p-Aminophenylalanine Involved in the Biosynthesis of Antitumor Dnacin B1 for Quinone Moiety Formation
Abstract
:1. Introduction
2. Results
2.1. Genome Sequencing Revealed a Circular Chromosome of Strain DSM 44131T
2.2. Comparative Genomic Analysis within Actinosynnema Resulted in the Discovery of Dnacin B1 BGC
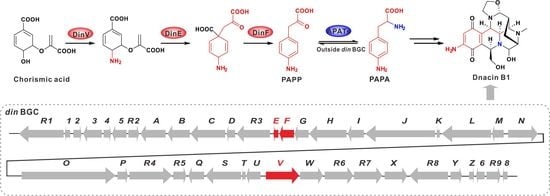
2.3. Functional Genes Involved in the Assembly of Dnacin B1 Skeleton
2.4. Reconstitution of PAPA Pathway Revealed TyrB Participating in the Mature of PAPA
2.5. Aminotransferases Responsible for the PAPA Maturation Dispersed within the Genome
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids and Reagents
4.2. General Experimental Procedures
4.3. DNA Sequencing for Strain DSM 44131T
4.4. Bioinformatics Analysis
4.5. Construction of the Dnacin B1 BGC Inactivation Mutant
4.6. Optimization of Fermentation Medium
4.7. Total RNA Extraction and q-PCR Analysis
4.8. Production and Detection of Dnacin B1 in Strain DSM 44131T
4.9. Reconstruction of PAPA Biosynthesis Pathway in E. coli
4.10. Protein Expression and Purification
4.11. Enzyme Activity Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hasegawa, T.; Lechevalier, M.P.; Lechevalier, H.A. New genus of the Actinomycetales: Actinosynnema gen. nov. Int. J. Syst. Evol. Microbiol. 1978, 28, 304–310. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Rainey, F.A.; Ward-Rainey, N.L. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Evol. Microbiol. 1997, 47, 479–491. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, T.; Tanida, S.; Hatano, K.; Higashide, E.; Yoneda, M. Motile Actinomycetes: Actinosynnema pretiosum subsp. pretiosum sp. nov., subsp. nov., and Actinosynnema pretiosum subsp. auranticum subsp. nov. Int. J. Syst. Evol. Microbiol. 1983, 33, 314–320. [Google Scholar] [CrossRef] [Green Version]
- Higashide, E.; Asai, M.; Ootsu, K.; Tanida, S.; Kozai, Y.; Hasegawa, T.; Kishi, T.; Sugino, Y.; Yoneda, M. Ansamitocin, a group of novel maytansinoid antibiotics with antitumour properties from Nocardia. Nature 1977, 270, 721–722. [Google Scholar] [CrossRef]
- Kang, Q.; Shen, Y.; Bai, L. Biosynthesis of 3,5-AHBA-derived natural products. Nat. Prod. Rep. 2012, 29, 243–263. [Google Scholar] [CrossRef] [PubMed]
- Siyu, M.; Hong, C.; Li, C.; Chuanxi, W.; Wei, J.; Xiaoming, C.; Huangjian, Y.; Wei, H.; Wei, Z. Two novel ansamitocin analogs from Actinosynnema pretiosum. Nat. Prod. Res. 2013, 27, 1532–1536. [Google Scholar] [CrossRef]
- Wei, G.Z.; Bai, L.Q.; Yang, T.; Ma, J.; Zeng, Y.; Shen, Y.M.; Zhao, P.J. A new antitumour ansamitocin from Actinosynnema pretiosum. Nat. Prod. Res. 2010, 24, 1146–1150. [Google Scholar] [CrossRef]
- Tanida, S.; Hasegawa, T.; Muroi, M.; Higashide, E. Dnacins, new antibiotics I. Producing organism, fermentation, and antimicrobial activities. J. Antibiot. 1980, 33, 1443–1448. [Google Scholar] [CrossRef]
- Lu, C.; Xie, F.; Shan, C.; Shen, Y. Two novel cyclic hexapeptides from the genetically engineered Actinosynnema pretiosum. Appl. Microbiol. Biotechnol. 2017, 101, 2273–2279. [Google Scholar] [CrossRef]
- Watanabe, K.; Okuda, T.; Yokose, K.; Furumai, T.; Maruyama, H.B. Actinosynnema mirum, a new producer of nocardicin antibiotics. J. Antibiot. 1983, 36, 321–324. [Google Scholar] [CrossRef]
