozkiewicz, F.; Karczmarski, J.; et al. Signatures of circulating microRNA in four sarcoma subtypes. J. Cancer 2020, 11, 874–882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gullu Amuran, G.; Tinay, I.; Filinte, D.; Ilgin, C.; Peker Eyuboglu, I.; Akkiprik, M. Urinary micro-RNA expressions and protein concentrations may differentiate bladder cancer patients from healthy controls. Int. Urol. Nephrol. 2020, 52, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huang, Z.; Chen, R. Microrna-136 promotes proliferation and invasion ingastric cancer cells through Pten/Akt/P-Akt signaling pathway. Oncol. Lett. 2018, 15, 4683–4689. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, D.; Jin, L.U.; Liu, J.; Li, Y.; Su, Z.; Qi, Z.; Shi, M.; Jiang, Z.; Yang, S.; et al. Oncogenic microRNA-142-3p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Oncol. Lett. 2016, 11, 1235–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Li, T.; Zheng, L.; Huang, X. Biomarker MicroRNAs for Diagnosis of Oral Squamous Cell Carcinoma Identified Based on Gene Expression Data and MicroRNA-mRNA Network Analysis. Comput. Math Methods Med. 2017, 2017, 9803018. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Kim, J.; Eom, K.; Oh, S.; Kim, S.; Kim, G.; Ahn, S.; Park, K.H.; Chung, D.; Lee, H. microRNA-944 overexpression is a biomarker for poor prognosis of advanced cervical cancer. Bmc Cancer 2019, 19, 419. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Tian, W.; Chen, H.; Jiang, K. MiR-944 functions as a novel oncogene and regulates the chemoresistance in breast cancer. Tumour Biol. 2016, 37, 1599–1607. [Google Scholar] [CrossRef]
- Ma, Z.; Liu, T.; Huang, W.; Liu, H.; Zhang, H.M.; Li, Q.; Chen, Z.; Guo, A.Y. MicroRNA regulatory pathway analysis identifies miR-142-5p as a negative regulator of TGF-beta pathway via targeting SMAD3. Oncotarget 2016, 7, 71504–71513. [Google Scholar] [CrossRef] [Green Version]
- Islam, F.; Gopalan, V.; Vider, J.; Lu, C.T.; Lam, A.K. MiR-142-5p act as an oncogenic microRNA in colorectal cancer: Clinicopathological and functional insights. Exp. Mol. Pathol. 2018, 104, 98–107. [Google Scholar] [CrossRef] [Green Version]
- Lamperska, K.M.; Kolenda, T.; Teresiak, A.; Kowalik, A.; Kruszyna-Mochalska, M.; Jackowiak, W.; Blizniak, R.; Przybyla, W.; Kapalczynska, M.; Kozlowski, P. Different levels of let-7d expression modulate response of FaDu cells to irradiation and chemotherapeutics. PLoS ONE 2017, 12, e0180265. [Google Scholar] [CrossRef] [PubMed]
- Rangan, S.R. A new human cell line (FaDu) from a hypopharyngeal carcinoma. Cancer 1972, 29, 117–121. [Google Scholar] [CrossRef]
- Chang, C.Y.; Chen, C.C.; Lin, L.T.; Chang, C.H.; Chen, L.C.; Wang, H.E.; Lee, T.W.; Lee, Y.J. PEGylated liposome-encapsulated rhenium-188 radiopharmaceutical inhibits proliferation and epithelial-mesenchymal transition of human head and neck cancer cells in vivo with repeated therapy. Cell Death Discov. 2018, 4, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonner, W.M.; Redon, C.E.; Dickey, J.S.; Nakamura, A.J.; Sedelnikova, O.A.; Solier, S.; Pommier, Y. GammaH2AX and cancer. Nat. Rev. Cancer 2008, 8, 957–967. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, J.W.; Li, M.; Cavenee, W.K.; Mitchell, P.S.; Zhou, X.; Tewari, M.; Furnari, F.B.; Taniguchi, T. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol. Cancer Res. 2011, 9, 1100–1111. [Google Scholar] [CrossRef] [Green Version]
- Xie, B.; Ding, Q.; Han, H.; Wu, D. miRCancer: A microRNA-cancer association database constructed by text mining on literature. Bioinformatics 2013, 29, 638–644. [Google Scholar] [CrossRef]
- Schuler, E.; Parris, T.Z.; Helou, K.; Forssell-Aronsson, E. Distinct microRNA expression profiles in mouse renal cortical tissue after 177Lu-octreotate administration. PLoS ONE 2014, 9, e112645. [Google Scholar] [CrossRef]
