FLIm and Raman Spectroscopy for Investigating Biochemical Changes of Bovine Pericardium upon Genipin Cross-Linking
Abstract
1. Introduction
2. Results
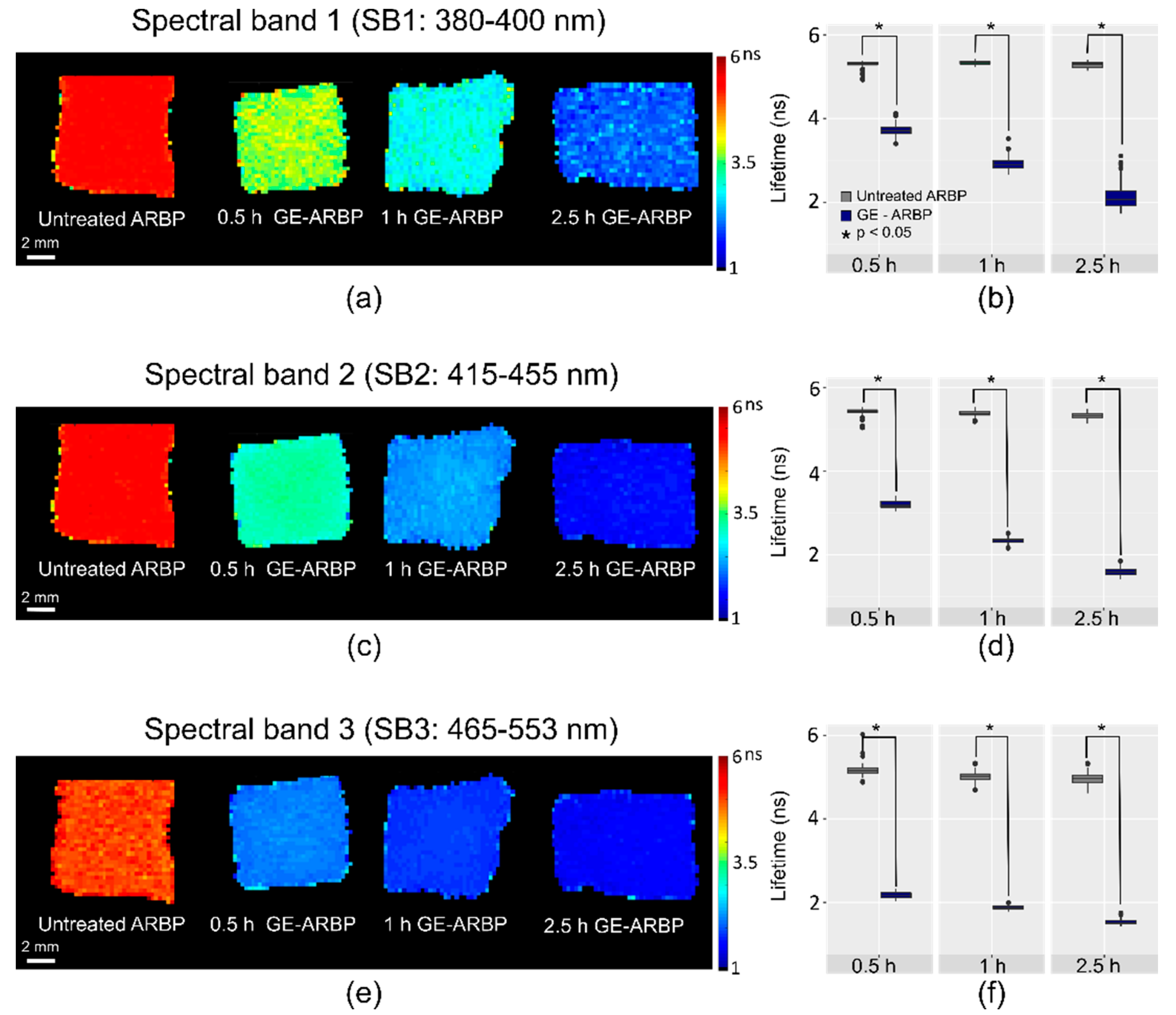
2.1. Fluorescence Lifetime Imaging (FLIm)
2.1.1. Lifetime Decrease upon GE Cross-Linking
2.1.2. Spectral Redshift upon GE Cross-Linking
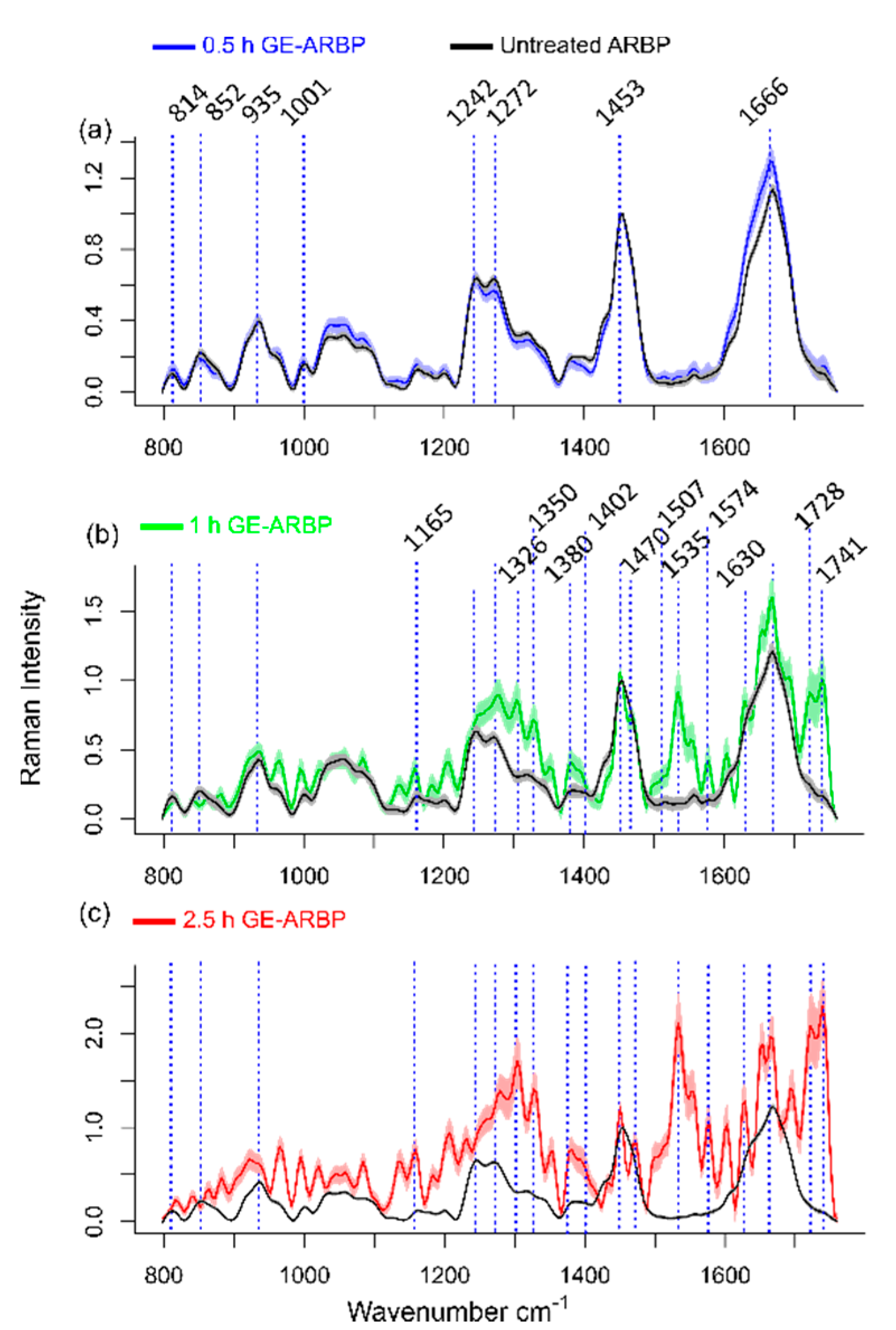
2.2. Raman Spectral Signature of GE Cross-Linked Tissue
2.3. Density Functional Theory (DFT) Simulations Validate the Raman Signature of GE Cross-Linked Tissue
2.4. Correlation between Raman Spectra and Fluorescence Parameters
3. Discussion
4. Materials and Methods
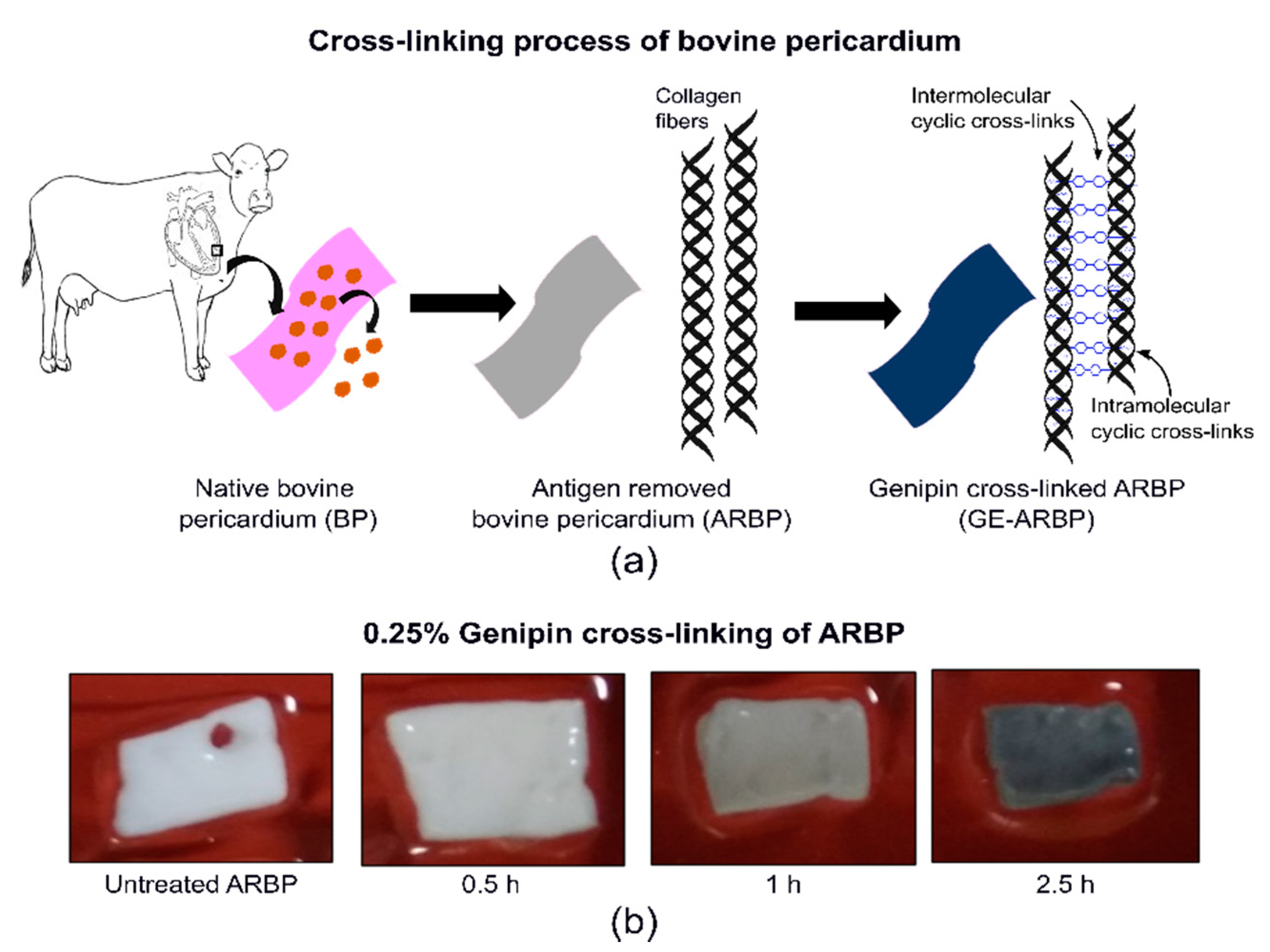
4.1. Pre-Processing of the Bovine Pericardium (BP)
Genipin (GE) Cross-Linking of ARBP
4.2. FLIm—Raman Multimodal Fiber Probe
4.3. FLIm—Raman Spectroscopy Instrument Design
4.3.1. FLIm System
4.3.2. Raman Spectrometer
4.3.3. Fluorescence Spectrometer
4.4. Data Analysis
4.4.1. FLIm Data Analysis
4.4.2. Raman Data Analysis
4.4.3. Multivariate Multiple Regression
4.5. DFT Calculations
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pradhan, S.; Farach-Carson, M.C. Mining the extracellular matrix for tissue engineering applications. Regen. Med. 2010, 5, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.A.; Jana, S.; Tefft, B.J.; Hennessy, R.S.; Go, J.; Morse, D.; Lerman, A.; Young, M.D. Biomaterial characterization of off-the-shelf decellularized porcine pericardial tissue for use in prosthetic valvular applications. J. Tissue Eng. Regen. Med. 2018, 12, 1608–1620. [Google Scholar] [CrossRef] [PubMed]
- Ikada, Y. Tissue Engineering: Fundamentals and Applications; Academic Press/Elsevier: Amsterdam, The Netherlands; Boston, MA, USA, 2006; ISBN 9780123705822. [Google Scholar]
- Ducheyne, P.; Grainger, D.W.; Healy, K.E.; Hutmacher, D.W.; Kirkpatrick, C.J. Comprehensive Biomaterials II; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Coentro, J.Q.; Capella-Monsonís, H.; Graceffa, V.; Wu, Z.; Mullen, A.M.; Raghunath, M.; Zeugolis, D.I. Collagen Quantification in Tissue Specimens BT—Fibrosis: Methods and Protocols; Rittié, L., Ed.; Springer: New York, NY, USA, 2017; pp. 341–350. ISBN 978-1-4939-7113-8. [Google Scholar]
