Analysis of Molecular Heterogeneity in Therapeutic IFNα2b from Different Manufacturers by LC/Q-TOF
Abstract
:1. Introduction
2. Results
2.1. Intact Protein Analysis
2.1.1. Purity Analysis
2.1.2. Identification of IFNα2b Variants
2.1.3. N-Terminal Heterogeneity
2.2. Evaluation of Modifications in IFNα2b by Peptide Mapping
2.2.1. Peptide Map
2.2.2. Modifications in IFNα2b from Different Manufacturers
2.2.3. Modifications in IFNα2b of Different Batches
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Sample Preparation
4.3. LC-MS/MS
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pestka, S. The Interferons: 50 Years after Their Discovery, There Is Much More to Learn. J. Biol. Chem. 2007, 282, 20047–20051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarhini, A.A.; Gogas, H.; Kirkwood, J.M. IFN-alpha in the treatment of melanoma. J. Immunol. 2012, 189, 3789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, D.; Ghosh, D.; Parida, P. Physiological Proteins in Therapeutics: A Current Review on Interferons. Mini Rev. Med. Chem. 2016, 16, 947. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, D.; Crowell, L.E.; Love, K.R.; Hancock, W.S. The application of HPLC/MS analysis with a multi-enzyme digest strategy to characterize different interferon product variants produced from Pichia pastoris. Amino Acids 2019, 51, 1353. [Google Scholar] [CrossRef]