- Zajic, G.; Nair, T.S.; Ptok, M.; Van Waes, C.; Altschuler, R.A.; Schacht, J.; Carey, T.E. Monoclonal antibodies to inner ear antigens: I. Antigens expressed by supporting cells of the guinea pig cochlea. Hear. Res. 1991, 52, 59–71. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Vasiliou, K.; Nebert, D.W. Analysis and update of the human solute carrier (SLC) gene superfamily. Hum. Genom. 2009, 3, 195–206. [Google Scholar] [CrossRef] [Green Version]
- De Maio, A.; Yalamanchili, H.K.; Adamski, C.J.; Gennarino, V.A.; Liu, Z.; Qin, J.; Jung, S.Y.; Richman, R.; Orr, H.; Zoghbi, H.Y. RBM17 Interacts with U2SURP and CHERP to Regulate Expression and Splicing of RNA-Processing Proteins. Cell Rep. 2018, 25, 726–736.e7. [Google Scholar] [CrossRef] [Green Version]
- Bramerson, A.; Nyman, J.; Nordin, S.; Bende, M. Olfactory loss after head and neck cancer radiation therapy. Rhinology 2013, 51, 206–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiss, M.; Stash, E.B.; Vellucci, V.F.; Zhou, Z.L. Activation of the autocrine transforming growth factor alpha pathway in human squamous carcinoma cells. Cancer Res. 1991, 51, 6254–6262. [Google Scholar] [PubMed]
- Nagy, A.; Lanczky, A.; Menyhart, O.; Gyorffy, B. Author Correction: Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018, 8, 11515. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.S.; Jiang, W.W.; Smith, I.; Poeta, L.M.; Begum, S.; Glazer, C.; Shan, S.; Westra, W.; Sidransky, D.; Califano, J.A. MicroRNA alterations in head and neck squamous cell carcinoma. Int. J. Cancer 2008, 123, 2791–2797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagar, S.M.; Thomas, R.J.; Loverock, L.T.; Spittle, M.F. Olfactory sensations produced by high-energy photon irradiation of the olfactory receptor mucosa in humans. Int. J. Radiat. Oncol. Biol. Phys. 1991, 20, 771–776. [Google Scholar] [CrossRef]
- Hua, M.S.; Chen, S.T.; Tang, L.M.; Leung, W.M. Olfactory function in patients with nasopharyngeal carcinoma following radiotherapy. Brain Inj. 1999, 13, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Ruo Redda, M.G.; Allis, S. Radiotherapy-induced taste impairment. Cancer Treat. Rev. 2006, 32, 541–547. [Google Scholar] [CrossRef]
- Epstein, J.B.; Smutzer, G.; Doty, R.L. Understanding the impact of taste changes in oncology care. Support. Care Cancer 2016, 24, 1917–1931. [Google Scholar] [CrossRef]
- Carlsson, G.; Gullberg, B.; Hafström, L. Estimation of liver tumor volume using different formulas—An experimental study in rats. J. Cancer Res. Clin. Oncol. 1983, 105, 20–23. [Google Scholar] [CrossRef]
- Andrews, S.; Krueger, F.; Segonds-Pichon, A.; Biggins, L.; Krueger, C.; Wingett, S.; Montgomery, J. Babraham Bioinformatics-Fastqc a Quality Control Tool For High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 22 January 2019).
- Creighton, C.J.; Reid, J.G.; Gunaratne, P.H. Expression profiling of microRNAs by deep sequencing. Brief. Bioinform. 2009, 10, 490–497. [Google Scholar] [CrossRef]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, C.H.; Lin, L.T.; Wang, C.Y.; Chiu, Y.W.; Chou, Y.T.; Chiu, S.J.; Wang, H.E.; Liu, R.S.; Wu, C.Y.; Chan, P.C.; et al. Over-expression of cofilin-1 suppressed growth and invasion of cancer cells is associated with up-regulation of let-7 microRNA. Biochim. Biophys. Acta 2015, 1852, 851–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saldanha, A.J. Java Treeview-extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef] [Green Version]
- Deng, M.; Bragelmann, J.; Kryukov, I.; Saraiva-Agostinho, N.; Perner, S. FirebrowseR: An R client to the Broad Institute’s Firehose Pipeline. Database 2017, 2017, baw160. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Yang, Z.; Wu, L.; Wang, A.; Tang, W.; Zhao, Y.; Zhao, H.; Teschendorff, A.E. dbDEMC 2.0: Updated database of differentially expressed miRNAs in human cancers. Nucleic. Acids Res. 2017, 45, D812–D818. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Lieven Sterck, V.d.P.Y. Draw Venn Diagram-Bioinformatics.psb.ugent.be. Available online: http://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 25 June 2020).
