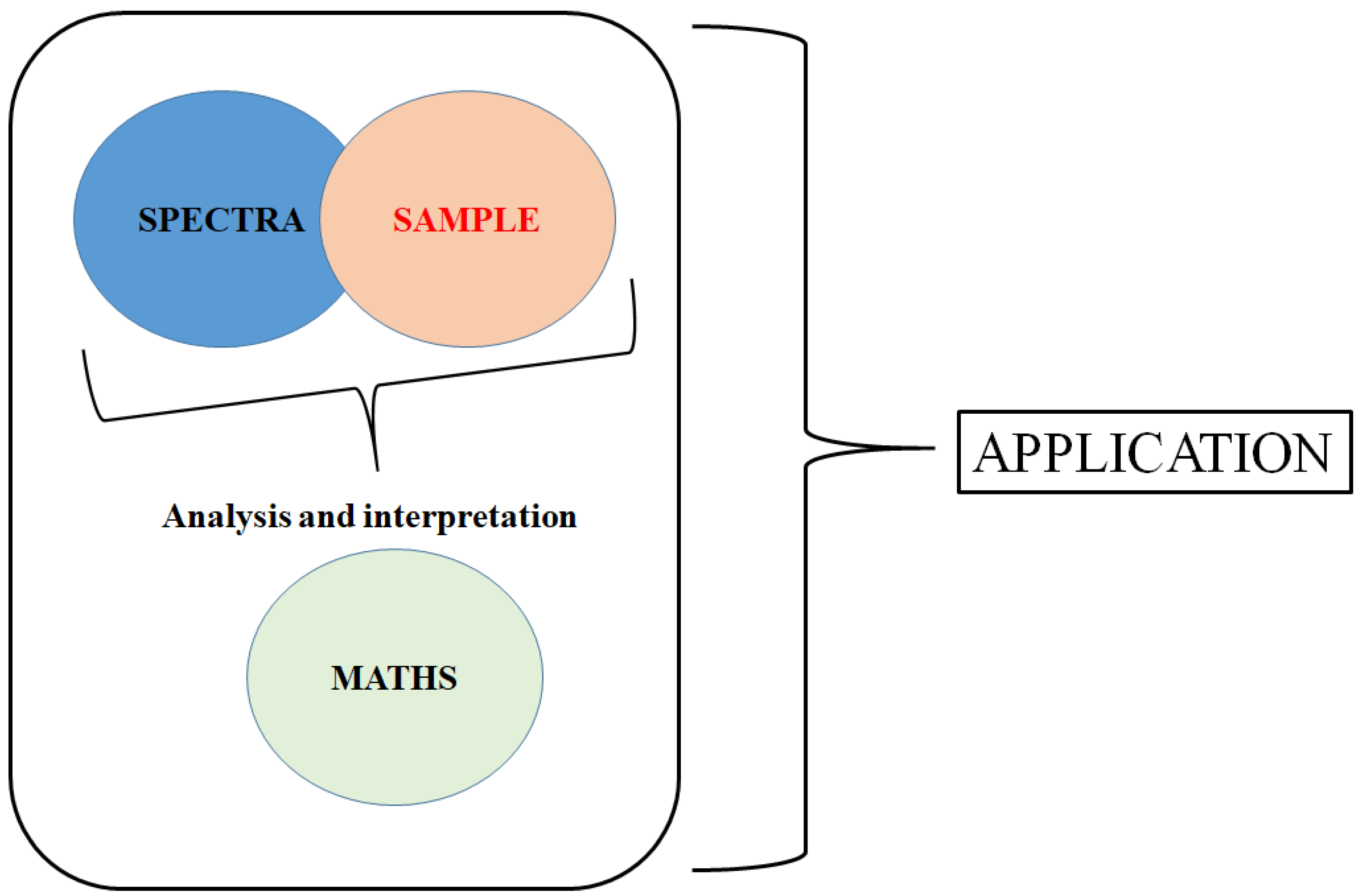
The Sample, the Spectra and the Maths—The Critical Pillars in the Development of Robust and Sound Applications of Vibrational Spectroscopy
Abstract
:1. Introduction
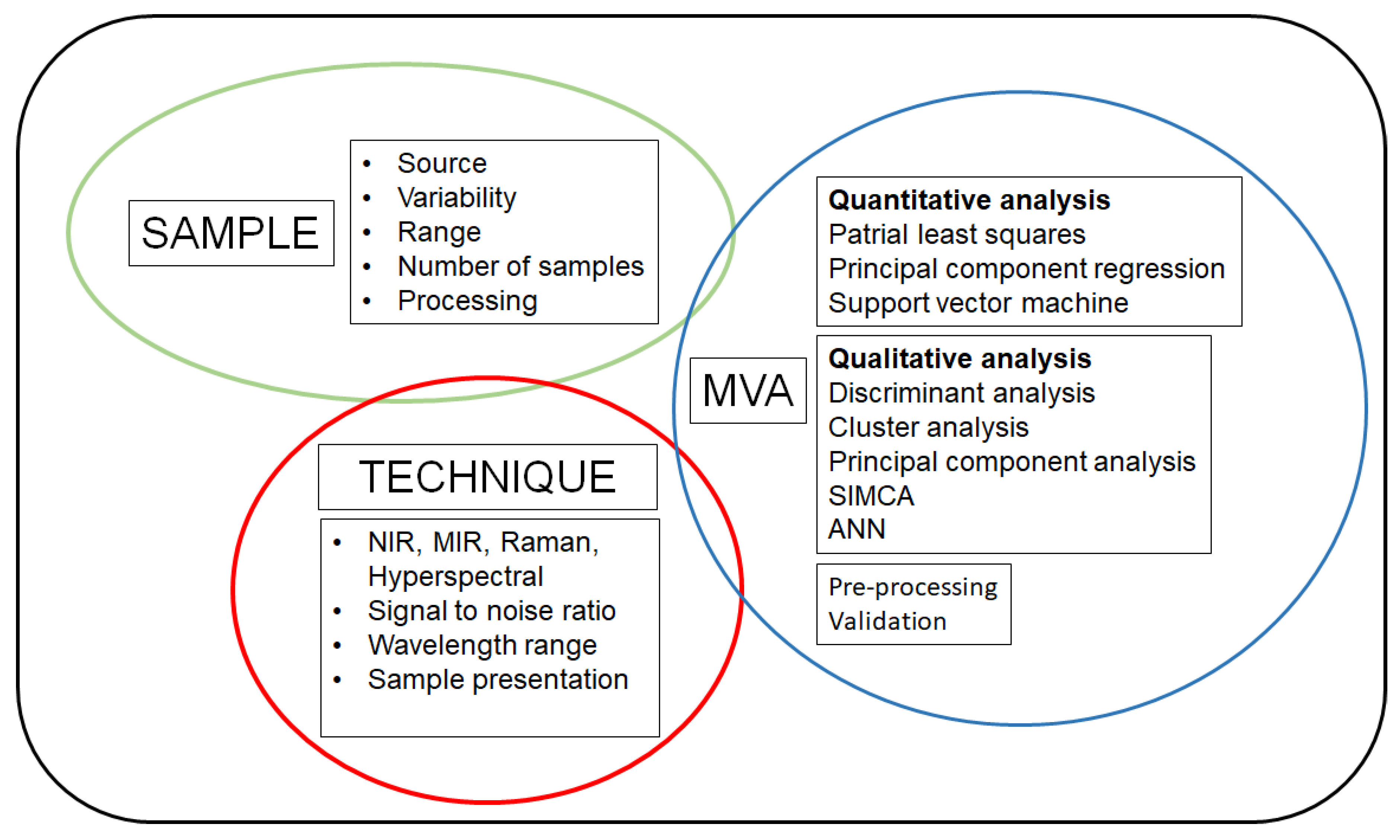
2. The Source of Information—The Experiment and the Sample
2.1. The Theory of Sampling and Uncertainty
2.2. Samples
2.3. Sample Properties and Pre-Processing
2.4. Sample Variability
3. Collecting the Information—The Spectra
4. Analysing and Interpreting the Information—The Maths
4.1. Data Pre-Processing
4.2. Mistakes and Error during Analysis and Interpretation of the Data
4.3. Algorithms Used to Develop Models
4.4. Validation
4.5. Data Interpretation
5. Outliers, Overfitting and Underfitting
6. Concluding Remarks
Funding
Conflicts of Interest
References
- Burlingame, B. Holistic and reductionist nutrition. J. Food Compos. Anal. 2004, 17, 585–586. [Google Scholar] [CrossRef]
- Castro-Puyana, M.; Mendiola, J.A.; Ibáñez, E. Strategies for a cleaner new scientific discipline of green foodomics. TrAC Trends Anal. Chem. 2013, 52, 23–35. [Google Scholar] [CrossRef]
- Capozzi, F.; Bordoni, A. Foodomics: A new comprehensive approach to food and nutrition. Genes Nutr. 2012, 8, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cevallos-Cevallos, J.M.; Reyes-De-Corcuera, J.I.; Etxeberria, E.; Danyluk, M.D.; Rodrick, G.E. Metabolomic analysis in food science: A review. Trends Food Sci. Technol. 2009, 20, 557–566. [Google Scholar] [CrossRef]
- Truong, V.K.; Dupont, M.; Elbourne, A.; Gangadoo, S.; Pathirannahalage, P.R.; Cheeseman, S.; Chapman, J.; Cozzolino, D. From Academia to Reality Check: A Theoretical Framework on the Use of Chemometric in Food Sciences. Foods 2019, 8, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallo, M.; Ferranti, P. The evolution of analytical chemistry methods in foodomics. J. Chromatogr. A 2016, 1428, 3–15. [Google Scholar] [CrossRef]
- Fardet, A. New Approaches to Studying the Potential Health Benefits of Cereals: From Reductionism to Holism. Cereal Foods World 2014, 59, 224–229. [Google Scholar] [CrossRef]
- Garcia-Cañas, V.; Simo, C.; Herrero, M.; Ibáñez, E.; Cifuentes, A. Present and Future Challenges in Food Analysis: Foodomics. Anal. Chem. 2012, 84, 10150–10159. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Peng, J.-Y.; Xie, C.; Bao, Y.; He, Y. Fruit Quality Evaluation Using Spectroscopy Technology: A Review. Sensors 2015, 15, 11889–11927. [Google Scholar] [CrossRef] [Green Version]
- Tahir, H.E.; Zou, X.; Jianbo, X.; Mahunu, G.K.; Jiyong, S.; Xu, J.-L.; Sun, D.-W. Recent Progress in Rapid Analyses of Vitamins, Phenolic, and Volatile Compounds in Foods Using Vibrational Spectroscopy Combined with Chemometrics: A Review. Food Anal. Methods 2019, 12, 2361–2382. [Google Scholar] [CrossRef]
- Amodio, M.L.; Chaudhry, M.M.A.; Colelli, G. Spectral and Hyperspectral Technologies as an Additional Tool to Increase Information on Quality and Origin of Horticultural Crops. Agronomy 2019, 10, 7. [Google Scholar] [CrossRef] [Green Version]