- Parker, B.S.; Rautela, J.; Hertzog, P. Antitumour actions of interferons: Implications for cancer therapy. Nat. Rev. Cancer 2016, 16, 131–144. [Google Scholar] [CrossRef]
- Carini, M.; Regazzoni, L.; Aldini, G. Mass spectrometric strategies and their applications for molecular mass determination of recombinant therapeutic proteins. Curr. Pharm. Biotechnol. 2011, 12, 1548. [Google Scholar] [CrossRef]
- Liu, H.; Nowak, C.; Shao, M.; Ponniah, G.; Neill, A. Impact of cell culture on recombinant monoclonal antibody product heterogeneity. Biotechnol. Prog. 2016, 32, 1103. [Google Scholar] [CrossRef]
- Hsiao, K.; Alves, J.; Patel, R.; Adams, M.; Nashine, V.; Goueli, S. A High-Throughput Bioluminescent Assay to Monitor the Deamidation of Asparagine and Isomerization of Aspartate Residues in Therapeutic Proteins and Antibodies. J. Pharm. Sci. 2017, 106, 1528. [Google Scholar] [CrossRef]
- Jefferis, R. Biologics: Structural heterogeneity and immunogenicity. Br. J. Hosp. Med. 2017, 78, 443. [Google Scholar] [CrossRef]
- Liu, Y.H.; Wylie, D.; Zhao, J.; Cure, R.; Cutler, C.; Cannon-Carlson, S.; Yang, X. Mass spectrometric characterization of the isoforms in Escherichia coli recombinant DNA-derived interferon alpha-2b. Anal. Biochem. 2011, 408, 105. [Google Scholar] [CrossRef]