- Asamizu, S.; Abugreen, M.; Mahmud, T. Comparative metabolomic analysis of an alternative biosynthetic pathway to Pseudosugars in Actinosynnema mirum DSM 43827. ChemBioChem 2013, 14, 1548–1551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murao, S.; Imafuku, S.; Oyama, H. Isolation of propioxatin A from Actinosynnema sp. SI-23 during a screening for serratia piscatorum metalloproteinase inhibitors. Biosci. Biotechnol. Biochem. 1997, 61, 561–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giessen, T.W.; Franke, K.B.; Knappe, T.A.; Kraas, F.I.; Bosello, M.; Xie, X.; Linne, U.; Marahiel, M.A. Isolation, structure elucidation, and biosynthesis of an unusual hydroxamic acid ester-containing siderophore from Actinosynnema mirum. J. Nat. Prod. 2012, 75, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Land, M.; Lapidus, A.; Mayilraj, S.; Chen, F.; Copeland, A.; Del Rio, T.G.; Nolan, M.; Lucas, S.; Tice, H.; Cheng, J.F.; et al. Complete genome sequence of Actinosynnema mirum type strain (101). Stand. Genomic. Sci. 2009, 1, 46–53. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Sun, R.; Ning, X.; Wang, X.; Wang, Z. Genome-scale metabolic model of Actinosynnema pretiosum ATCC 31280 and its application for ansamitocin P-3 production improvement. Genes 2018, 9, 364. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Hu, X.; Sun, G.; Li, L.; Jiang, B.; Li, S.; Bai, L.; Liu, H.; Yu, L.; Wu, L. Genome-guided discovery of pretilactam from Actinosynnema pretiosum ATCC 31565. Molecules 2019, 24, 2281. [Google Scholar] [CrossRef] [Green Version]
- Zhong, C.; Zong, G.; Qian, S.; Liu, M.; Fu, J.; Zhang, P.; Li, J.; Cao, G. Complete genome sequence of Actinosynnema pretiosum X47, an industrial strain that produces the antibiotic ansamitocin AP-3. Curr. Microbiol. 2019, 76, 954–958. [Google Scholar] [CrossRef]
- Muroi, M.; Tanida, S.; Asai, M.; Kishi, T. Dnacins, new antibiotics II. Isolation and characterization. J. Antibiot. 1980, 33, 1449–1456. [Google Scholar] [CrossRef] [Green Version]
- Scott, J.D.; Williams, R.M. Chemistry and biology of the tetrahydroisoquinoline antitumor antibiotics. Chem. Rev. 2002, 102, 1669–1730. [Google Scholar] [CrossRef]
- Le, V.H.; Inai, M.; Williams, R.M.; Kan, T. Ecteinascidins. A review of the chemistry, biology and clinical utility of potent tetrahydroisoquinoline antitumor antibiotics. Nat. Prod. Rep. 2015, 32, 328–347. [Google Scholar] [CrossRef] [Green Version]
- Tang, G.; Tang, M.; Song, L.; Zhang, Y. Biosynthesis of tetrahydroisoquinoline antibiotics. Curr. Top. Med. Chem. 2016, 16, 1717–1726. [Google Scholar] [CrossRef] [PubMed]
- Jeedigunta, S.; Krenisky, J.; Kerr, R. Diketopiperazines as advanced intermediates in the biosynthesis of ecteinascidins. Tetrahedron 2000, 56, 3303–3307. [Google Scholar] [CrossRef]
- Mikami, Y.; Takahashi, K.; Yazawa, K.; Arai, T.; Namikoshi, M.; Iwasaki, S.; Okuda, S. Biosynthetic studies on saframycin A, a quinone antitumor antibiotic produced by Streptomyces lavendulae. J. Biol. Chem. 1985, 260, 344–348. [Google Scholar] [PubMed]
- Velasco, A.; Acebo, P.; Gomez, A.; Schleissner, C.; Rodriguez, P.; Aparicio, T.; Conde, S.; Munoz, R.; de la Calle, F.; Garcia, J.L.; et al. Molecular characterization of the safracin biosynthetic pathway from Pseudomonas fluorescens A2-2: Designing new cytotoxic compounds. Mol. Microbiol. 2005, 56, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Zmijewski, M.J.; Mikolajczak, M.; Viswanatha, V.; Hruby, V.J. Biosynthesis of the antitumor antibiotic naphthyridinomycin. J. Am. Chem. Soc. 1982, 104, 4969–4971. [Google Scholar] [CrossRef]