- Cortés, V.; Blasco, J.; Aleixos, N.; Cubero, S.; Talens, P. Monitoring strategies for quality control of agricultural products using visible and near-infrared spectroscopy: A review. Trends Food Sci. Technol. 2019, 85, 138–148. [Google Scholar] [CrossRef]
- Roberts, J.; Cozzolino, D. An Overview on the Application of Chemometrics in Food Science and Technology—An Approach to Quantitative Data Analysis. Food Anal. Methods 2016, 9, 3258–3267. [Google Scholar] [CrossRef]
- Wang, L.; Sun, D.-W.; Pu, H.; Cheng, J.-H. Quality analysis, classification, and authentication of liquid foods by near-infrared spectroscopy: A review of recent research developments. Crit. Rev. Food Sci. Nutr. 2016, 57, 1524–1538. [Google Scholar] [CrossRef] [PubMed]
- Alander, J.T.; Bochko, V.; Martinhauppi, B.; Saranwong, S.; Mantere, T. A review of optical non-destructive visual and near infrared methods for food quality and safety. Int. J. Spectrosc. 2013, 2013, 341402. [Google Scholar] [CrossRef]
- Manley, M. Near-infrared spectroscopy and hyperspectral imaging: Non-destructive analysis of biological materials. Chem. Soc. Rev. 2014, 43, 8200–8214. [Google Scholar] [CrossRef] [Green Version]
- Sorak, D.; Herberholz, L.; Iwascek, S.; Altinpinar, S.; Pfeifer, F.; Siesler, H.W. New Developments and Applications of Handheld Raman, Mid-Infrared, and Near-Infrared Spectrometers. Appl. Spectrosc. Rev. 2012, 47, 83–115. [Google Scholar] [CrossRef]
- Callao, M.; Ruisánchez, I. An overview of multivariate qualitative methods for food fraud detection. Food Control 2018, 86, 283–293. [Google Scholar] [CrossRef]
- Cozzolino, D. The role of vibrational spectroscopy as a tool to assess economically motivated fraud and counterfeit issues in agricultural products and foods. Anal. Methods 2015, 7, 9390–9400. [Google Scholar] [CrossRef]
- Power, A.; Cozzolino, D. How Fishy Is Your Fish? Authentication, Provenance and Traceability in Fish and Seafood by Means of Vibrational Spectroscopy. Appl. Sci. 2020, 10, 4150. [Google Scholar] [CrossRef]
- Eisen, K.; Eifert, T.; Herwig, C.; Maiwald, M. Current and future requirements to industrial analytical infrastructure—Part 1: Process analytical laboratories. Anal. Bioanal. Chem. 2020, 412, 2027–2035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eifert, T.; Eisen, K.; Maiwald, M.; Herwig, C. Current and future requirements to industrial analytical infrastructure—Part 2: Smart sensors. Anal. Bioanal. Chem. 2020, 412, 2037–2045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herwig, C. Applied basic science in process analytics and control technology. Anal. Bioanal. Chem. 2020, 412, 2025–2026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolinger, L.; Rudt, N.; Hubbuch, J. A critical review of recent trends, and a future perspective of optical spectroscopy as PAT in biopharmaceutical downstream processing. Anal. Bioanal. Chem. 2020, 412, 2047–2064. [Google Scholar] [CrossRef] [Green Version]
- Bendell, A.; Disney, J.; McCollin, C. The Future Role of Statistics in Quality Engineering and Management (with Discussion). J. R. Stat. Soc. A. Stat. 1999, 48, 299–326. [Google Scholar] [CrossRef]
- Brereton, R.G. Applied Chemometrics for Scientists; Johns Wiley & Sons Ltd.: Chichester, UK, 2007. [Google Scholar]
