Discovery of New Secondary Metabolites by Epigenetic Regulation and NMR Comparison from the Plant Endophytic Fungus Monosporascus eutypoides
Abstract
:1. Introduction
2. Results
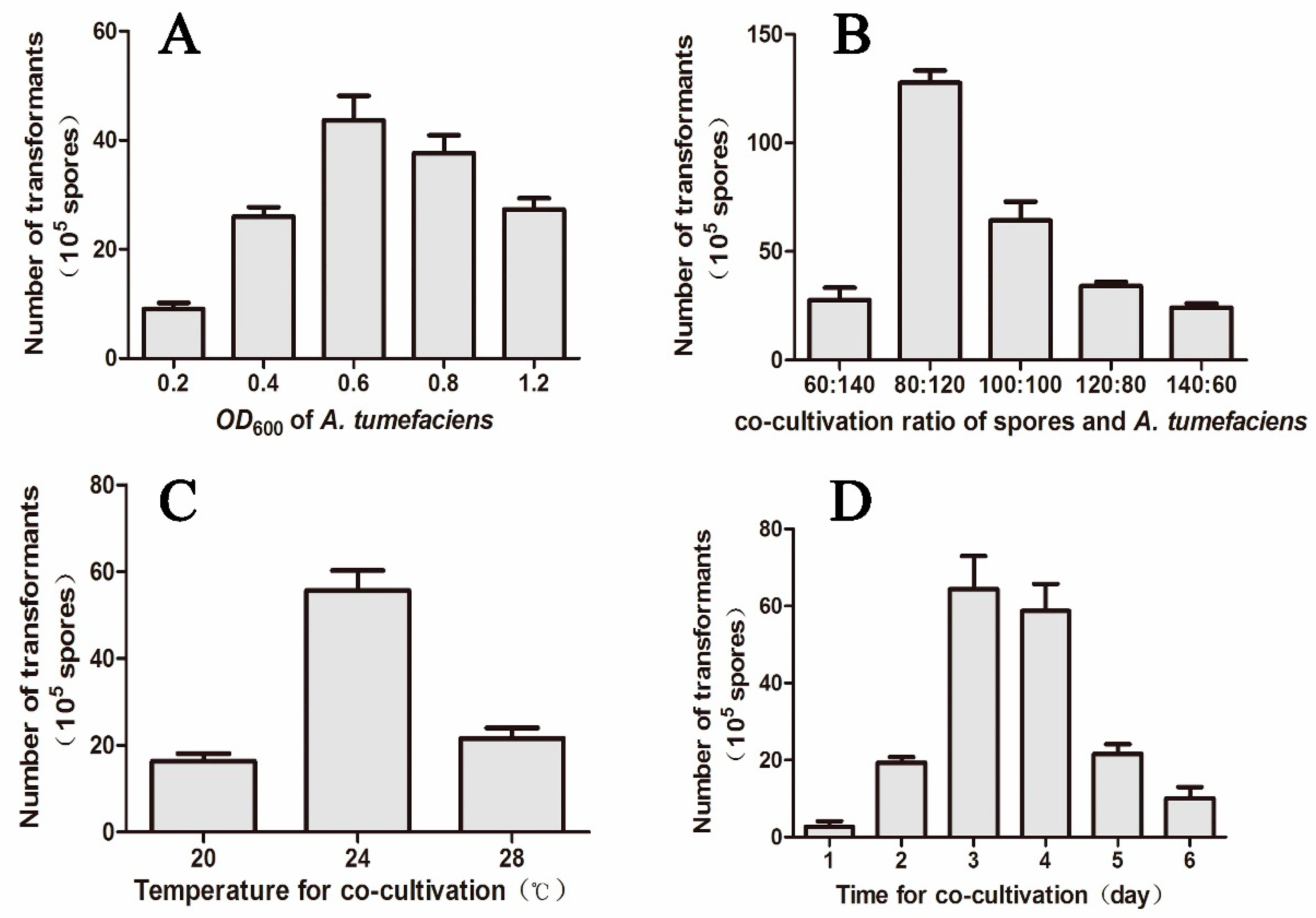
2.1. The ATMT Is an Effective Manipulation of Gene Expression in M. eutypoides
2.2. Identification of MeHat and the Overexpression of MeHat in M. eutypoides
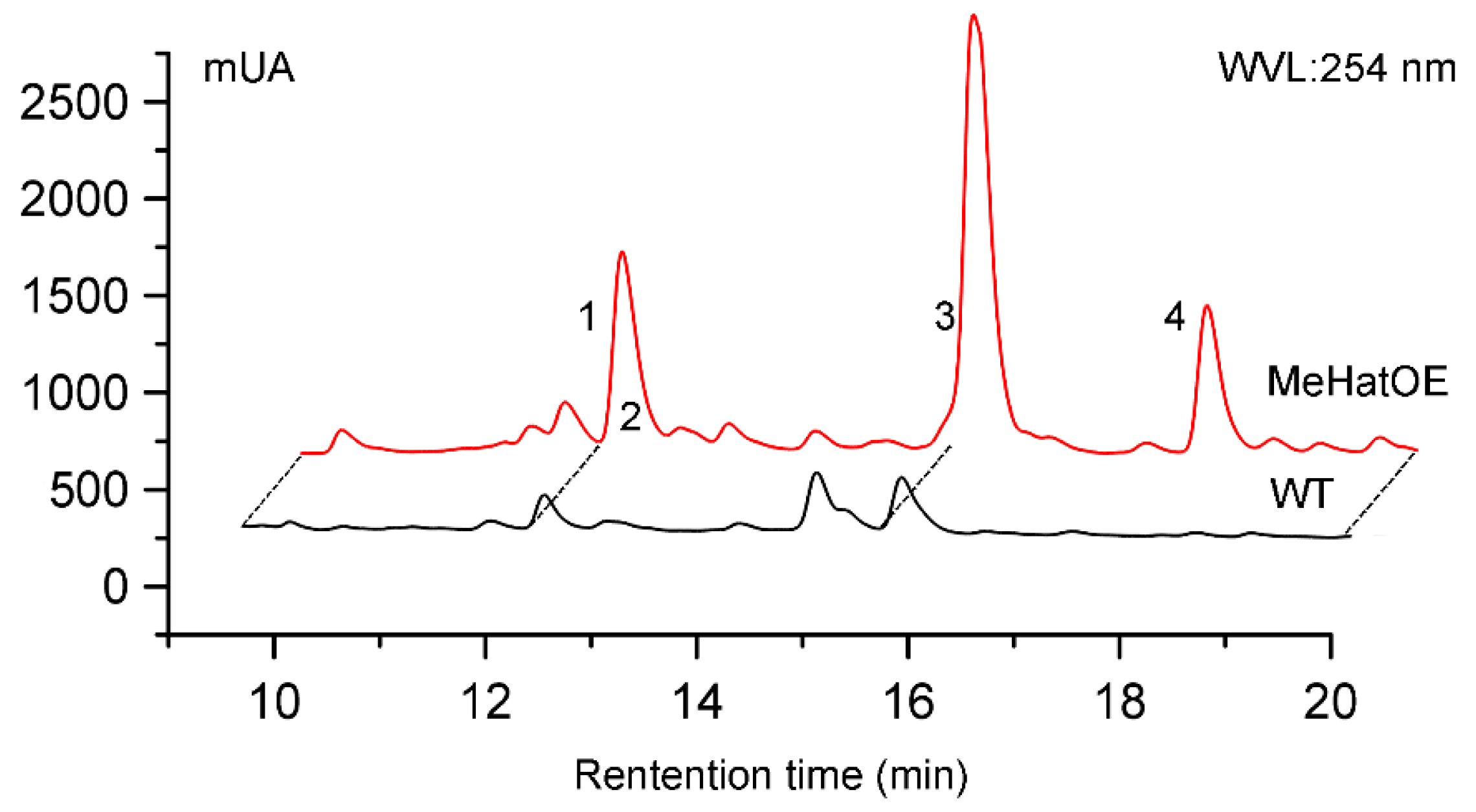
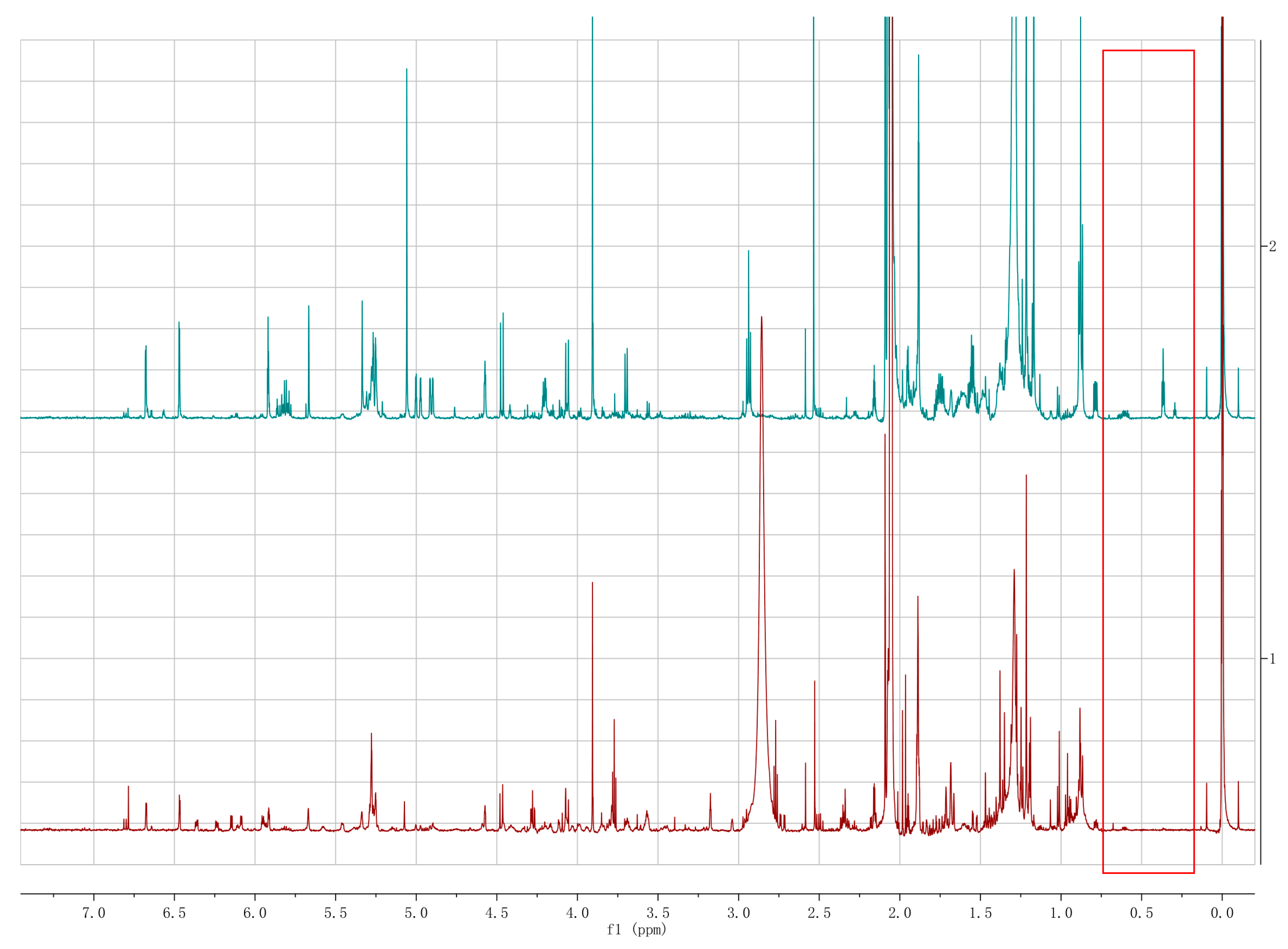
2.3. Secondary Metabolite Assessment by HPLC Chromatograms and 1H NMR Analysis
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedure
4.2. Biology Materials
4.3. Construction of ATMT System for M. eutypoides
4.4. Construction of the MeHatOE
4.5. Fermentation and Isolation
4.6. Computational Details
4.7. MTT Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brakhage, A.A.; Volker, S. Fungal secondary metabolites—Strategies to activate silent gene clusters. Fungal Genet. Biol. 2011, 48, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhou, H.; Zhang, P.; Wang, X.; Li, W.; Zhang, W.; Liu, X.; Liu, H.-W.; Keller, N.P.; An, Z.; et al. Polyketide production of pestaloficiols and macrodiolide ficiolides revealed by manipulations of epigenetic regulators in an endophytic fungus. Org. Lett. 2016, 18, 1832–1835. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Dominguez, Y.; Boedi, S.; Sulyok, M.; Wiesenberger, G.; Stoppacher, N.; Krska, R.; Strauss, J. Heterochromatin influences the secondary metabolite profile in the plant pathogen Fusarium graminearum. Fungal Genet. Biol. 2012, 49, 39–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giles, S.S.; Soukup, A.A.; Lauer, C.; Shaaban, M.; Lin, A.; Oakley, B.R.; Wang, C.C.C.; Keller, N.P. Cryptic Aspergillus nidulans antimicrobials. Appl. Environ. Microbiol. 2011, 77, 3669–3675. [Google Scholar] [CrossRef] [Green Version]