- Hiratsuka, T.; Koketsu, K.; Minami, A.; Kaneko, S.; Yamazaki, C.; Watanabe, K.; Oguri, H.; Oikawa, H. Core assembly mechanism of quinocarcin/SF-1739: Bimodular complex nonribosomal peptide synthetases for sequential mannich-type reactions. Chem. Biol. 2013, 20, 1523–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes—A review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef] [Green Version]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends. Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef]
- Feyereisen, M.; Mahony, J.; Kelleher, P.; Roberts, R.J.; O’Sullivan, T.; Geertman, J.-M.A.; van Sinderen, D. Comparative genome analysis of the Lactobacillus brevis species. BMC Genom. 2019, 20, 416. [Google Scholar] [CrossRef] [Green Version]
- Yao, T.; Liu, J.; Liu, Z.; Li, T.; Li, H.; Che, Q.; Zhu, T.; Li, D.; Gu, Q.; Li, W. Genome mining of cyclodipeptide synthases unravels unusual tRNA-dependent diketopiperazine-terpene biosynthetic machinery. Nat. Commun. 2018, 9, 4091. [Google Scholar] [CrossRef]
- Zhang, Y.; Wen, W.; Pu, J.; Tang, M.; Zhang, L.; Peng, C.; Xu, Y.; Tang, G. Extracellularly oxidative activation and inactivation of matured prodrug for cryptic self-resistance in naphthyridinomycin biosynthesis. Proc. Natl. Acad. Sci. USA 2018, 115, 11232–11237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pu, J.; Peng, C.; Tang, M.; Zhang, Y.; Guo, J.; Song, L.; Hua, Q.; Tang, G. Naphthyridinomycin biosynthesis revealing the use of leader peptide to guide nonribosomal peptide assembly. Org. Lett. 2013, 15, 3674–3677. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Pu, J.; Song, L.; Jian, X.; Tang, M.; Tang, G. Hijacking a hydroxyethyl unit from a central metabolic ketose into a nonribosomal peptide assembly line. Proc. Natl. Acad. Sci. USA 2012, 109, 8540–8545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prija, F.; Prasad, R. DrrC protein of Streptomyces peucetius removes daunorubicin from intercalated dnrI promoter. Microbiol. Res. 2017, 202, 30–35. [Google Scholar] [CrossRef]
- Chang, Z.; Sun, Y.; He, J.; Vining, L.C. p-Aminobenzoic acid and chloramphenicol biosynthesis in Streptomyces venezuelae: Gene sets for a key enzyme, 4-amino-4-deoxychorismate synthase. Microbiology 2001, 147, 2113–2126. [Google Scholar] [CrossRef] [Green Version]
- Yanai, K.; Sumida, N.; Okakura, K.; Moriya, T.; Watanabe, M.; Murakami, T. Para-position derivatives of fungal anthelmintic cyclodepsipeptides engineered with Streptomyces venezuelae antibiotic biosynthetic genes. Nat. Biotechnol. 2004, 22, 848–855. [Google Scholar] [CrossRef]
- Grishin, N.V.; Phillips, M.A.; Goldsmith, E.J. Modeling of the spatial structure of eukaryotic ornithine decarboxylases. Protein Sci. Publ. Protein Soc. 1995, 4, 1291–1304. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, A.; Kato, R.; Masui, R.; Yamagishi, A.; Oshima, T.; Kuramitsu, S. An aspartate aminotransferase from an extremely thermophilic bacterium, Thermus thermophilus HB8. J. Biochem. 1996, 119, 135–144. [Google Scholar] [CrossRef]
- Son, H.F.; Kim, K.J. Structural insights into a novel class of aspartate aminotransferase from Corynebacterium glutamicum. PLoS ONE 2016, 11, e0158402. [Google Scholar] [CrossRef] [Green Version]