- Brereton, R.G. Introduction to multivariate calibration in analytical chemistry. Analyst 2000, 125, 2125–2154. [Google Scholar] [CrossRef]
- Buco, S.M. How good are your results? An approach to qualitative and quantitative statistical analysis for food monitoring and process control systems. Food Control 1990, 1, 40–46. [Google Scholar] [CrossRef]
- Granato, D.; Calado, V.M.D.A.; Jarvis, B. Observations on the use of statistical methods in Food Science and Technology. Food Res. Int. 2014, 55, 137–149. [Google Scholar] [CrossRef]
- Nunes, C.A.; Alvarenga, V.; Sant’Ana, A.D.S.; Santos, J.S.; Granato, D. The use of statistical software in food science and technology: Advantages, limitations and misuses. Food Res. Int. 2015, 75, 270–280. [Google Scholar] [CrossRef] [Green Version]
- Wold, S. Chemometrics; what do we mean with it, and what do we want from it? Chemom. Intell. Lab. Syst. 1995, 30, 109–115. [Google Scholar] [CrossRef]
- Bronson, K.; Knezevic, I. Big Data in food and agriculture. Big Data Soc. 2016, 3, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Ahearn, M.C.; Armbruster, W.; Young, R. Big Data’s Potential to Improve Food Supply Chain Environmental Sustainability and Food Safety. Int. Food Agribus. Man. 2016, 19, 155–171. [Google Scholar] [CrossRef]
- Szymańska, E. Modern data science for analytical chemical data—A comprehensive review. Anal. Chim. Acta 2018, 1028, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Esbensen, K.H.; Wagner, C. Theory of sampling (TOS) versus measurement uncertainty (MU)—A call for integration. Trends Analyt. Chem. 2014, 57, 93–106. [Google Scholar] [CrossRef]
- Wagner, C.; Esbensen, K.H. Theory of Sampling: Four Critical Success Factors before Analysis. J. AOAC Int. 2015, 98, 275–281. [Google Scholar] [CrossRef]
- Esbensen, K.H.; Swarbrick, B. Sampling for spectroscopic analysis: Consequences for multivariate calibration. Spectrosc. Eur. 2019, 3, 22–28. [Google Scholar]
- Analytical Methods Committee, AMCTB No. 71. Sampling theory and sampling uncertainty. Anal. Methods 2015, 7, 10085–10087. [Google Scholar] [CrossRef]
- Saeys, W.; Trong, N.N.D.; Van Beers, R.; Nicolaï, B.M. Multivariate calibration of spectroscopic sensors for postharvest quality evaluation: A review. Postharvest Biol. Technol. 2019, 158, 110981. [Google Scholar] [CrossRef]
- Cozzolino, D.; Shah, N.; Cynkar, W.; Smith, P. A practical overview of multivariate data analysis applied to spectroscopy. Food Res. Int. 2011, 44, 1888–1896. [Google Scholar] [CrossRef]
- Badertscher, M.; Pretsch, E. Bad results from good data. TrAC Trends Anal. Chem. 2006, 25, 1131–1138. [Google Scholar] [CrossRef]
- Murray, I. Forage analysis by near infrared spectroscopy. In Sward Management Handbook; Davies, A., Baker, R.D., Grant, S.A., Laidlaw, A.S., Eds.; British Grassland Society: Cheshire, UK, 1993; pp. 285–312. [Google Scholar]
- Murray, I. Hirschfeld Award. NIR spectroscopy of food: Simple things, subtle things and spectra. NIR News 1999, 10, 10–12. [Google Scholar] [CrossRef]
