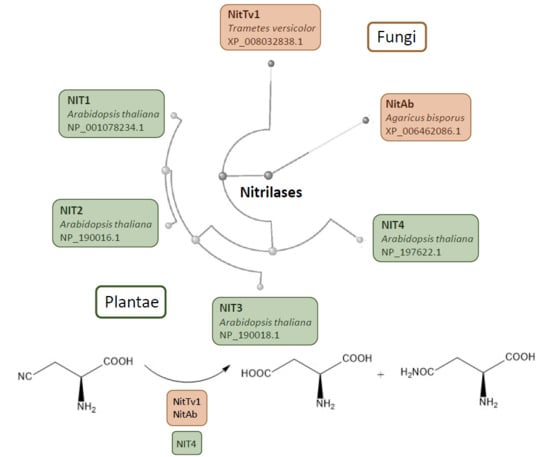
Plant Nitrilase Homologues in Fungi: Phylogenetic and Functional Analysis with Focus on Nitrilases in Trametes versicolor and Agaricus bisporus
Abstract
:1. Introduction
2. Results
2.1. Analysis of Published Sequences
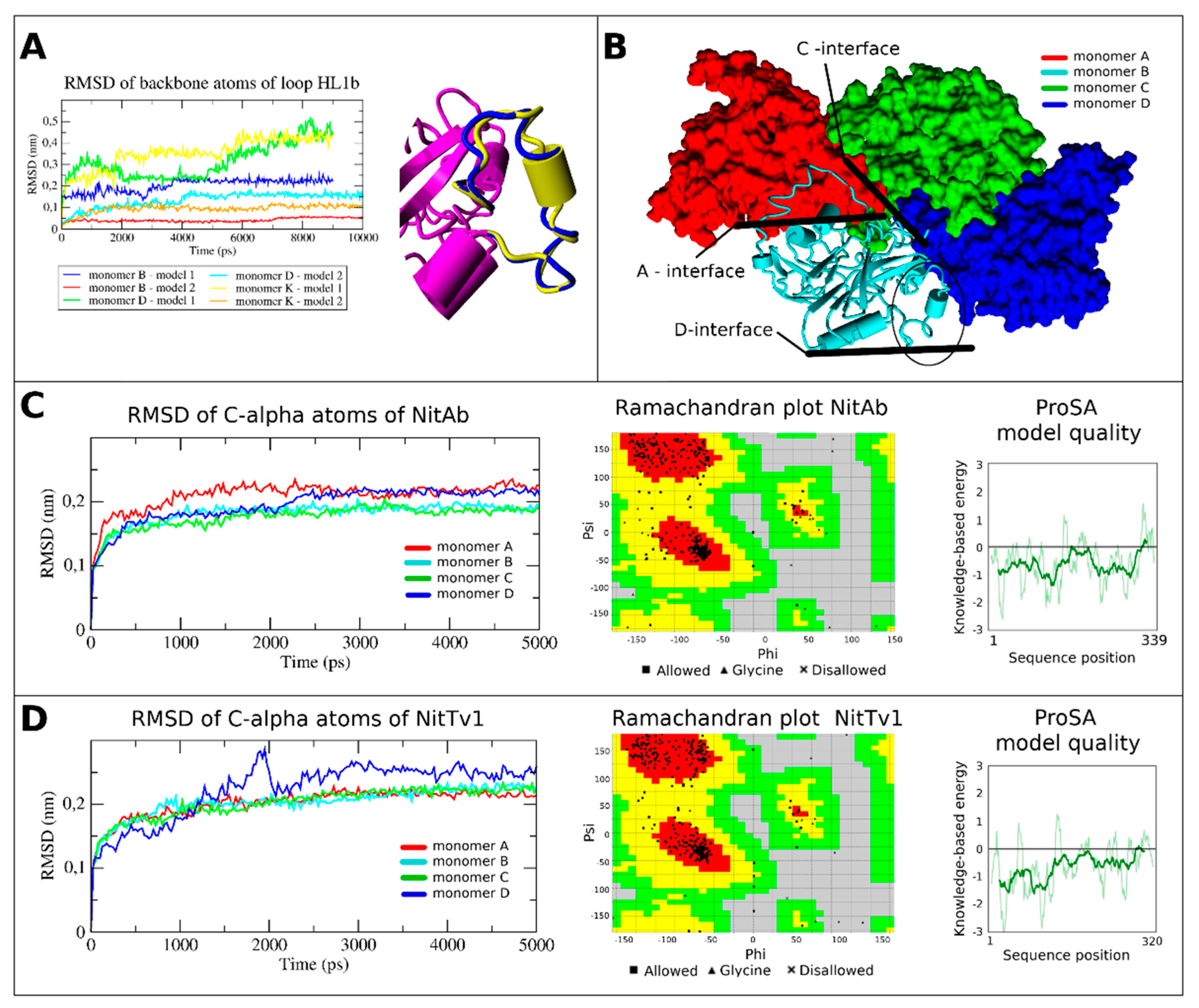
2.2. Overproduction and Characterization of Fungal Nitrilases
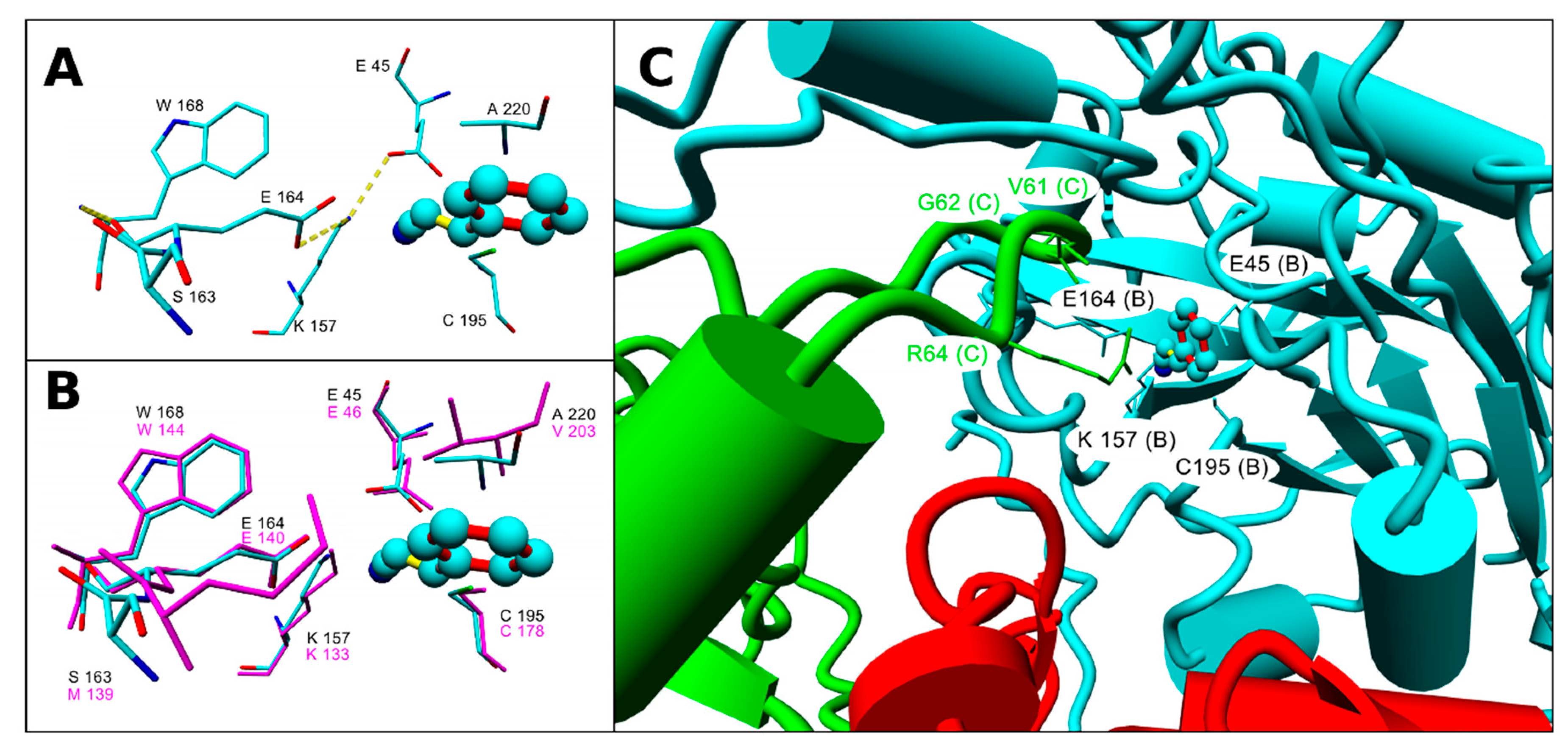
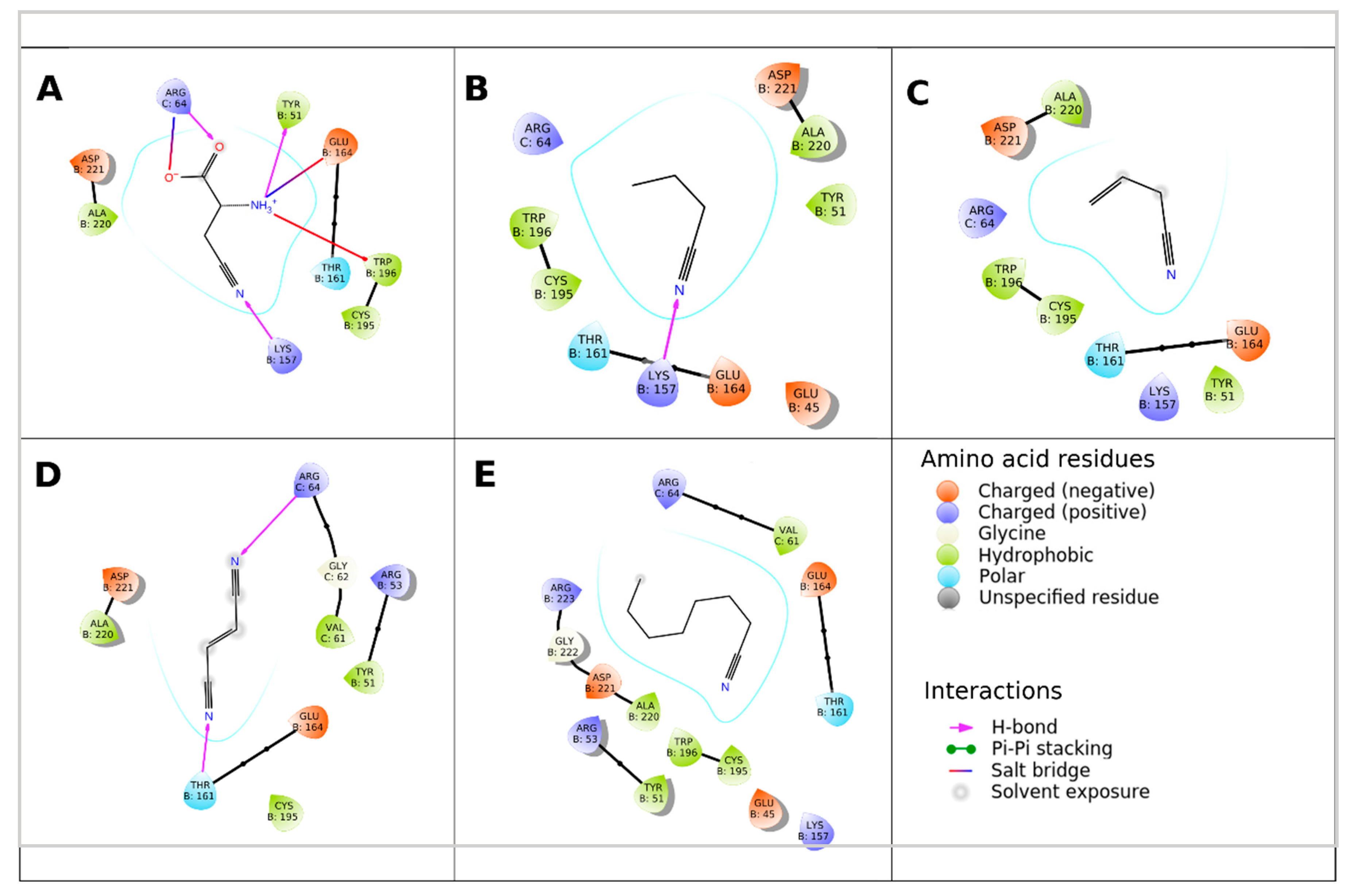
2.3. Assessment of the Enzyme Affinities to Various Substrates Using Homology Modeling and Substrate Docking