- Tao, L.; Pei, D.N.; Han, C.M.; Chen, W.; Rao, C.M.; Wang, J.Z. Characterization and comparison of interferon reference standards using UPLC-MS. Yao Xue Xue Bao Acta Pharm. Sin. 2015, 50, 75. [Google Scholar]
- Sherman, F.; Stewart, J.W.; Tsunasawa, S. Methionine or not methionine at the beginning of a protein. Bioessays 1985, 3, 27. [Google Scholar] [CrossRef] [PubMed]
- Ben-Bassat, A.; Bauer, K.; Chang, S.Y.; Myambo, K.; Boosman, A.; Chang, S. Processing of the initiation methionine from proteins: Properties of the Escherichia coli methionine aminopeptidase and its gene structure. J. Bacteriol. 1987, 169, 751–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.D.; Jeng, J.C.; Wang, C.F.; Wang, S.C.; Chang, S.T. Removal of N-terminal methionine from recombinant proteins by engineered E. coli methionine aminopeptidase. Protein Sci. 2004, 13, 1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirel, P.H.; Schmitter, M.J.; Dessen, P.; Fayat, G.; Blanquet, S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. USA 1989, 86, 8247–8251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradshaw, R.A.; Brickey, W.W.; Walker, K.W. N-terminal processing: The methionine aminopeptidase and N alpha-acetyl transferase families. Trends Biochem. Sci. 1998, 23, 263. [Google Scholar] [CrossRef]
- Ahsan, F.; Arif, A.; Mahmood, N.; Afza Gardner, Q.-T.-A.; Rashid, N.; Akhtar, M. Characterization and bioassay of post-translationally modified interferon alpha-2b expressed in Escherichia coli. J. Biotechnol. 2014, 184, 11. [Google Scholar] [CrossRef]