- Murray, I.; Cowe, I. Sample preparation. In Near Infrared Spectroscopy in Agriculture; Roberts, C.A., Workman, J., Reeves, J.B., Eds.; American Society of Agronomy, Crop Science Society of America, Soil Science Society of America: Madison, WI, USA, 2004; pp. 75–115. [Google Scholar]
- Cozzolino, D. Sample presentation, sources of error and future perspectives on the application of vibrational spectroscopy in the wine industry. J. Sci. Food Agric. 2014, 95, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.C. Implementation of Near-Infrared technology. In Near Infrared Technology in the Agricultural and Food Industries; Williams, P.C., Norris, K.H., Eds.; American Association of Cereal Chemist: St. Paul, MN, USA, 2001; pp. 145–169. [Google Scholar]
- Nicolai, B.M.; Beullens, K.; Bobelyn, E.; Peirs, A.; Saeys, W.; Theron, K.I.; Lammertyn, J. Non-destructive measurement of fruit and vegetable quality by means of NIR spectroscopy: A review. Postharvest Biol. Tec. 2007, 46, 99–118. [Google Scholar] [CrossRef]
- Brereton, R.G.; Jansen, J.; Lopes, J.; Marini, F.; Pomerantsev, A.L.; Rodionova, O.Y.; Roger, J.-M.; Walczak, B.; Tauler, R. Chemometrics in analytical chemistry—Part II: Modeling, validation, and applications. Anal. Bioanal. Chem. 2018, 410, 6691–6704. [Google Scholar] [CrossRef]
- Gani, W.; Limam, M. A kernel distance-based representative subset selection method. J. Stat. Comput. Simul. 2015, 86, 135–148. [Google Scholar] [CrossRef]
- Morais, C.L.M.; Santos, M.C.D.; Lima, K.M.G.; Martin, F.L. Improving data splitting for classification applications in spectrochemical analyses employing a random-mutation Kennard-Stone algorithm approach. Bioinformatics 2019, 35, 5257–5263. [Google Scholar] [CrossRef]
- Munck, L. A new holistic exploratory approach to Systems Biology by Near Infrared Spectroscopy evaluated by chemometrics and data inspection. J. Chemom. 2007, 21, 406–426. [Google Scholar] [CrossRef]
- Munck, L.; Nørgaard, L.; Engelsen, S.B.; Bro, R.; Andersson, C. Chemometrics in food science—A demonstration of the feasibility of a highly exploratory, inductive evaluation strategy of fundamental scientific significance. Chemom. Intell. Lab. Syst. 1998, 44, 31–60. [Google Scholar] [CrossRef]
- Munck, L.; Møller, J.B.; Rinnan, Å.; Fast, S.H.; Møller, E.M.; Nørgaard, L.; Balling, E.S. A physiochemical theory on the applicability of soft mathematical models—Experimentally interpreted. J. Chemom. 2010, 24, 481–495. [Google Scholar] [CrossRef]
- Khakimov, B.; Bak, S.; Engelsen, S.B. High-throughput cereal metabolomics: Current analytical technologies, challenges and perspectives. J. Cereal Sci. 2014, 59, 393–418. [Google Scholar] [CrossRef]
- Cattaneo, T.; Stellari, A. Review: NIR Spectroscopy as a Suitable Tool for the Investigation of the Horticultural Field. Agronomy 2019, 9, 503. [Google Scholar] [CrossRef] [Green Version]
- Pasquini, C. Near infrared spectroscopy: A mature analytical technique with new perspectives—A review. Anal. Chim. Acta 2018, 1026, 8–36. [Google Scholar] [CrossRef] [PubMed]