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Analysis of Protein Sequences
4.3. Homology Models and Ligand Docking
4.4. Genes and Strains of Microorganisms
4.5. Enzyme Overproduction and Extraction
4.6. Enzyme Purification
4.7. Enzyme Assays
4.8. LC-MS Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
Homology Modeling and Loop Modeling
References
- Pace, H.C.; Brenner, C. The nitrilase superfamily: Classification, structure and function. Genome Biol. 2001, 2, REVIEWS0001. [Google Scholar] [CrossRef] [PubMed]
- Brenner, C. Catalysis in the nitrilase superfamily. Curr. Opin. Struct. Biol. 2002, 12, 775–782. [Google Scholar] [CrossRef]
- Thuku, R.N.; Brady, D.; Benedik, M.J.; Sewell, B.T. Microbial nitrilases: Versatile, spiral forming, industrial enzymes. J. Appl. Microbiol. 2009, 106, 703–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piotrowski, M.; Schönfelder, S.; Weiler, E.W. The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode β-cyano-L-alanine hydratase/nitrilase. J. Biol. Chem. 2001, 276, 2616–2621. [Google Scholar] [CrossRef] [Green Version]
- Martínková, L.; Rucká, L.; Nešvera, J.; Pátek, M. Recent advances and challenges in the heterologous production of microbial nitrilases for biocatalytic applications. World J. Microbiol. Biotechnol. 2017, 33, 8. [Google Scholar] [CrossRef]
- Prozomix. Available online: http://www.prozomix.com/products/view?product=1831 (accessed on 14 August 2020).
- Codexis 3. Available online: https://www.codexis-estore.com/product-page/codex-nitrilase-nit-screening-kit (accessed on 14 August 2020).
- Merck KGaA. Available online: https://www.sigmaaldrich.com/catalog/product/sigma/04529 (accessed on 14 August 2020).