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds BMEDA and liposomes are available from the authors. |






| MicroRNA | Fold Change (188Re-lipo./Ctrl.) a | Ctrl. Norm. by TMM b | 188Re-lipo. Norm.by TMM | Ctrl. RPM c | 188Re-lipo. RPM |
|---|---|---|---|---|---|
| Hsa-miR-206-3p | 40.19 | 76.05 | 3056.53 | 2.92 | 90.71 |
| Hsa-miR-668-3p | 11.16 | 2.45 | 27.38 | 0.09 | 0.81 |
| Hsa-miR-485-3p | 9.77 | 2.45 | 23.96 | 0.09 | 0.71 |
| Hsa-miR-382-5p | 9.15 | 22.08 | 201.94 | 0.85 | 5.99 |
| Hsa-miR-1268b-5p | 8.37 | 2.45 | 20.54 | 0.09 | 0.61 |
| Hsa-miR-193a-5p | 6.28 | 4.91 | 30.8 | 0.19 | 0.91 |
| Hsa-miR-7-1-5p | 6.28 | 9.81 | 61.61 | 0.38 | 1.83 |
| Hsa-miR-378a-5p | 5.78 | 51.52 | 297.78 | 1.98 | 8.84 |
| Hsa-miR-1266-5p | 5.58 | 2.45 | 13.69 | 0.09 | 0.41 |
| Hsa-miR-4510-5p | 5.58 | 2.45 | 13.69 | 0.09 | 0.41 |
| Hsa-miR-370-3p | 5.58 | 4.91 | 27.38 | 0.19 | 0.81 |
| Hsa-miR-34a-5p | 5.15 | 127.57 | 657.17 | 4.9 | 19.5 |
| Hsa-miR-342-5p | 5.12 | 7.36 | 37.65 | 0.28 | 1.12 |
| Hsa-miR-3960-3p | −5.73 | 117.76 | 20.54 | 4.52 | 0.61 |
| Hsa-miR-143-5p | −5.73 | 19.63 | 3.42 | 0.75 | 0.1 |
| Hsa-miR-194-2-5p | −5.73 | 19.63 | 3.42 | 0.75 | 0.1 |
| Hsa-miR-151b-3p | −6.45 | 22.08 | 3.42 | 0.85 | 0.1 |
| Hsa-miR-136-3p | −7.17 | 24.53 | 3.42 | 0.94 | 0.1 |
| Hsa-miR-142-3p | −7.88 | 26.99 | 3.42 | 1.04 | 0.1 |
| Hsa-miR-944-3p | −9.32 | 31.89 | 3.42 | 1.22 | 0.1 |
| Hsa-miR-6723-5p | −10.03 | 34.35 | 3.42 | 1.32 | 0.1 |
| Hsa-miR-142-5p | −10.14 | 242.88 | 23.96 | 9.33 | 0.7 |
| MiRNAs up-Regulated in HPC | Original Change in HNSCC a | Role Prediction | Functional Importance | GEO ID | References b |
|---|---|---|---|---|---|
| Hsa-miR-206-3p | down-regulation | Tumor suppressor | Weaks cell proliferation, migration, invasion, promotes S phase cell arrest. Inhibits cell aggressiveness | TCGA_HNSC | [34,35] |
| Hsa-miR-668-3p | down-regulation | Tumor suppressor | Induces growth arrest and premature senescence | - c | [36,37] |
| Hsa-miR-485-3p | down-regulation | Tumor suppressor | Inhibits mitochondrial biogenesis or promotes cancer growth and migration | GSE75630 | [38,39,40] |
| Hsa-miR-382-5p | down-regulation | Tumor Suppressor | Promotes lymph node metastasis and TNM stage; inhibition of proliferation and EMT in glioma cells | TCGA_HNSC | [41,42] |
| Hsa-miR-1268b-5p | - | Tumor suppressor | Increases chemosensitivity | - | [43] |
| Hsa-miR-193a-5p | up-regulation | Tumor Suppressor | Suppresses the growth and the metastasis of cancer cells | TCGA_HNSC | [44,45] |
| Hsa-miR-7-1-5p | up-regulation | Tumor suppressor | Inhibits proliferation, invasion and induces apoptosis in cancer cells | TCGA_HNSC | [46,47] |
| Hsa-miR-378a-5p | down-regulation | Tumor suppressor | Inhibits cellular proliferation and colony formation. | TCGA_HNSC | [48,49] |
| Hsa-miR-1266-5p | down-regulation | Tumor suppressor | Induces apoptosis and reduces proliferation | TCGA_HNSC | [50,51] |
| Hsa-miR-4510-5p | - | Tumor suppressor | Down-regulation in recurrent cancer and a potential cancer biomarker | - | [52,53] |