- Karoui, R.; Downey, G.; Blecker, C. Mid-Infrared Spectroscopy Coupled with Chemometrics: A Tool for the Analysis of Intact Food Systems and the Exploration of Their Molecular Structure−Quality Relationships—A Review. Chem. Rev. 2010, 110, 6144–6168. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.; Robards, K. Analytical Chemistry Considerations in Plant Metabolomics. Sep. Purif. Rev. 2006, 35, 319–356. [Google Scholar] [CrossRef]
- Beć, K.B.; Grabska, J.E.; Huck, C.W. Near-Infrared Spectroscopy in Bio-Applications. Molecules 2020, 25, 2948. [Google Scholar] [CrossRef]
- Cozzolino, D.; Roberts, J. Applications and Developments on the Use of Vibrational Spectroscopy Imaging for the Analysis, Monitoring and Characterisation of Crops and Plants. Molecules 2016, 21, 755. [Google Scholar] [CrossRef] [Green Version]
- Walsh, K.; McGlone, V.; Han, D. The uses of near infra-red spectroscopy in postharvest decision support: A review. Postharvest Biol. Technol. 2020, 163, 111139. [Google Scholar] [CrossRef]
- Beć, K.B.; Huck, C.W. Breakthrough Potential in Near-Infrared Spectroscopy: Spectra Simulation. A Review of Recent Developments. Front. Chem. 2019, 7. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Chen, J. A Review of Hyperspectral Imaging for Chicken Meat Safety and Quality Evaluation: Application, Hardware, and Software. Compr. Rev. Food Sci. Food Saf. 2019, 18, 535–547. [Google Scholar] [CrossRef] [Green Version]
- Crocombe, R.A. Portable Spectroscopy. Appl. Spectrosc. 2018, 72, 1701–1751. [Google Scholar] [CrossRef]
- Adams, M.J. Chemometrics in Analytical Spectroscopy. In Chemometrics in Analytical Spectroscopy; Royal Society of Chemistry (RSC): Cambridge, UK, 2004; p. 216. [Google Scholar]
- Blanco, M.; Bernardez, M. Multivariate calibration for quantitative analysis. In Infrared Spectroscopy for Food Quality Analysis and Control; Sun, D.W., Ed.; Elsevier: Oxford, UK, 2009; p. 448. [Google Scholar]
- Esbensen, K.H. Multivariate Data Analysis in Practice; CAMO Process AS: Oslo, Norway, 2002. [Google Scholar]
- Frank, I.E.; Friedman, J.H. A Statistical View of Some Chemometrics Regression Tools. Technometrics 1993, 35, 109. [Google Scholar] [CrossRef]
- Geladi, P.; Kowalski, B.R. Partial least-squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
- Geladi, P. Chemometrics in spectroscopy. Part 1. Classical chemometrics. Spectrochim. Acta Part B: At. Spectrosc. 2003, 58, 767–782. [Google Scholar] [CrossRef]
- Kumar, N.; Bansal, A.; Sarma, G.; Rawal, R.K. Chemometrics tools used in analytical chemistry: An overview. Talanta 2014, 123, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Haaland, D.M.; Thomas, E.V. Partial least-squares methods for spectral analyses. 1. Relation to other quantitative calibration methods and the extraction of qualitative information. Anal. Chem. 1988, 60, 1193–1202. [Google Scholar] [CrossRef]
- Mark, H.; Workman, J. Statistics in Spectroscopy, 2nd ed.; Elsevier BV: London, UK, 2003; 342p. [Google Scholar]
- Martens, H.M.A.M. Multivariate Analysis of Quality. An Introduction. Meas. Sci. Technol. 2001, 12, 1746. [Google Scholar] [CrossRef]
- Massart, D.L.; Vandegiste, B.G.M.; Deming, S.N.; Michotte, Y.; Kaufman, L. Chemometrics: A Textbook; Elsevier: Amsterdam, The Netherlands, 1988; 500p. [Google Scholar]