- Palazzotto, E.; Tong, Y.; Lee, S.Y.; Weber, T. Synthetic biology and metabolic engineering of actinomycetes for natural product discovery. Biotechnol. Adv. 2019, 37, 107366. [Google Scholar] [CrossRef]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2014, 31, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids. Res. 2019, 47, 81–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishikawa, J.; Hotta, K. FramePlot: A new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 1999, 174, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, B.O.; Ravel, J. Methods for In Silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. In Methods in Enzymology; Academic Press: San Diego, CA, USA, 2009; Volume 458, pp. 181–217. [Google Scholar]
- Ning, X.; Wang, X.; Wu, Y.; Kang, Q.; Bai, L. Identification and engineering of post-PKS modification bottlenecks for Ansamitocin P-3 titer improvement in Actinosynnema pretiosum subsp. pretiosum ATCC 31280. Biotechnol. J. 2017, 12, 1700484. [Google Scholar] [CrossRef]
- Masuo, S.; Zhou, S.; Kaneko, T.; Takaya, N. Bacterial fermentation platform for producing artificial aromatic amines. Sci. Rep. 2016, 6, 25764. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Toda, K.; Maeda, H.A. Biochemical properties and subcellular localization of tyrosine aminotransferases in Arabidopsis thaliana. Phytochemistry 2016, 132, 16–25. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |





| ORF | Number of Amino Acids | Proposed Function | Sequence Similarity (Protein, Origin) | % Identity/ Similarity | Accession No. |
|---|---|---|---|---|---|
| DinR1 | 951 | ABC transporter | IQ63_41860, Streptomyces acidiscabies | 64/75 | KND24720.1 |
| ORF1 | 117 | unknown | SD37_10880, Amycolatopsis orientalis | 55/75 | ANN21703.1 |
| ORF2 | 175 | protease inhibitor | AC230_04780, Streptomyces caatingaensis | 38/54 | KNB54254.1 |
| ORF3 | 395 | histidine kinase | A9W97_15640, Mycobacterium gordonae | 51/60 | OBJ88517.1 |
| ORF4 | 159 | ATP-binding protein | RKT69099.1, Saccharothrix variisporea | 45/51 | DFJ66_2293 |
| ORF5 | 306 | epimerase | DI639_00155, Leifsonia xyli | 74/83 | PZO61411.1 |
| DinR2 | 228 | TetR family transcriptional regulator | STRAU_2618, Streptomyces aurantiacus | 58/71 | EPH44178.1 |
| DinA | 534 | flavin adenine dinucleotide (FAD)-linked oxidase | NapU, Streptomyces lusitanus | 64/75 | AGD80628.1 |
| DinB | 471 | gamma-aminobutyraldehyde dehydrogenase | MCBG_02966, Micromonospora sp. M42 | 53/65 | EWM65833.1 |
| DinC | 741 | peptidase | NapG, Streptomyces lusitanus | 48/56 | AGD80614.1 |
| DinD | 183 | flavin mononucleotide (FMN) reductase | SsuE, Streptomyces noursei | 58/67 | WP_067345371.1 |
| DinR3 | 707 | regulatory protein | NapR3, Streptomyces lusitanus | 55/68 | AGD80615.1 |
| DinE | 103 | 4-amino-4-deoxychorismate mutase | PapB, Streptomyces venezuelae | 34/53 | BAD21142.1 |
| DinF | 307 | 4-amino-4-deoxyprephenate dehydrogenase | PapC, Streptomyces venezuelae | 40/51 | BAD21141.1 |
| DinG | 300 | unknown | CLV43_102760, Umezawaea tangerina | 47/57 | PRY45195.1 |
| DinH | 796 | adenylation domain | NapH, Streptomyces lusitanus | 51/60 | AGD80616.1 |
| DinI | 342 | non-heme iron hydroxylase | NapI, Streptomyces lusitanus | 63/75 | AGD80617.1 |
| DinJ | 1482 | NRPS | NapJ, Streptomyces lusitanus | 68/79 | AGD80618.1 |
| Module 5 | C-A-PCP-RE | ||||