| Hsa-miR-370-3p | down-regulation | Tumor suppressor | Potential cancer biomarker | TCGA_HNSC | [54] |
| Hsa-miR-34a-5p | down-regulation | Tumor suppressor | Inhibits tumorigenesis and progression | TCGA_HNSC | [55,56] |
| Hsa-miR-342-5p | up-regulation | Tumor suppressor | Reduces cell cycle progression | TCGA_HNSC | [57,58] |
| MiRNAs Down-Regulated in HPC | Original Change in HNSCC | Role Prediction | Functional Importance | GEO ID | Reference |
|---|---|---|---|---|---|
| Hsa-miR-3960-3p | - | - | - | - | - |
| Hsa-miR-143-5p | - | Both | cell viability, colony formation | - | [59,60] |
| Hsa-miR-194-2-5p | - | Oncogene | cell proliferation, migration and invasion | - | [61,62] |
| Hsa-miR-151b-3p | - | Oncogene? a | Biomarker of sarcoma | - | [63] |
| Hsa-miR-136-3p | down-regulation | Oncogene | Biomarker of bladder cancer, promote cancer growth and migration | TCGA-HNSC | [64,65] |
| Hsa-miR-142-3p | up-regulation | Oncogene | Over-expression in OSCC, association with cancer growth and migration | TCGA-HNSC | [66,67] |
| Hsa-miR-944-3p | up-regulation | Oncogene | A biomarker of poor prognosis/Regulation of chemoresistance | TCGA-HNSC | [68,69] |
| Hsa-miR-6723-5p | - | - | - | - | - |
| Hsa-miR-142-5p | up-regulation | Oncogene | Deregulation of cell proliferation; SMAD3/TGF-β | GSE31277 | [70,71] |
| Pathways Description | No. of DEGs with Annotated Pathways (4498) a | Percentage of DEGs with Annotated Pathways (4498) | Down Regulated Gene | Up Regulated Gene | Unknown Regulated Gene | No. of All Genes with Annotated Pathways (6883) | Percentage of All Genes with Annotated Pathways (6883) | p-Value b |
|---|---|---|---|---|---|---|---|---|
| Olfactory transduction | 51 | 1.13% | 14 | 33 | 4 | 415 | 6.03% | 6.37 × 10−45 |
| Pathways in cancer | 329 | 7.31% | 95 | 184 | 50 | 397 | 5.77% | 0.00127 |
| Taste transduction | 16 | 0.36% | 2 | 11 | 3 | 52 | 0.76% | 0.00373 |
| HTLV-I infection | 217 | 4.82% | 61 | 117 | 39 | 258 | 3.75% | 0.00623 |
| Proteoglycans in cancer | 176 | 3.91% | 60 | 93 | 23 | 203 | 2.95% | 0.00739 |
| Neuroactive ligand-receptor interaction | 139 | 3.09% | 40 | 84 | 15 | 277 | 4.02% | 0.00779 |
| MicroRNAs in cancer | 151 | 3.36% | 41 | 89 | 21 | 297 | 4.31% | 0.00885 |
| Viral carcinogenesis | 175 | 3.89% | 47 | 90 | 38 | 205 | 2.98% | 0.0103 |
| Chemical carcinogenesis | 32 | 0.71% | 4 | 21 | 7 | 81 | 1.18% | 0.01124 |
| Hippo signaling pathway | 135 | 3.00% | 34 | 82 | 19 | 154 | 2.24% | 0.01459 |
| Focal adhesion | 174 | 3.87% | 55 | 90 | 29 | 207 | 3.01% | 0.01626 |
| MAPK signaling pathway | 209 | 4.65% | 53 | 128 | 28 | 255 | 3.70% | 0.01732 |
| Drug metabolism - cytochrome P450 | 27 | 0.60% | 5 | 17 | 5 | 68 | 0.99% | 0.01947 |
| Metabolism of xenobiotics by cytochrome P450 | 30 | 0.67% | 4 | 20 | 6 | 74 | 1.08% | 0.01954 |
| ErbB signaling pathway | 82 | 1.82% | 30 | 40 | 12 | 87 | 1.26% | 0.02121 |
| Signaling pathways regulating pluripotency of stem cells | 124 | 2.76% | 29 | 80 | 15 | 142 | 2.06% | 0.02202 |