- Eyre, D.R.; Koob, T.J.; Van Ness, K.P. Quantitation of hydroxypyridinium crosslinks in collagen by high-performance liquid chromatography. Anal. Biochem. 1984, 137, 380–388. [Google Scholar] [CrossRef]
- Li, C.; Shklover, J.; Parvizi, M.; Sherlock, B.E.; Alfonso Garcia, A.; Haudenschild, A.K.; Griffiths, L.G.; Marcu, L. Label-Free Assessment of Collagenase Digestion on Bovine Pericardium Properties by Fluorescence Lifetime Imaging. Ann. Biomed. Eng. 2018, 46, 1870–1881. [Google Scholar] [CrossRef]
- Alfonso-Garcia, A.; Haudenschild, A.K.; Marcu, L. Label-free assessment of carotid artery biochemical composition using fiber-based fluorescence lifetime imaging. Biomed. Opt. Express. 2018, 9, 4064. [Google Scholar] [CrossRef]
- Sherlock, B.E.; Harvestine, J.N.; Mitra, D.; Haudenschild, A.; Hu, J.; Athanasiou, K.A.; Leach, J.K.; Marcu, L. Nondestructive assessment of collagen hydrogel cross-linking using time-resolved autofluorescence imaging. J. Biomed. Opt. 2018, 23, 1. [Google Scholar] [CrossRef]
- Alfonso-Garcia, A.; Shklover, J.; Sherlock, B.E.; Panitch, A.; Griffiths, L.G.; Marcu, L. Fiber-based fluorescence lifetime imaging of recellularization processes on vascular tissue constructs. J. Biophotonics 2018, 11, e201700391. [Google Scholar] [CrossRef]
- Haudenschild, A.K.; Sherlock, B.E.; Zhou, X.; Hu, J.C.; Leach, J.K.; Marcu, L.; Athanasiou, K.A. Non-destructive detection of matrix stabilization correlates with enhanced mechanical properties of self-assembled articular cartilage. J. Tissue Eng. Regen. Med. 2019, 13, 637–648. [Google Scholar] [CrossRef]
- Miller, D.R.; Jarrett, J.W.; Hassan, A.M.; Dunn, A.K. Deep Tissue Imaging with Multiphoton Fluorescence Microscopy. Curr. Opin. Biomed. Eng. 2017, 4, 32–39. [Google Scholar] [CrossRef]
- Jastrzebska, M.; Wrzalik, R.; Kocot, A.; Zalewska-Rejdak, J.; Cwalina, B. Raman spectroscopic study of glutaraldehyde-stabilized collagen and pericardium tissue. J. Biomater. Sci. Polym. Ed. 2003, 14, 185–197. [Google Scholar] [CrossRef]