- Vorwerk, S.; Biernacki, S.; Hillebrand, H.; Janzik, I.; Müller, A.; Weiler, E.W.; Piotrowski, M. Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta 2001, 212, 508–516. [Google Scholar] [CrossRef]
- Osswald, S.; Wajant, H.; Effenberger, F. Characterization and synthetic applications of recombinant AtNIT1 from Arabidopsis thaliana. Eur. J. Biochem. 2002, 269, 680–687. [Google Scholar] [CrossRef]
- Piotrowski, M. Primary or secondary? Versatile nitrilases in plant metabolism. Phytochemistry 2008, 69, 2655–2667. [Google Scholar] [CrossRef]
- Howden, A.J.M.; Preston, G.M. Nitrilase enzymes and their role in plant–microbe interactions. Microb. Biotechnol. 2009, 2, 441–451. [Google Scholar] [CrossRef] [Green Version]
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef] [Green Version]
- Martínková, L. Nitrile metabolism in fungi: A review of its key enzymes nitrilases with focus on their biotechnological impact. Fungal Biol. Rev. 2019, 33, 149–157. [Google Scholar] [CrossRef]
- Basile, L.J.; Willson, R.C.; Sewell, B.T.; Benedik, M.J. Genome mining of cyanide degrading nitrilases from filamentous fungi. Appl. Microbiol. Biotechnol. 2008, 80, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Rinágelová, A.; Kaplan, O.; Veselá, A.B.; Chmátal, M.; Křenková, A.; Plíhal, O.; Pasquarelli, F.; Cantarella, M.; Martínková, L. Cyanide hydratase from Aspergillus niger K10: Overproduction in Escherichia coli, purification, characterization and use in continuous cyanide degradation. Process Biochem. 2014, 49, 445–450. [Google Scholar] [CrossRef]
- Veselá, A.B.; Rucká, L.; Kaplan, O.; Pelantová, H.; Nešvera, J.; Pátek, M.; Martínková, L. Bringing nitrilase sequences from databases to life: The search for novel substrate specificities with a focus on dinitriles. Appl. Microbiol. Biotechnol. 2016, 100, 2193–2202. [Google Scholar] [CrossRef]
- Rucká, L.; Chmátal, M.; Kulik, N.; Petrásková, L.; Pelantová, H.; Novotný, P.; Příhodová, R.; Pátek, M.; Martínková, L. Genetic and functional diversity of nitrilases in Agaricomycotina. Int. J. Mol. Sci. 2019, 20, 5990. [Google Scholar] [CrossRef] [Green Version]
- Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 30 June 2020).
- Petříčková, A.; Veselá, A.B.; Kaplan, O.; Kubáč, D.; Uhnáková, B.; Malandra, A.; Felsberg, J.; Rinágelová, A.; Weyrauch, P.; Křen, V.; et al. Purification and characterization of heterologously expressed nitrilases from filamentous fungi. Appl. Microbiol. Biotechnol. 2012, 93, 1553–1561, Erratum in 2013, 97, 9263–9264. [Google Scholar]
- Mulelu, A.E.; Kirykowicz, A.M.; Woodward, J.D. Cryo-EM and directed evolution reveal how Arabidopsis nitrilase specificity is influenced by its quaternary structure. Commun. Biol. 2019, 2, 260. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yin, B.; Wang, C.; Jiang, S.; Wang, H.; Yuan, Y.A.; Wei, D. Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Synechocystis sp. PCC6803. J. Struct. Biol. 2014, 188, 93–101. [Google Scholar] [CrossRef]
- Cuff, J.A.; Clamp, M.E.; Siddiqui, A.S.; Finlay, M.; Barton, G.J. JPred: A consensus secondary structure prediction server. Bioinformatics 1998, 14, 892–893. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Jiang, S.; Wang, H.; Wang, L.; Wei, D. Switching the regioselectivity of two nitrilases toward succinonitrile by mutating the active center pocket key residues through a semi-rational engineering. Chem. Commun. 2019, 55, 2948–2951. [Google Scholar] [CrossRef]