- Johnson, M.; Coulton, A.T.; Geeves, M.A.; Mulvihill, D.P. Targeted Amino-Terminal Acetylation of Recombinant Proteins in E. coli. PLoS ONE 2010, 5, e15801. [Google Scholar] [CrossRef] [Green Version]
- Arnesen, T.; Van Damme, P.; Polevoda, B.; Helsens, K.; Evjenth, R.; Colaert, N.; Varhaug, J.E.; Vandekerckhove, J.; Lillehaug, J.R.; Sherman, F.; et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. USA 2009, 106, 8157. [Google Scholar] [CrossRef] [Green Version]
- Ahsan, F.; Gardner, Q.A.; Rashid, N.; Towers, G.J.; Akhtar, M. Preventing the N-terminal processing of human interferon α-2b and its chimeric derivatives expressed in Escherichia coli. Bioorg. Chem. 2018, 76, 294–302. [Google Scholar] [CrossRef] [Green Version]
- Bandi, S.; Singh, S.M.; Shah, D.D.; Upadhyay, V.; Mallela, K.M. 2D NMR Analysis of the Effect of Asparagine Deamidation Versus Methionine Oxidation on the Structure, Stability, Aggregation, and Function of a Therapeutic Protein. Mol. Pharm. 2019, 16, 4621–4635. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of rhIFNα2b are available from the authors. |
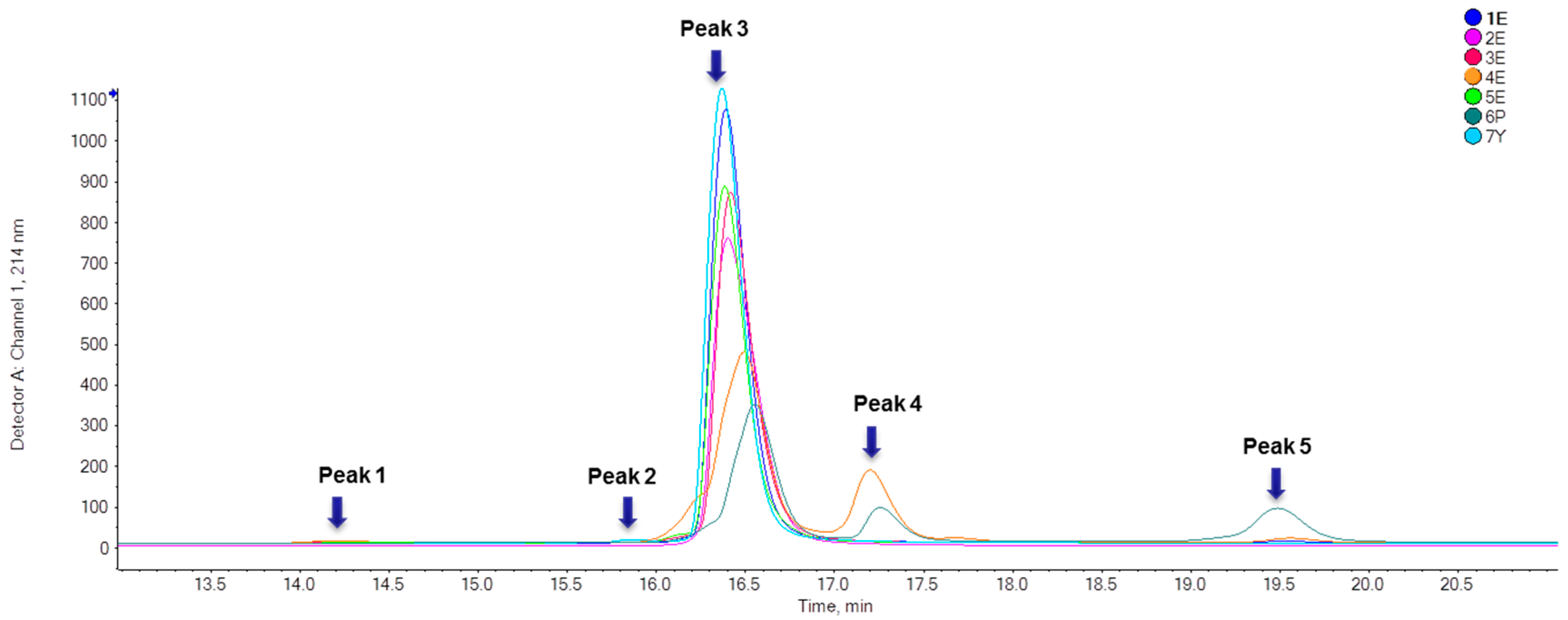

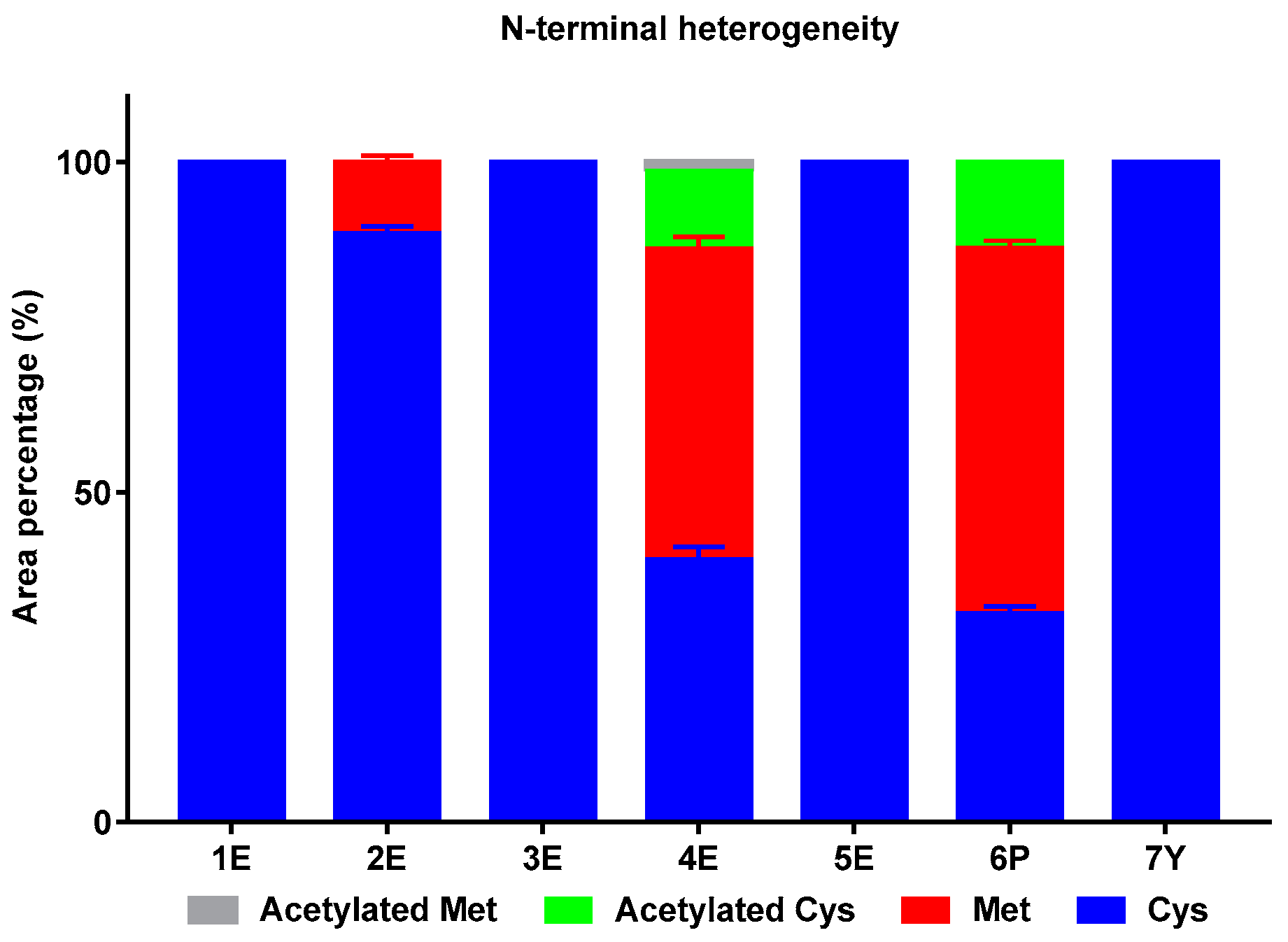

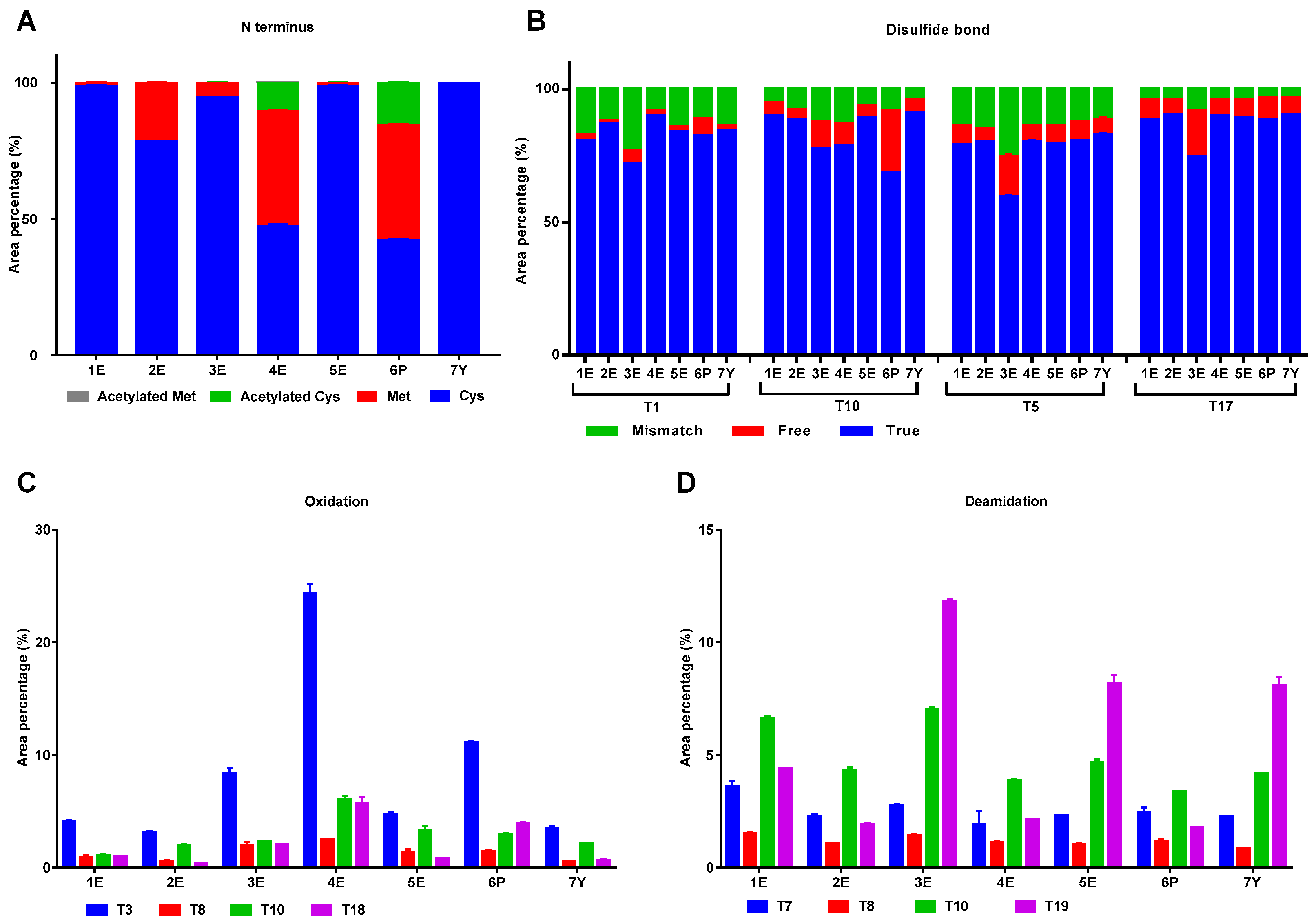
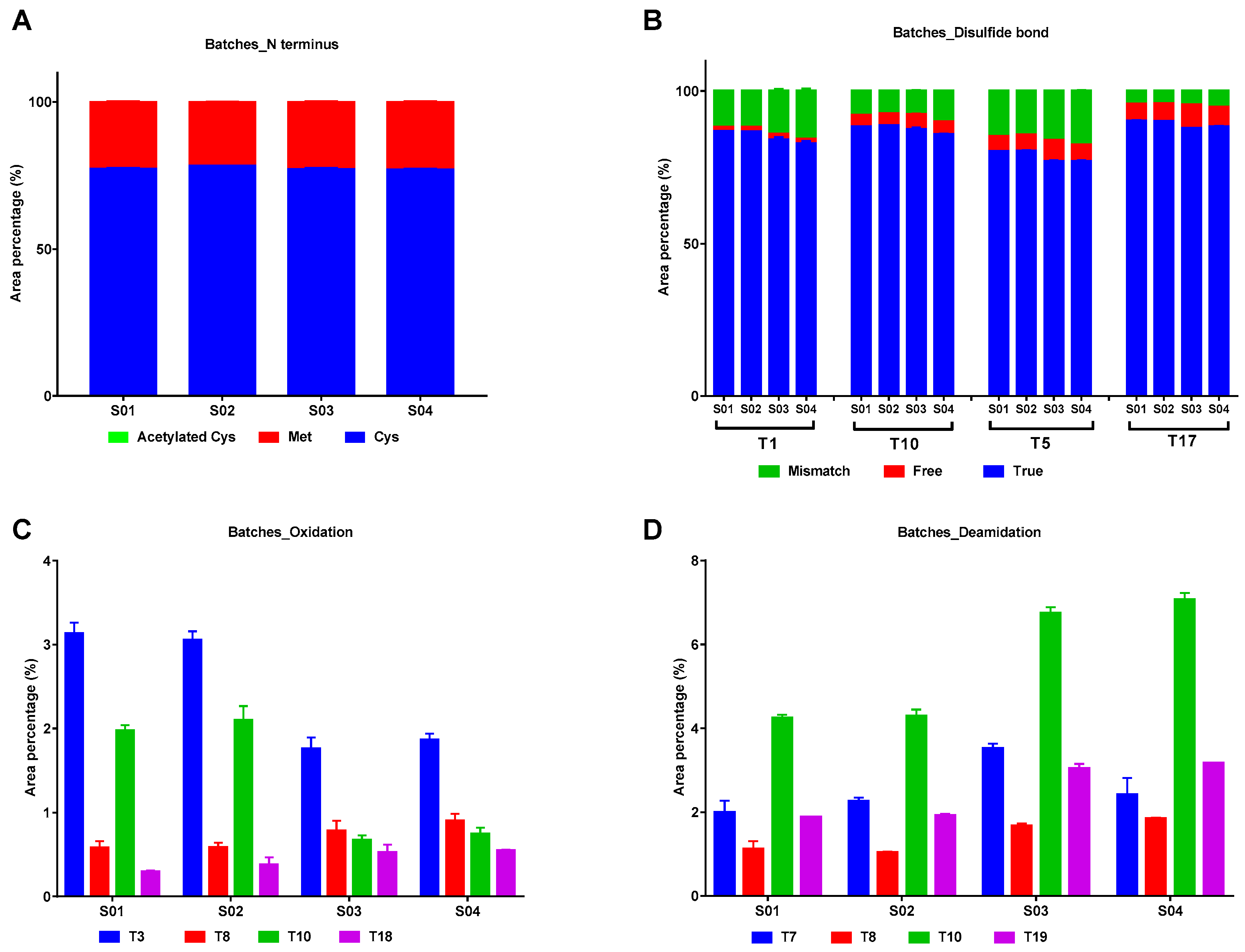
| Sample No. | Peak 1 (%) | Peak 2 (%) | Peak 3 (%) | Peak 4 (%) | Peak 5 (%) |