- Xu, C.; Wang, Y. Collagen cross linking increases its biodegradation resistance in wet dentin bonding. J. Adhes. Dent. 2012, 14, 11–18. [Google Scholar] [CrossRef]
- Gamsjaeger, S.; Robins, S.P.; Tatakis, D.N.; Klaushofer, K.; Paschalis, E.P. Identification of Pyridinoline Trivalent Collagen Cross-Links by Raman Microspectroscopy. Calcif. Tissue Int. 2017, 100, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Votteler, M.; Carvajal Berrio, D.A.; Pudlas, M.; Walles, H.; Stock, U.A.; Schenke-Layland, K. Raman spectroscopy for the non-contact and non-destructive monitoring of collagen damage within tissues. J. Biophotonics 2012, 5, 47–56. [Google Scholar] [CrossRef]
- Dochow, S.; Fatakdawala, H.; Phipps, J.E.; Ma, D.; Bocklitz, T.; Schmitt, M.; Bishop, J.W.; Margulies, K.B.; Marcu, L.; Popp, J. Comparing Raman and fluorescence lifetime spectroscopy from human atherosclerotic lesions using a bimodal probe. J. Biophotonics 2016, 9, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Shaik, T.A.; Alfonso-García, A.; Zhou, X.; Arnold, K.M.; Haudenschild, A.K.; Krafft, C.; Griffiths, L.G.; Popp, J.; Marcu, L. FLIm-guided Raman imaging to study cross-linking and calcification of bovine pericardium. Anal. Chem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Nam, H.S.; Kang, W.J.; Lee, M.W.; Song, J.W.; Kim, J.W.; Oh, W.-Y.; Yoo, H. Multispectral analog-mean-delay fluorescence lifetime imaging combined with optical coherence tomography. Biomed. Opt. Express 2018, 9, 1930. [Google Scholar] [CrossRef]
- Baria, E.; Brehm, B.R.; Lattermann, A.; Popp, J.; Pavone, F.S.; Cicchi, R.; Matthäus, C.; Lange, M. Non-linear imaging and characterization of atherosclerotic arterial tissue using combined SHG and FLIM microscopy. J. Biophotonics 2015, 8, 347–356. [Google Scholar] [CrossRef]
- Bec, J.; Phipps, J.E.; Gorpas, D.; Ma, D.; Fatakdawala, H.; Margulies, K.B.; Southard, J.A.; Marcu, L. In vivo label-free structural and biochemical imaging of coronary arteries using an integrated ultrasound and multispectral fluorescence lifetime catheter system. Sci. Rep. 2017, 7, 8960. [Google Scholar] [CrossRef]