- Howden, A.J.; Harrison, J.; Preston, G.M. A conserved mechanism for nitrile metabolism in bacteria and plants. Plant J. 2009, 57, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Kiziak, C.; Conradt, D.; Stolz, A.; Mattes, R.; Klein, J. Nitrilase from Pseudomonas fluorescens EBC191: Cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme. Microbiology 2005, 151, 3639–3648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Mukherjee, C.; Yang, Y.; Rios, B.E.; Gallagher, D.T.; Smith, N.N.; Biehl, E.R.; Hua, L. A new nitrilase from Bradyrhizobium japonicum USDA 110—Gene cloning, biochemical characterization and substrate specificity. J. Biotechnol. 2008, 133, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, B.C.M.; Mateo, C.; Kiziak, C.; Chmura, A.; Wacker, J.; van Rantwijk, F.; Stolz, A.; Sheldon, R.A. Nitrile hydratase activity of a recombinant nitrilase. Adv. Synth. Catal. 2006, 348, 2597–2603. [Google Scholar] [CrossRef]
- Sosedov, O.; Stolz, A. Random mutagenesis of the arylacetonitrilase from Pseudomonas fluorescens EBC191 and identification of variants, which form increased amounts of mandeloamide from mandelonitrile. Appl. Microbiol. Biotechnol. 2014, 98, 1595–1607. [Google Scholar] [CrossRef]
- Xu, C.; Tang, L.; Liang, Y.; Jiao, S.; Yu, H.; Luo, H. Novel chaperones RrGroEL and RrGroES for activity and stability enhancement of nitrilase in Escherichia coli and Rhodococcus ruber. Molecules 2020, 25, 1002. [Google Scholar] [CrossRef] [Green Version]
- Jamwal, S.; Dautoo, U.K.; Ranote, S.; Dharela, R.; Chauhan, G.S. Enhanced catalytic activity of new acryloyl crosslinked cellulose dialdehyde-nitrilase Schiff base and its reduced form for nitrile hydrolysis. Int. J. Biol. Macromol. 2019, 131, 117–126. [Google Scholar] [CrossRef]
- Talley, K.; Alexov, E. On the pH-optimum of activity and stability of proteins. Proteins 2010, 78, 2699–2706. [Google Scholar] [CrossRef] [Green Version]
- Jandhyala, D.; Berman, M.; Meyers, P.R.; Sewell, B.T.; Willson, R.C.; Benedik, M.J. CynD, the cyanide dehydratase from Bacillus pumilus: Gene cloning and structural studies. Appl. Environ. Microbiol. 2003, 69, 4794–4805. [Google Scholar] [CrossRef] [Green Version]
- Effenberger, F.; Osswald, S. Enantioselective hydrolysis of (RS)-2-fluoroarylacetonitriles using nitrilase from Arabidopsis thaliana. Tetrahedron Asymmetry 2001, 12, 279–285. [Google Scholar] [CrossRef]
- Constraint-Based Multiple Alignment Tool. Available online: https://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi (accessed on 30 June 2020).
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2018–2; Schrödinger, LLC: New York, NY, USA, 2018.
- Krieger, E.; Darden, T.; Nabuurs, S.B.; Finkelstein, A.; Vriend, G. Making optimal use of empirical energy functions: Force-field parameterization in crystal space. Proteins 2004, 57, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Dunbrack, R.L., Jr.; Hooft, R.W.W.; Krieger, B. Assignment of protonation states in proteins and ligands: Combining pKa prediction with hydrogen bonding network optimization. Methods Mol. Biol. 2012, 819, 405–421. [Google Scholar] [CrossRef]
- Brunner, S.; Eppinger, E.; Fischer, S.; Gröning, J.; Stolz, A. Conversion of aliphatic nitriles by the arylacetonitrilase from Pseudomonas fluorescens EBC191. World J. Microbiol. Biotechnol. 2018, 34, 91. [Google Scholar] [CrossRef]
- Kriechbaumer, V.; Park, W.J.; Piotrowski, M.; Meeley, R.B.; Gierl, A.; Glawischnig, E. Maize nitrilases have a dual role in auxin homeostasis and β-cyanoalanine hydrolysis. J. Exp. Bot. 2007, 58, 4225–4233. [Google Scholar] [CrossRef]
- MyCurveFit. Available online: https://mycurvefit.com (accessed on 7 July 2020).