- Bok, J.W.; Chiang, Y.; Szewczyk, E.; Reyes-Dominguez, Y.; Davidson, A.D.; Sanchez, J.F.; Lo, H.-C.; Watanabe, K.; Strauss, J.; Oakley, B.R.; et al. Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 2009, 5, 462–464. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.Y.; Yu, J.H. Epigenetics of fungal secondary metabolism related genes. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Springer: New York, NY, USA, 2015. [Google Scholar]
- Soukup, A.A.; Chiang, Y.M.; Bok, J.W.; Reyes-Dominguez, Y.; Oakley, B.R.; Wang, C.C.; Strauss, J.; Keller, N.P. Overexpression of the Aspergillus nidulans histone 4 acetyltransferase Esaa increases activation of secondary metabolite production. Mol. Microbiol. 2012, 86, 314–330. [Google Scholar] [CrossRef] [Green Version]
- Asai, T.; Otsuki, S.; Sakurai, H.; Yamashita, K.; Ozeki, T.; Oshima, Y. Benzophenones from an endophytic fungus, Graphiopsis chlorocephala, from Paeonia lactiflora cultivated in the presence of an NAD+-dependent hdac inhibitor. Org. Lett. 2013, 15, 2058–2061. [Google Scholar] [CrossRef]
- Dallery, J.-F.; Le Goff, G.; Adelin, E.; Iorga, B.I.; Pigné, S.; O’Connell, R.J.; Ouazzani, J. Deleting a chromatin remodeling gene increases the diversity of secondary metabolites produced by Colletotrichum higginsianum. J. Nat. Prod. 2019, 82, 813–822. [Google Scholar] [CrossRef]
- De Groot, M.J.; Bundock, P.; Hooykaas, P.J.; Beijersbergen, A.G. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 1998, 16, 839–842. [Google Scholar] [CrossRef]