- Madhavan, K.; Belchenko, D.; Motta, A.; Tan, W. Evaluation of composition and crosslinking effects on collagen-based composite constructs. Acta Biomater. 2010, 6, 1413–1422. [Google Scholar] [CrossRef]
- Jayakrishnan, A.; Jameela, S.R. Glutaraldehyde as a fixative in bioprostheses and drug delivery matrices. Biomaterials 1996, 17, 471–484. [Google Scholar] [CrossRef]
- Chang, Y.; Lee, M.-H.; Liang, H.-C.; Hsu, C.-K.; Sung, H.-W. Acellular bovine pericardia with distinct porous structures fixed with genipin as an extracellular matrix. Tissue Eng. 2004, 10, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.W.; Liang, I.L.; Chen, C.N.; Huang, R.N.; Liang, H.F. Stability of a biological tissue fixed with a naturally occurring crosslinking agent (genipin). J. Biomed. Mater. Res. 2001, 55, 538–546. [Google Scholar] [CrossRef]
- Sung, H.-W.; Chang, W.-H.; Ma, C.-Y.; Lee, M.-H. Crosslinking of biological tissues using genipin and/or carbodiimide. J. Biomed. Mater. Res. A 2003, 64, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.W.; Chang, Y.; Chiu, C.T.; Chen, C.N.; Liang, H.C. Crosslinking characteristics and mechanical properties of a bovine pericardium fixed with a naturally occurring crosslinking agent. J. Biomed. Mater. Res. 1999, 47, 116–126. [Google Scholar] [CrossRef]
- Muzzarelli, R.A.A.; El Mehtedi, M.; Bottegoni, C.; Aquili, A.; Gigante, A. Genipin-crosslinked chitosan gels and scaffolds for tissue engineering and regeneration of cartilage and bone. Mar. Drugs 2015, 13, 7314–7338. [Google Scholar] [CrossRef]
- Sung, H.W.; Huang, D.M.; Chang, W.H.; Huang, R.N.; Hsu, J.C. Evaluation of gelatin hydrogel crosslinked with various crosslinking agents as bioadhesives: In vitro study. J. Biomed. Mater. Res. 1999, 46, 520–530. [Google Scholar] [CrossRef]
- Wang, G.-L.; Lim, J.-M.; Bhoo, S.-H.; Paik, Y.-S.; Hahn, T.-R. Colorimetric determination of amino acids using genipin from Gardenia jasminoides. Anal. Chim. Acta 2003, 480, 267–274. [Google Scholar] [CrossRef]