|---|---|---|---|---|---|
| 1E | / | / | 99.31 ± 0.33 | 0.31 ± 0.003 | 0.38 ± 0.33 |
| 2E | / | / | 100.00 | / | / |
| 3E | 0.51 ± 0.02 | / | 99.49 ± 0.02 | / | / |
| 4E | / | 74.89 ± 0.05 | 22.18 ± 0.02 | 1.89 ± 0.07 | |
| 5E | 0.19 ± 0.002 | / | 99.81 ± 0.002 | / | / |
| 6P | / | / | 65.97 ± 0.06 | 13.67 ± 0.02 | 20.36 ± 0.06 |
| 7Y | / | 0.74 ± 0.01 | 99.27 ± 0.004 | / | / |
| IFNα2b Variants | Retention Time (min) | Shift of Average MW (Da) | Theoretical Average MW (Da) | Measured Average MW (Da) | Error (Da) |
|---|---|---|---|---|---|
| Oxi | ~14.2 | +16.00 | 19,280.91 | 19,280.68 | −0.23 |
| +32.00 | 19,296.91 | 19,296.50 | −0.41 | ||
| dOx | ~16.0 | −16.00 | 19,248.16 | 19,248.66 | 0.5 |
| Native * | ~16.4 | / | 19,264.91 | 19,264.19 | −0.72 |
| +Met | +131.20 | 19,396.11 | 19,395.33 | −0.78 | |
| ACE | ~17.2 | +42.04 | 19,306.95 | 19,306.49 | −0.46 |
| IFNα2b Variant | Sample 4E | Sample 6P | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | Average | 1 | 2 | 3 | Average | |
| IFNα2b + M, Oxidation-1 | 8690.8 | 8628.6 | 8580.7 | 8580.7 | 3604.8 | 3567.6 | 3609.5 | 3594.0 |
| IFNα2b + M | 24,271.3 | 22,641.7 | 22,905.7 | 22,905.7 | 23,047.2 | 21,887.2 | 23,409.9 | 22,781.4 |
| IFNα2b, Oxidation-2 | 0.0 | 0.0 | 0.0 | 0.0 | 897.2 | 864.6 | 888.4 | 883.4 |
| IFNα2b, Oxidation-1 | 4722.9 | 4960.8 | 4913.6 | 4913.6 | 1599.2 | 1727.5 | 1717.8 | 1681.5 |