- Naes, T.; Isaksson, T.; Fearn, T.; Davies, T. A User-Friendly Guide to Multivariate Calibration and Classification; NIR Publications: Chichester, UK, 2002; 420p. [Google Scholar]
- Skov, T.; Honore, A.H.; Jensen, H.M.; Næs, T.; Engelsen, S.B. Chemometrics in foodomics: Handling data structures from multiple analytical platforms. TrAC Trends Anal. Chem. 2014, 60, 71–79. [Google Scholar] [CrossRef]
- Bureau, S.; Cozzolino, D.; Clark, C. Contributions of Fourier-transform mid infrared (FT-MIR) spectroscopy to the study of fruit and vegetables: A review. Postharvest Biol. Technol. 2019, 148, 1–14. [Google Scholar] [CrossRef]
- Duckworth, J. Mathematical data processing. In Near Infrared Spectroscopy in Agriculture; Roberts, C.A., Workman, J., Reeves, J.B., Eds.; American Society of Agronomy, Crop Science Society of America, Soil Science Society of America: Madison, WI, USA, 2004; pp. 115–132. [Google Scholar]
- Engel, J.; Gerretzen, J.; Szymanska, E.; Jansen, J.J.; Downey, G.; Blanchet, L.; Buydens, L.M. Breaking with trends in pre-processing? TrAC Trends Anal. Chem. 2013, 50, 96–106. [Google Scholar] [CrossRef]
- Mehmood, T.; Liland, K.H.; Snipen, L.; Sæbø, S. A review of variable selection methods in Partial Least Squares Regression. Chemom. Intell. Lab. Syst. 2012, 118, 62–69. [Google Scholar] [CrossRef]
- Rinnan, Å.; Berg, F.V.D.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Mutihac, L.; Mutihac, R. Mining in chemometrics. Anal. Chim. Acta 2008, 612, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Leardi, R. Experimental design in chemistry: A tutorial. Anal. Chim. Acta 2009, 652, 161–172. [Google Scholar] [CrossRef] [PubMed]
- DiFoggio, R. Guidelines for Applying Chemometrics to Spectra: Feasibility and Error Propagation. Appl. Spectrosc. 2000, 54, 94A–113A. [Google Scholar] [CrossRef]
- Ahmadvand, M.; Parastar, H.; Sereshti, H.; Olivieri, A.; Tauler, R. A systematic study on the effect of noise and shift on multivariate figures of merit of second-order calibration algorithms. Anal. Chim. Acta 2017, 952, 18–31. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Wold, S.; Josefson, M.; Gottfries, J.; Linusson, A. The utility of multivariate design in PLS regression. J. Chemom. 2004, 18, 156–165. [Google Scholar] [CrossRef]
- Hopke, P.K. The evolution of chemometrics. Anal. Chim. Acta 2003, 500, 365–377. [Google Scholar] [CrossRef]
- Møller, S.F.; Von Frese, J.; Bro, R. Robust methods for multivariate data analysis. J. Chemom. 2005, 19, 549–563. [Google Scholar] [CrossRef]
- Vogt, F. Information fusion via constrained principal component regression for robust quantification with incomplete calibrations. Anal. Chim. Acta 2013, 797, 20–29. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef] [Green Version]
- Szymanska, E.; Gerretzen, J.; Engel, J.; Geurts, B.; Blanchet, L.; Buydens, L.M. Chemometrics and qualitative analysis have a vibrant relationship. TrAC Trends Anal. Chem. 2015, 69, 34–51. [Google Scholar] [CrossRef]
- Bevilacqua, M.; Bro, R.; Marini, F.; Rinnan, Å.; Rasmussen, M.A.; Skov, T. Recent chemometrics advances for foodomics. TrAC Trends Anal. Chem. 2017, 96, 42–51. [Google Scholar] [CrossRef]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis—A marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef]