- Butler, M.F.; Ng, Y.F.; Pudney, P.D.A. Mechanism and kinetics of the crosslinking reaction between biopolymers containing primary amine groups and genipin. J. Polym. Sci. Part A Polym. Chem. 2003, 41, 3941–3953. [Google Scholar] [CrossRef]
- Hwang, Y.-J.; Larsen, J.; Krasieva, T.B.; Lyubovitsky, J.G. Effect of genipin crosslinking on the optical spectral properties and structures of collagen hydrogels. ACS Appl. Mater. Interfaces 2011, 3, 2579–2584. [Google Scholar] [CrossRef]
- Sun, W.; Incitti, T.; Migliaresi, C.; Quattrone, A.; Casarosa, S.; Motta, A. Genipin-crosslinked gelatin–silk fibroin hydrogels for modulating the behaviour of pluripotent cells. J. Tissue Eng. Regen. Med. 2014, 10. [Google Scholar] [CrossRef]
- Sundararaghavan, H.G.; Monteiro, G.A.; Lapin, N.A.; Chabal, Y.J.; Miksan, J.R.; Shreiber, D.I. Genipin-induced changes in collagen gels: Correlation of mechanical properties to fluorescence. J. Biomed. Mater. Res. Part A 2008, 87, 308–320. [Google Scholar] [CrossRef]
- Krafft, C.; Codrich, D.; Pelizzo, G.; Sergo, V. Raman and FTIR microscopic imaging of colon tissue: A comparative study. J. Biophotonics 2008, 1, 154–169. [Google Scholar] [CrossRef]
- Heidari, A.; Esposito, J.; Caissutti, A. Time–resolved resonance FT–IR and raman spectroscopy and density functional theory investigation of vibronic–mode coupling structure in vibrational spectra of nano-polypeptide macromolecule beyond the multi–dimensional franck–condon integrals approximatio. Glob. Imaging Insights 2020, 5, 1–14. [Google Scholar] [CrossRef][Green Version]
- Stepanić, V.; Baranović, G.; Smrečki, V. Structure and vibrational spectra of conjugated acids of trans- and cis-azobenzene. J. Mol. Struct. 2001, 569, 89–109. [Google Scholar] [CrossRef]
- Staniszewska, M.; Kupfer, S.; Łabuda, M.; Guthmuller, J. Theoretical Assessment of Excited State Gradients and Resonance Raman Intensities for the Azobenzene Molecule. J. Chem. Theory Comput. 2017, 13, 1263–1274. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, S.; Fukui, Y.; Koga, K.; Iwashita, T.; Komura, H.; Nomoto, K. Structure of genipocyanin G1, a spontaneous reaction product between genipin and glycine. Tetrahedron Lett. 1987, 28, 4699–4700. [Google Scholar] [CrossRef]