| IFNα2b | 18,739.6 | 19,652.7 | 19,395.4 | 19,395.4 | 12,413.0 | 12,756.8 | 13,133.5 | 12,767.8 |
| Oxidation in IFNα2b +M (%) | 26.4 | 27.6 | 27.9 | 27.3 | 14.0 | 13.4 | 13.6 | 13.5 |
| Oxidation in IFNα2b (%) | 20.1 | 20.2 | 20.3 | 20.2 | 20.3 | 19.8 | 20.1 | 20.1 |
| Oxidation in total IFNα2b (%) | 23.8 | 24.3 | 24.5 | 24.2 | 15.1 | 14.5 | 14.8 | 14.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, L.; Tao, L.; Zhao, Y.; Li, Y.; Pei, D.; Rao, C. Analysis of Molecular Heterogeneity in Therapeutic IFNα2b from Different Manufacturers by LC/Q-TOF. Molecules 2020, 25, 3965. https://doi.org/10.3390/molecules25173965
Yu L, Tao L, Zhao Y, Li Y, Pei D, Rao C. Analysis of Molecular Heterogeneity in Therapeutic IFNα2b from Different Manufacturers by LC/Q-TOF. Molecules. 2020; 25(17):3965. https://doi.org/10.3390/molecules25173965
Chicago/Turabian StyleYu, Lei, Lei Tao, Yinghua Zhao, Yonghong Li, Dening Pei, and Chunming Rao. 2020. "Analysis of Molecular Heterogeneity in Therapeutic IFNα2b from Different Manufacturers by LC/Q-TOF" Molecules 25, no. 17: 3965. https://doi.org/10.3390/molecules25173965
APA StyleYu, L., Tao, L., Zhao, Y., Li, Y., Pei, D., & Rao, C. (2020). Analysis of Molecular Heterogeneity in Therapeutic IFNα2b from Different Manufacturers by LC/Q-TOF. Molecules, 25(17), 3965. https://doi.org/10.3390/molecules25173965