- Tange, R.I.; Rasmussen, M.A.; Taira, E.; Bro, R. Benchmarking support vector regression against partial least squares regression and artificial neural network: Effect of sample size on model performance. J. Near Infrared Spectrosc. 2017, 25, 381–390. [Google Scholar] [CrossRef]
- Balabin, R.M.; Lomakina, E.I. Support vector machine regression (SVR/LS-SVM)—An alternative to neural networks (ANN) for analytical chemistry? Comparison of nonlinear methods on near infrared (NIR) spectroscopy data. Analyst 2011, 136, 1703–1712. [Google Scholar] [CrossRef]
- Balabin, R.M.; Smirnov, S.V. Interpolation and extrapolation problems of multivariate regression in analytical chemistry: Benchmarking the robustness on near-infrared (NIR) spectroscopy data. Analyst 2012, 137, 1604. [Google Scholar] [CrossRef]
- Ni, W.; Nørgaard, L.; Mørup, M. Non-linear calibration models for near infrared spectroscopy. Anal. Chim. Acta 2014, 813, 1–14. [Google Scholar] [CrossRef]
- Tange, R.; Rasmussen, M.A.; Taira, E.; Bro, R. Application of Support Vector Regression for Simultaneous Modelling of near Infrared Spectra from Multiple Process Steps. J. Near Infrared Spectrosc. 2015, 23, 75–84. [Google Scholar] [CrossRef]
- Souihi, N.; Lindegren, A.; Eriksson, L.; Trygg, J. OPLS in batch monitoring—Opens up new opportunities. Anal. Chim. Acta 2015, 857, 28–38. [Google Scholar] [CrossRef]
- Funes, E.; Allouche, Y.; Beltrán, G.; Jiménez, A. A Review: Artificial Neural Networks as Tool for Control Food Industry Process. J. Sens. Technol. 2015, 5, 28–43. [Google Scholar] [CrossRef] [Green Version]
- Zupan, J.; Gasteiger, J. Neural networks: A new method for solving chemical problems or just a passing phase? Anal. Chim. Acta 1991, 248, 1–30. [Google Scholar] [CrossRef]
- Liu, A.; Li, G.; Fu, Z.; Guan, Y.; Lin, L. Non-linearity correction in NIR absorption spectra by grouping modeling according to the content of analyte. Sci. Rep. 2018, 8, 8564. [Google Scholar] [CrossRef] [PubMed]
- Brereton, R.G. Pattern recognition in chemometrics. Chemom. Intell. Lab. Syst. 2015, 149, 90–96. [Google Scholar] [CrossRef]
- Fearn, T. Assessing calibrations: SEP, RPD, RER and R2. NIR News 2002, 13, 12–13. [Google Scholar] [CrossRef]
- Fearn, T. Validation. NIR News 1997, 8, 7–8. [Google Scholar] [CrossRef]
- Norris, K.H.; Ritchie, G.E. Assuring specificity for a multivariate near-infrared (NIR) calibration: The example of the Chambersburg Shoot-out 2002 data set. J. Pharm. Biomed. Anal. 2008, 48, 1037–1041. [Google Scholar] [CrossRef]
- Walsh, K.B.; Kawano, S. Near infrared spectroscopy. In Optical Monitoring of Fresh and Processed Agricultural Crops; Zude, M., Ed.; CRC Press: Boca Raton, FL, USA, 2009; pp. 192–239. [Google Scholar]
- Westad, F.; Marini, F. Validation of chemometric models—A tutorial. Anal. Chim. Acta 2015, 893, 14–24. [Google Scholar] [CrossRef]
- Asuero, A.G.; Sayago, A.; Gustavo, A.G. The Correlation Coefficient: An Overview. Crit. Rev. Anal. Chem. 2006, 36, 41–59. [Google Scholar] [CrossRef]