- Touyama, R.; Takeda, Y.; Inoue, K.; Kawamura, I.; Yatsuzuka, M.; Ikumoto, T.; Shingu, T.; Yokoi, T.; Inouye, H. Studies on the Blue Pigments Produced from Genipin and Methylamine. I. Structures of the Brownish-Red Pigments, Intermediates Leading to the Blue Pigments. Chem. Pharm. Bull. 1994, 42, 668–673. [Google Scholar] [CrossRef]
- Fujikawa, S.; Nakamura, S.; Kooa, K. Genipin, a New Type of Protein Crosslinking Reagent from Gardenia Fruits. Agric. Biol. Chem. 1988, 52, 869–870. [Google Scholar] [CrossRef]
- Gebrekidan, M.T.; Knipfer, C.; Stelzle, F. A shifted-excitation Raman difference spectroscopy (SERDS) evaluation strategy for the efficient isolation of Raman spectra from extreme fluorescence interference. J. Raman Spectrosc. 2016, 47, 198–209. [Google Scholar] [CrossRef]
- Cordero, E.; Korinth, F.; Stiebing, C.; Krafft, C.; Schie, I.W.; Popp, J. Evaluation of Shifted Excitation Raman Difference Spectroscopy and Comparison to Computational Background Correction Methods Applied to Biochemical Raman Spectra. Sensors 2017, 17, 1724. [Google Scholar] [CrossRef]
- Guo, S.; Chernavskaia, O.; Popp, J.; Bocklitz, T. Spectral reconstruction for shifted-excitation Raman difference spectroscopy (SERDS). Talanta 2018, 186, 372–380. [Google Scholar] [CrossRef]
- Sinha, P.; Zurakowski, D.; Kumar, T.K.S.; He, D.; Rossi, C.; Jonas, R.A. Effects of glutaraldehyde concentration, pretreatment time, and type of tissue (porcine versus bovine) on postimplantation calcification. J. Thorac. Cardiovasc. Surg. 2012, 143, 224–227. [Google Scholar] [CrossRef] [PubMed]
- de Leval, M.R. Comprehensive Surgical Management of Congenital Heart Disease. J. R. Soc. Med. 2004, 97, 407–408. [Google Scholar] [CrossRef][Green Version]
- Wong, M.L.; Wong, J.L.; Athanasiou, K.A.; Griffiths, L.G. Stepwise solubilization-based antigen removal for xenogeneic scaffold generation in tissue engineering. Acta Biomater. 2013, 9, 6492–6501. [Google Scholar] [CrossRef] [PubMed]