- King, R.D.; Sternberg, M.J.E. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 1996, 5, 2298–2310. [Google Scholar] [CrossRef]
- Rost, B.; Sander, C. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 1993, 232, 584–599. [Google Scholar] [CrossRef] [Green Version]
- Guermeur, Y.; Geourjon, C.; Gallinari, P.; Deléage, G. Improved performance in protein secondary structure prediction by inhomogeneous score combination. Bioinformatics 1999, 15, 413–421. [Google Scholar] [CrossRef] [Green Version]
- Fiser, A.; Sali, A. ModLoop: Automated modeling of loops in protein structures. Bioinformatics 2003, 19, 2500–2501. [Google Scholar] [CrossRef] [PubMed]
- Canutescu, A.A.; Dunbrack, R.L. Cyclic coordinate descent: A robotics algorithm for protein loop closure. Protein Sci. 2003, 12, 963–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canutescu, A.A.; Shelenkov, A.A.; Dunbrack, R.L. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003, 12, 2001–2014. [Google Scholar] [CrossRef] [PubMed]
- Konagurthu, A.S.; Whisstock, J.C.; Stuckey, P.J.; Lesk, A.M. MUSTANG: A multiple structural alignment algorithm. Proteins 2006, 64, 559–574. [Google Scholar] [CrossRef]
- Willard, L.; Ranjan, A.; Zhang, H.Y.; Monzavi, H.; Boyko, R.F.; Sykes, B.D.; Wishart, D.S. VADAR: A web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003, 31, 3316–3319. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |



| NIT1 | NIT2 | NIT3 | NIT4 | NitTv1 | NitAb | |
|---|---|---|---|---|---|---|
| NIT1 | - | 90.00 (98) | 83.82 (100) | 68.42 (93) | 47.35 (89) | 40.36 (86) |
| NIT2 | - | 83.82 (100) | 70.15 (95) | 49.37 (90) | 39.70 (88) | |
| NIT3 | - | 68.92 (93) | 48.44 (88) | 39.58 (86) | ||
| NIT4 | - | 52.20 (86) | 39.88 (84) | |||
| NitTv1 | - | 41.96 (96) | ||||
| NitAb | - |
| Enzyme 1 | Substrate β-CA | β-CA:PPN Activity | Reference | ||||
|---|---|---|---|---|---|---|---|
| Specific Activity [U mg−1 protein] | Vmax [U mg−1 rotein)] KM (mM) | NHase:NLase Activity | |||||
| Asparaginase | |||||||
| + (Total Activity) | − (NLase Activity) | NLase Activity 2 | NHase Activity 2 | ||||
| NitTv1 | 131.5 ± 0.5 3 | 94.2 ± 1.0 3 | 129.8 ± 11.4 7.72 ± 1.82 | 53.2 ± 9.2 6.74 ± 3.35 | 0.40 ± 0.02 3 | 91 ± 3 3 | This work |
| NitAb | 40.1 ± 0.1 3 | 26.8 ± 0.4 3 | 34.8 ± 2.0 7.38 ± 1.40 | 19.6 ± 2.5 4.97 ± 2.12 | 0.50 ± 0.02 3 | 56 ± 4.0 3 | This work |
| NIT4 | n.d. | 31.8 4 | 110.4 ± 9.6 0.74 ± 0.25 | 153.0 ± 25.8 0.70 ± 0.25 | 1.36 ± 0.21 4 | 119 ± 18 4 | [4] |
| TNIT4A | n.d. | n.d. | n.d. | n.d. | 0.87 ± 0.04 4 | 28 ± 8 4 | [4] |
| TNIT4B | n.d. | n.d. | n.d. | n.d. | 1.06 ± 0.12 4 | 20 ± 4 4 | [4] |
| Enzyme | Relative Activity [%] | Optima; Stabilities | Reference |
|---|---|---|---|
| NitTv1 (purified) 1 | β-CA (100), FN (8.6), 4CP (1.7), CN (1.6), PPN (1.1), PAN (1.0), PTAN (<1), IAN (<1) 3 | pH 7.5–8.5/30–35 °C; pH 5.2–9.3/≤35 °C | This work |
| NitAb (purified) 1 | β-CA (100), CN (3.3), PPN (1.8), PAN (1.1), FN (<1), PTAN (<1), 4CP (<1), IAN (<1) 3 | pH 6–8/25–30 °C; pH 5–7/≤25 °C | This work |