- Frandsen, R.J.N. A guide to binary vectors and strategies for targeted genome modification in fungi using Agrobacterium tumefaciens -mediated transformation. J. Microbiol. Methods 2011, 87, 247–262. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Liu, L.; Guan, F.; Li, E.; Jin, J.; Li, J.; Che, Y.; Liu, G. Characterization of a prenyltransferase for iso-a82775c biosynthesis and generation of new congeners of chloropestolides. ACS Chem. Biol. 2018, 13, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, X.; Zhang, X.; Li, W.; Liu, G.; Wang, S.; Yan, X.; Zou, H.; Yin, W.B. Cop9 signalosome subunit pfcsne regulates secondary metabolism and conidial formation in Pestalotiopsis fici. Sci. China Life Sci. 2017, 60, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ma, K.; Lyu, H.; Huang, Y.; Liu, H.; Liu, L.; Che, Y.; Liu, X.; Zou, H.; Yin, W.-B. Genetic manipulation of the Cop9 signalosome subunit pfcsne leads to the discovery of pestaloficins in Pestalotiopsis fici. Org. Lett. 2017, 19, 4700–4703. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, B.; Timári, I.; Somogyi, Á.; Li, D.-W.; Adcox, H.E.; Gunn, J.S.; Bruschweiler-Li, L.; Brüschweiler, R. Accurate and efficient determination of unknown metabolites in metabolomics by NMR-based molecular motif identification. Anal. Chem. 2019, 91, 15686–15693. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, D.; Sun, W.; Yang, B.; Li, F.; Liu, M.; Wang, J.; Xue, Y.; Hu, Z.; Zhang, Y. Hplc-dad-directed isolation of linearly fused prenylated indole alkaloids from a soil-derived Aspergillus versicolor. J. Nat. Prod. 2019, 82, 2181–2188. [Google Scholar] [CrossRef] [PubMed]
- Su, J.-C.; Cheng, W.; Song, J.-G.; Zhong, Y.-L.; Huang, X.-J.; Jiang, R.-W.; Li, Y.-L.; Li, M.-M.; Ye, W.-C.; Wang, Y. Macrocyclic diterpenoids from Euphorbia helioscopia and their potential anti-inflammatory activity. J. Nat. Prod. 2019, 82, 2818–2827. [Google Scholar] [CrossRef]