- Yankelevich, D.R.; Ma, D.; Liu, J.; Sun, Y.; Sun, Y.; Bec, J.; Elson, D.S.; Marcu, L. Design and evaluation of a device for fast multispectral time-resolved fluorescence spectroscopy and imaging. Rev. Sci. Instrum. 2014, 85. [Google Scholar] [CrossRef]
- Liu, J.; Sun, Y.; Qi, J.; Marcu, L. A novel method for fast and robust estimation of fluorescence decay dynamics using constrained least-squares deconvolution with Laguerre expansion. Phys. Med. Biol. 2012, 57, 843–865. [Google Scholar] [CrossRef]
- Ryan, C.G.; Clayton, E.; Griffin, W.L.; Sie, S.H.; Cousens, D.R. SNIP, a statistics-sensitive background treatment for the quantitative analysis of PIXE spectra in geoscience applications. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 1988, 34, 396–402. [Google Scholar] [CrossRef]
- Gibb, S.; Strimmer, K. MALDIquant: A versatile R package for the analysis of mass spectrometry data. Bioinformatics 2012, 28, 2270–2271. [Google Scholar] [CrossRef]
- Beleites, C.; Sergo, V. hyperSpec: A Package to Handle Hyperspectral Data Sets in R. 2018. Available online: http://hyperspec.r-forge.r-project.org (accessed on 26 July 2019).
- Shao, Y.; Gan, Z.; Epifanovsky, E.; Gilbert, A.T.B.; Wormit, M.; Kussmann, J.; Lange, A.W.; Behn, A.; Deng, J.; Feng, X.; et al. Advances in molecular quantum chemistry contained in the Q-Chem 4 program package. Mol. Phys. 2015, 113, 184–215. [Google Scholar] [CrossRef]
- Kesharwani, M.K.; Brauer, B.; Martin, J.M.L. Frequency and Zero-Point Vibrational Energy Scale Factors for Double-Hybrid Density Functionals (and Other Selected Methods): Can Anharmonic Force Fields Be Avoided? J. Phys. Chem. A 2015, 119, 1701–1714. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples were prepared at the Biophotonics Laboratory led by Dr. Marcu, at the Biomedical Engineering Department, University of California, Davis. Tissue samples can be prepared following the information on the Methods section. Additional information is available from the authors upon request. |