| Endocytosis | 208 | 4.62% | 65 | 116 | 27 | 258 | 3.75% | 0.02587 |
| FoxO signaling pathway | 117 | 2.60% | 32 | 75 | 10 | 134 | 1.95% | 0.02612 |
| Ras signaling pathway | 185 | 4.11% | 50 | 109 | 26 | 227 | 3.30% | 0.0272 |
| Neurotrophin signaling pathway | 106 | 2.36% | 28 | 64 | 14 | 120 | 1.74% | 0.02764 |
| Regulation of actin cytoskeleton | 175 | 3.89% | 49 | 98 | 28 | 214 | 3.11% | 0.03031 |
| Chronic myeloid leukemia | 70 | 1.56% | 22 | 40 | 8 | 73 | 1.06% | 0.03053 |
| Transcriptional misregulation in cancer | 149 | 3.31% | 43 | 80 | 26 | 179 | 2.60% | 0.03365 |
| Glioma | 63 | 1.40% | 23 | 30 | 10 | 65 | 0.94% | 0.03554 |
| Pancreatic cancer | 64 | 1.42% | 19 | 35 | 10 | 66 | 0.96% | 0.03682 |
| Colorectal cancer | 60 | 1.33% | 17 | 30 | 13 | 62 | 0.90% | 0.03971 |
| Acute myeloid leukemia | 56 | 1.24% | 17 | 28 | 11 | 57 | 0.83% | 0.04123 |
| Protein processing in endoplasmic reticulum | 140 | 3.11% | 40 | 78 | 22 | 169 | 2.46% | 0.04447 |
| Prostate cancer | 80 | 1.78% | 26 | 38 | 16 | 89 | 1.29% | 0.04703 |
| TGF-beta signaling pathway | 76 | 1.69% | 23 | 46 | 7 | 84 | 1.22% | 0.04999 |
| MicroRNA | Stem Loop Primer Sequence |
| miR-206-3p | 5′-GTCGTATCCAGTGCAGGGTCCGAG |
| GTATTCGCACTGGATACGACCCACAC-3′ | |
| miR-382-5p | 5′-GTCGTATCCAGTGCAGGGTCCGA |
| GGTATTCGCACTGGATACGACCGAATC-3′ | |
| miR-378a-5p | 5′-GTCGTATCCAGTGCAGGGTCCGAG |
| GTATTCGCACTGGATACGACACACAG-3′ | |
| miR-3960-3p | 5′-GTCGTATCCAGTGCAGGGTCCGAG |
| GTATTCGCACTGGATACGACCCCCCG-3′ | |
| miR-142-5p | 5′-GTCGTATCCAGTGCAGGGTCCGAG |
| GTATTCGCACTGGATACGACAGTAGT-3′ | |
| MicroRNA | Forward Primer Sequence |
| miR-206-3p | 5′-CACGCATGGAATGTAAGGAAGT-3′ |
| miR-382-5p | 5′-CACGCAGAAGTTGTTCGTGGTG-3′ |
| miR-378a-5p | 5′-TGATTACTCCTGACTCCAGGTC-3′ |
| miR-3960-3p | 5′-TAATTATGGCGGCGGCGGAG-3′ |
| miR-142-5p | 5′-CACGCGCATAAAGTAGAAAGCA-3′ |
| MicroRNA | Reverse Primer Sequence |
| Universal reverse primer | 5′-CCAGTGCAGGGTCCGAGGT-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, B.-Z.; Wan, S.-Y.; Lin, M.-Y.; Chang, C.-H.; Chen, T.-W.; Yang, M.-H.; Lee, Y.-J. Involvement of Differentially Expressed microRNAs in the PEGylated Liposome Encapsulated 188Rhenium-Mediated Suppression of Orthotopic Hypopharyngeal Tumor. Molecules 2020, 25, 3609. https://doi.org/10.3390/molecules25163609
Lin B-Z, Wan S-Y, Lin M-Y, Chang C-H, Chen T-W, Yang M-H, Lee Y-J. Involvement of Differentially Expressed microRNAs in the PEGylated Liposome Encapsulated 188Rhenium-Mediated Suppression of Orthotopic Hypopharyngeal Tumor. Molecules. 2020; 25(16):3609. https://doi.org/10.3390/molecules25163609
Chicago/Turabian StyleLin, Bing-Ze, Shen-Ying Wan, Min-Ying Lin, Chih-Hsien Chang, Ting-Wen Chen, Muh-Hwa Yang, and Yi-Jang Lee. 2020. "Involvement of Differentially Expressed microRNAs in the PEGylated Liposome Encapsulated 188Rhenium-Mediated Suppression of Orthotopic Hypopharyngeal Tumor" Molecules 25, no. 16: 3609. https://doi.org/10.3390/molecules25163609