| DinK | 66 | MbtH-like protein | NapK, Streptomyces lusitanus | 73/84 | AGD80619.1 |
| DinL | 1125 | NRPS | NapL, Streptomyces lusitanus | 54/64 | AGD80620.1 |
| Module 4 | C-A-PCP | ||||
| DinM | 252 | thioesterase II | NapM, Streptomyces lusitanus | 51/64 | AGD80621.1 |
| DinN | 632 | AMP-dependent synthetase and ligase | NapN, Streptomyces lusitanus | 64/74 | AGD80622.1 |
| Loading | AL-PCP | ||||
| DinO | 3049 | NRPS | NapO, Streptomyces lusitanus | 52/63 | AGD80623.1 |
| Module 1 | C-A-PCP | ||||
| Module 2 | C-A-PCP | ||||
| Module 3 | C-A-PCP | ||||
| DinP | 223 | hypothetical protein | NapP, Streptomyces lusitanus | 45/59 | AGD80633.1 |
| DinR4 | 909 | ABC transporter | B0I31_10111, Saccharothrix carnea | 56/67 | PSL57800.1 |
| DinR5 | 280 | ABC transporter | CLV69_102810, Yuhushiella deserti | 53/69 | TDX97704.1 |
| DinQ | 336 | 3-oxoacyl-synthase III (KS) | NapE, Streptomyces lusitanus | 72/80 | AGD80612.1 |
| DinS | 754 | Transketolase | NapD, Streptomyces lusitanus | 59/68 | AGD80611.1 |
| DinT | 76 | acyl carrier protein (ACP) | NapC, Streptomyces lusitanus | 63/75 | AGD80633.1 |
| DinU | 306 | transketolase | NapB, Streptomyces lusitanus | 68/77 | AGD80609.1 |
| DinV | 706 | 4-amino-4-deoxychorismate synthase | PapA, Streptomyces venezuelae | 48/58 | BAD21140.1 |
| DinW | 486 | 4-hydroxyphenylacetate 3-monooxygenase | EWI31_24745, Streptomyces tsukubensis | 56/69 | TAI42134.1 |
| DinR6 | 613 | ABC transporter | CLV43_102777, Umezawaea tangerina | 70/78 | PRY45212.1 |
| DinR7 | 601 | ABC transporter | DIU55_13605, Firmicutes bacterium | 51/66 | PZN68908.1 |
| DinX | 477 | monooxygenase | NapA, Streptomyces lusitanus | 69/76 | AGD80608.1 |
| DinR8 | 818 | UV-repair protein | NapR1, Streptomyces lusitanus | 75/84 | AGD80606.1 |
| DinY | 244 | methyltransferase | QncJ, Streptomyces melanovinaceus | 47/65 | AGD95052.1 |
| DinZ | 118 | long-chain-fatty-acid--CoA ligase | NCTC13184_06982, Nocardia africana | 60/81 | SUA48430.1 |
| ORF6 | 182 | sugar O-acetyltransferase | E1091_12580, Micromonospora fluostatini | 64/72 | TDB92781.1 |
| DinR9 | 304 | LysR family transcriptional regulator | AFR_16085, Actinoplanes friuliensis | 57/69 | AGZ41499.1 |
| ORF7 | 104 | transposase | CLV43_10550, Umezawaea tangerina | 56/61 | PRY41292.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Li, X.; Sheng, Y.; Wang, H.; Li, X.; Ou, Y.; Deng, Z.; Bai, L.; Kang, Q. p-Aminophenylalanine Involved in the Biosynthesis of Antitumor Dnacin B1 for Quinone Moiety Formation. Molecules 2020, 25, 4186. https://doi.org/10.3390/molecules25184186
Hu X, Li X, Sheng Y, Wang H, Li X, Ou Y, Deng Z, Bai L, Kang Q. p-Aminophenylalanine Involved in the Biosynthesis of Antitumor Dnacin B1 for Quinone Moiety Formation. Molecules. 2020; 25(18):4186. https://doi.org/10.3390/molecules25184186
Chicago/Turabian StyleHu, Xiaojing, Xing Li, Yong Sheng, Hengyu Wang, Xiaobin Li, Yixin Ou, Zixin Deng, Linquan Bai, and Qianjin Kang. 2020. "p-Aminophenylalanine Involved in the Biosynthesis of Antitumor Dnacin B1 for Quinone Moiety Formation" Molecules 25, no. 18: 4186. https://doi.org/10.3390/molecules25184186
APA StyleHu, X., Li, X., Sheng, Y., Wang, H., Li, X., Ou, Y., Deng, Z., Bai, L., & Kang, Q. (2020). p-Aminophenylalanine Involved in the Biosynthesis of Antitumor Dnacin B1 for Quinone Moiety Formation. Molecules, 25(18), 4186. https://doi.org/10.3390/molecules25184186