- Johansen, K.T.; Wubshet, S.G.; Nyberg, N.T.; Jaroszewski, J.W. From retrospective assessment to prospective decisions in natural product isolation: HPLC-SPE-NMR analysis of Carthamus oxyacantha. J. Nat. Prod. 2011, 74, 2454–2461. [Google Scholar] [CrossRef]
- Casasflores, S.; Rosalessaavedra, T.; Herreraestrella, A. Three decades of fungal transformation: Novel technologies. Methods Mol. Biol. 2004, 267, 315–325. [Google Scholar]
- Diedrich, C.; Grimme, S. Systematic investigation of modern quantum chemical methods to predict electronic circular dichroism spectra. J. Phys. Chem. A 2003, 107, 2524–2539. [Google Scholar] [CrossRef]
- Crawford, T.D.; Tam, M.C.; Abrams, M.L. The current state of ab initio calculations of optical rotation and electronic circular dichroism spectra. J. Phys. Chem. A 2008, 111, 12057–12068. [Google Scholar] [CrossRef]
- Ding, Y.; Li, X.-C.; Ferreira, D. Theoretical calculation of electronic circular dichroism of the rotationally restricted 3,8’’-biflavonoid morelloflavone. J. Org. Chem. 2007, 72, 9010–9017. [Google Scholar] [CrossRef] [PubMed]
- Berova, N.; Bari, L.D.; Pescitelli, G. Application of electronic circular dichroism in configurational and conformational analysis of organic compounds. Chem. Soc. Rev. 2007, 36, 914–931. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, G.; Bruhn, T.; Maksimenka, K.; Hemberger, Y. The assignment of absolute stereostructures through quantum chemical circular dichroism calculations. Eur. J. Org. Chem. 2009, 2009, 2717–2727. [Google Scholar] [CrossRef]