- Williams, P.; Dardenne, P.; Flinn, P. Tutorial: Items to be included in a report on a near infrared spectroscopy project. J. Near Infrared Spectrosc. 2017, 25, 85–90. [Google Scholar] [CrossRef]
- Agelet, L.E.; Hurburgh, C.R. A Tutorial on Near Infrared Spectroscopy and Its Calibration. Crit. Rev. Anal. Chem. 2010, 40, 246–260. [Google Scholar] [CrossRef]
- Oliveri, P.; Malegori, C.; Simonetti, R.; Casale, M. The impact of signal pre-processing on the final interpretation of analytical outcomes—A tutorial. Anal. Chim. Acta 2019, 1058, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Alewijn, M.; van der Voet, H.; van Ruth, S. Validation of multivariate classification methods using analytical fingerprints—Concept and case study on organic feed for laying hens. J. Food Compost. Anal. 2016, 51, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Perez-Guaita, D.; Kuligowski, J.; Lendl, B.; Wood, B.R.; Quint, G. Assessment of discriminant models in infrared imaging using constrained repeated random sampling-Cross validation. Anal. Chim. Acta 2018, 1033, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Kjeldahl, K.; Bro, R. Some common misunderstandings in chemometrics. J. Chemom. 2010, 24, 558–564. [Google Scholar] [CrossRef]
- Hawkins, D.M. The problem of overfitting. J. Chem. Inf. Comput. Sci. 2004, 44, 1–12. [Google Scholar] [CrossRef]
- Wise, B.M.; Gallagher, N.B. The process chemometrics approach to process monitoring and fault detection. J. Process. Control 1996, 6, 329–348. [Google Scholar] [CrossRef]
- Hibbert, D.B.; Minkkinen, P.; Faber, N.; Wise, B.M. IUPAC project: A glossary of concepts and terms in chemometrics. Anal. Chim. Acta 2009, 642, 3–5. [Google Scholar] [CrossRef]
- Martens, H. Quantitative Big Data: Where chemometrics can contribute. J. Chemom. 2015, 29, 563–581. [Google Scholar] [CrossRef] [Green Version]
- Oliveri, P. Class-Modelling Approaches: Advantages over Discriminant Methods for Compliance Verifications. NIR News 2016, 27, 29–30. [Google Scholar] [CrossRef]
- Oliveri, P. Class-modelling in food analytical chemistry: Development, sampling, optimisation and validation issues—A tutorial. Anal. Chim. Acta 2017, 982, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Riedl, J.; Esslinger, S.; Fauhl-Hassek, C. Review of validation and reporting of non-targeted fingerprinting approaches for food authentication. Anal. Chim. Acta 2015, 885, 17–32. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cozzolino, D. The Sample, the Spectra and the Maths—The Critical Pillars in the Development of Robust and Sound Applications of Vibrational Spectroscopy. Molecules 2020, 25, 3674. https://doi.org/10.3390/molecules25163674
Cozzolino D. The Sample, the Spectra and the Maths—The Critical Pillars in the Development of Robust and Sound Applications of Vibrational Spectroscopy. Molecules. 2020; 25(16):3674. https://doi.org/10.3390/molecules25163674
Chicago/Turabian StyleCozzolino, Daniel. 2020. "The Sample, the Spectra and the Maths—The Critical Pillars in the Development of Robust and Sound Applications of Vibrational Spectroscopy" Molecules 25, no. 16: 3674. https://doi.org/10.3390/molecules25163674