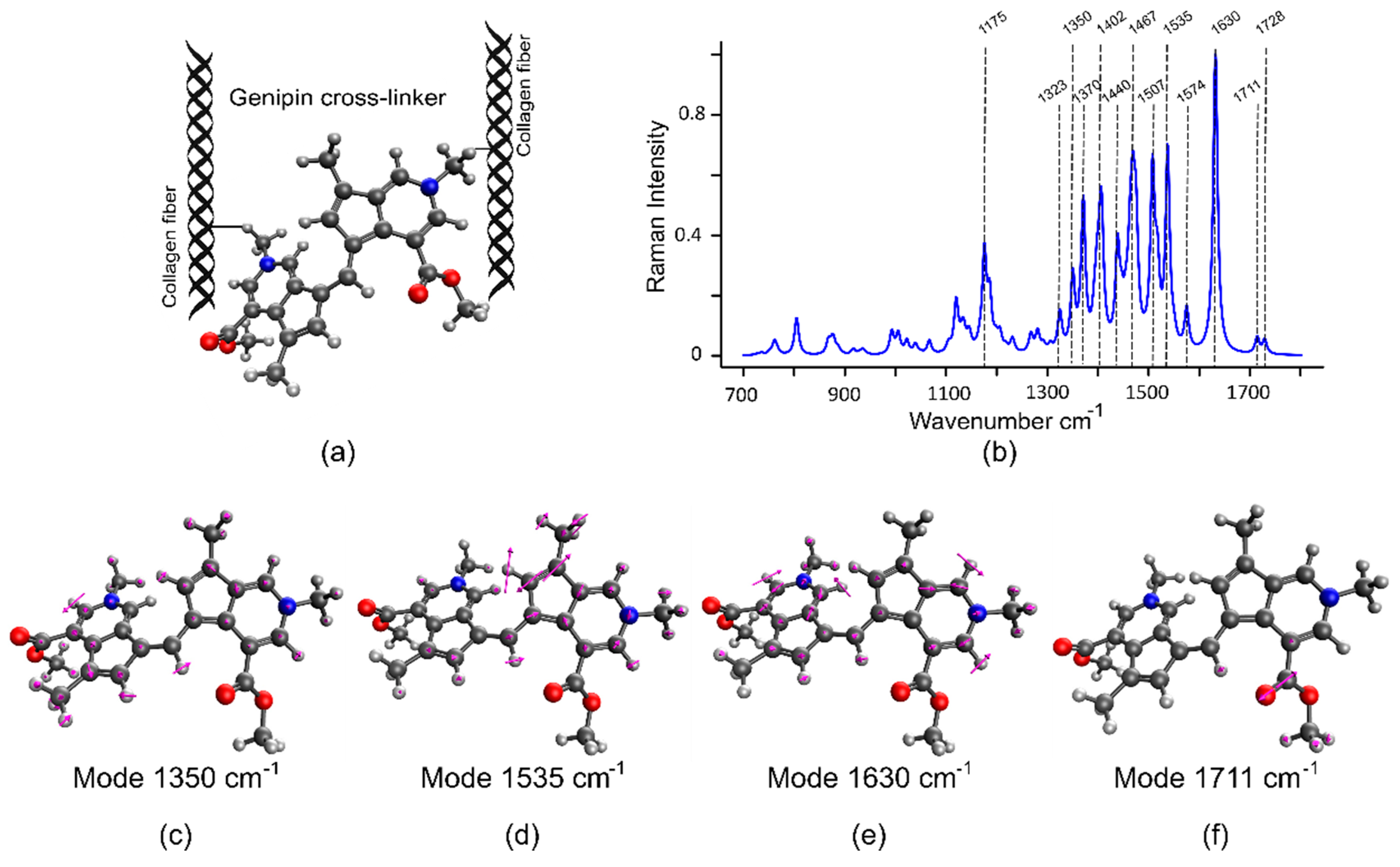

| Wavenumber (cm−1) | Band Assignment | Reference |
|---|---|---|
| 814 | Proline | [18,35] |
| 852 | Hydroxy-proline | |
| 935 | C-C α stretch | |
| 1001 | Phenylalanine | |
| 1242 | Amide III | |
| 1272 | ||
| 1165 | In-plane vibrations of the ring system of GE-cross-links | DFT |
| 1175 | ||
| 1323 | ||
| 1350 | ||
| 1370 | ||
| 1380 | ||
| 1400 | In-plane ring system vibration and bending of motion of CH3 group | DFT |
| 1402 | ||
| 1440 | ||
| 1467 | ||
| 1470 | ||
| 1453 | CH2 collagen and elastin | [35] |
| 1507 | In-plane motion of atoms in the 5-membered ring system | DFT |
| 1535 | ||
| 1574 | Strong ring distortion of the 5-membered ring system. | |
| 1630 | Strong in-plane motion of the 6-membered ring | |
| 1666 | Amide I | [18,35] |
| 1711 | C=O | DFT |
| 1728 | ||
| 1741 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaik, T.A.; Alfonso-Garcia, A.; Richter, M.; Korinth, F.; Krafft, C.; Marcu, L.; Popp, J. FLIm and Raman Spectroscopy for Investigating Biochemical Changes of Bovine Pericardium upon Genipin Cross-Linking. Molecules 2020, 25, 3857. https://doi.org/10.3390/molecules25173857
Shaik TA, Alfonso-Garcia A, Richter M, Korinth F, Krafft C, Marcu L, Popp J. FLIm and Raman Spectroscopy for Investigating Biochemical Changes of Bovine Pericardium upon Genipin Cross-Linking. Molecules. 2020; 25(17):3857. https://doi.org/10.3390/molecules25173857
Chicago/Turabian StyleShaik, Tanveer Ahmed, Alba Alfonso-Garcia, Martin Richter, Florian Korinth, Christoph Krafft, Laura Marcu, and Jürgen Popp. 2020. "FLIm and Raman Spectroscopy for Investigating Biochemical Changes of Bovine Pericardium upon Genipin Cross-Linking" Molecules 25, no. 17: 3857. https://doi.org/10.3390/molecules25173857
APA StyleShaik, T. A., Alfonso-Garcia, A., Richter, M., Korinth, F., Krafft, C., Marcu, L., & Popp, J. (2020). FLIm and Raman Spectroscopy for Investigating Biochemical Changes of Bovine Pericardium upon Genipin Cross-Linking. Molecules, 25(17), 3857. https://doi.org/10.3390/molecules25173857