- Stephens, P.; Devlin, F.; Gasparrini, F.; Ciogli, A.; Spinelli, D.; Cosimelli, B. Determination of the absolute configuration of a chiral oxadiazol-3-one calcium channel blocker, resolved using chiral chromatography, via concerted density functional theory calculations of its vibrational circular dichroism, electronic circular dichroism. J. Org. Chem. 2007, 72, 4707–4715. [Google Scholar] [CrossRef] [PubMed]
- Qian-Cutrone, J.; Gao, Q.; Huang, S.; Klohr, S.E.; Veitch, J.A.; Shu, Y.-Z. Arthrinone, a novel fungal metabolite from Arthrinium sp. Fa 1744. J. Nat. Prod. 1994, 57, 1656–1660. [Google Scholar] [CrossRef]
- Wu, H.M.; Lin, L.P.; Xu, Q.L.; Han, W.B.; Zhang, S.; Liu, Z.W.; Mei, Y.N.; Yao, Z.J.; Tan, R.X. Nodupetide, a potent insecticide and antimicrobial from Nodulisporium sp. Associated with riptortus pedestris. Tetrahedron Lett. 2017, 58. [Google Scholar] [CrossRef]
- Shen, Y.; Xu, Q.L.; Cheng, P.; Liu, C.L.; Lu, Z.Y.; Li, W.; Wang, T.T.; Lu, Y.H.; Tan, R.X.; Ge, H.M. Aromatic polyketides from a caterpillar associated Alternaria sp. Tetrahedron Lett. 2017, 58, 3069–3072. [Google Scholar] [CrossRef]
- Tian, J.; Lai, D.; Zhou, L. Secondary metabolites from acremonium fungi: Diverse structures and bioactivities. Mini Rev. Med. Chem. 2016, 17, 603–632. [Google Scholar] [CrossRef]
- Pusztahelyi, T.; Holb, I.J.; Pócsi, I. Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 2015, 6, 573. [Google Scholar] [CrossRef] [Green Version]
- Daniel, C.; Gardner, D.R.; Pfister, J.A. Swainsonine-containing plants and their relationship to endophytic fungi. J. Agric. Food Chem. 2014, 62, 7326–7334. [Google Scholar]