| NIT1 (cell extract) 1 | PPN (100), AC (94), PTAN (68), PAN (5), 4CP (<1), β-CA (<1) 4 | n.d.; n.d. | [9] |
| NIT1 (purified) 2 | PPN (100), ON (40), PBEN (26), PBN (21), BN (14), CN (6.5) 5 | pH 9, 35 °C; <35 °C | [10] |
| NIT2 (cell extract) 1 | PPN (100), AC (100), PTAN (80), PAN (13), 4CP (<1), β-CA (<1) 4 | n.d.; n.d. | [9] |
| NIT3 (cell extract) 1 | PPN (100), PTAN (63), AC (43), PAN (2), 4CP (<1), β-CA (<1) 4 | n.d.; n.d. | [9] |
| NIT4 (purified) 1 | β-CA (100), PPN (0.75), PAN (0.23), MTAN (0.22) 6 | pH 7–9/40 °C; n.d. | [4] |
| Ligand | Glide SP Score [kcal/mol] | |||
|---|---|---|---|---|
| NitTv1 | NitAb | |||
| Monomer | Tetramer | Monomer | Tetramer | |
| Cinnamonitrile | n.i. 1 | −2.569 | −2.409 | −2.569 |
| Fumaronitrile | −0.150 | −0.085 | −1.118 2 | −0.870 2 |
| Phenylacetonitrile | −3.838 | −4.639 | −4.170 | −4.669 |
| 3-Phenylpropionitrile | −3.490 | −3.571 | −3.146 | −4.099 |
| 4-Cyanopyridine | −3.260 | n.i. 1 | n.i. 1 | −3.057 |
| Phenylthioacetonitrile | −3.591 | −3.602 | −3.453 | −4.343 |
| Methylthioacetonitrile | n.i. 1 | −3.897 | −3.694 | −3.250 |
| β-Cyano-L-alanine | −2.713 | −2.758 | −2.416 | −2.557 |
| Indole-3-acetonitrile | −4.310 | −5.798 | −4.112 | −5.718 |
| 4-Phenylbutyronitrile | −4.201 | −3.873 | −3.072 | −2.932 |
| 4-Cyanophenylacetonitrile | −3.144 | −4.528 | −3.467 | −4.389 |
| Allylcyanide | n.i. 1 | −1.004 | −1.487 | −1.355 |
| Butyronitrile | −2.978 | −3.477 | −3.299 | −3.538 |
| Octanenitrile | n.i. 1 | −1.073 | −0.651 | −0.312 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rucká, L.; Kulik, N.; Novotný, P.; Sedova, A.; Petrásková, L.; Příhodová, R.; Křístková, B.; Halada, P.; Pátek, M.; Martínková, L. Plant Nitrilase Homologues in Fungi: Phylogenetic and Functional Analysis with Focus on Nitrilases in Trametes versicolor and Agaricus bisporus. Molecules 2020, 25, 3861. https://doi.org/10.3390/molecules25173861
Rucká L, Kulik N, Novotný P, Sedova A, Petrásková L, Příhodová R, Křístková B, Halada P, Pátek M, Martínková L. Plant Nitrilase Homologues in Fungi: Phylogenetic and Functional Analysis with Focus on Nitrilases in Trametes versicolor and Agaricus bisporus. Molecules. 2020; 25(17):3861. https://doi.org/10.3390/molecules25173861
Chicago/Turabian StyleRucká, Lenka, Natalia Kulik, Petr Novotný, Anastasia Sedova, Lucie Petrásková, Romana Příhodová, Barbora Křístková, Petr Halada, Miroslav Pátek, and Ludmila Martínková. 2020. "Plant Nitrilase Homologues in Fungi: Phylogenetic and Functional Analysis with Focus on Nitrilases in Trametes versicolor and Agaricus bisporus" Molecules 25, no. 17: 3861. https://doi.org/10.3390/molecules25173861
APA StyleRucká, L., Kulik, N., Novotný, P., Sedova, A., Petrásková, L., Příhodová, R., Křístková, B., Halada, P., Pátek, M., & Martínková, L. (2020). Plant Nitrilase Homologues in Fungi: Phylogenetic and Functional Analysis with Focus on Nitrilases in Trametes versicolor and Agaricus bisporus. Molecules, 25(17), 3861. https://doi.org/10.3390/molecules25173861