- Mousa, W.K.; Raizada, M.N. The diversity of anti-microbial secondary metabolites produced by fungal endophytes: An interdisciplinary perspective. Front. Microbiol. 2013, 4, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Shan, T.; Mou, Y.; Zhou, L. Plant-derived bioactive compounds produced by endophytic fungi. Mini Rev. Med. Chem. 2011, 11, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, D.A. Antibiotics: Opportunities for genetic manipulation. Philos. Trans. R. Soc. Lond. 1989, 324, 549–562. [Google Scholar]
- Michielse, C.B.; Ca, H.P.D.H.; Ram, A.F. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 2005, 48, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Piyan, Z.; Bin, X.; Yuezhu, W.; Yunqian, L.; Zhen, Q.; Shengrong, T.; Shen, H.; Shuangxi, R. Agrobacterium tumefaciens-mediated transformation as a tool for insertional mutagenesis in the fungus Penicillium marneffei. Mycol. Res. 2008, 112, 943–949. [Google Scholar]
- Combier, J.P.; Melayah, D.; Raffier, C.; Gay, G.; Marmeisse, R. Agrobacterium tumefaciens-mediated transformation as a tool for insertional mutagenesis in the symbiotic ectomycorrhizal fungus Hebeloma cylindrosporum. Fems Microbiol. Lett. 2010, 220, 141–148. [Google Scholar] [CrossRef] [Green Version]
- Michielse, C.B.; Ram, A.F.J.; Hooykaas, P.J.J.; Van den Hondel, C.A.M.J.J. Role of bacterial virulence proteins in Agrobacterium-mediated transformation of Aspergillus awamori. Fungal Genet. Biol. 2004, 41, 571–578. [Google Scholar] [CrossRef]
- Meyer, V.; Mueller, D.; Strowig, T.; Stahl, U. Comparison of different transformation methods for Aspergillus giganteus. Curr. Genet. 2003, 43, 371–377. [Google Scholar] [CrossRef]
- Shwab, E.K.; Bok, J.W.; Tribus, M.; Galehr, J.; Graessle, S.; Keller, N.P. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 2007, 6, 1656–1664. [Google Scholar] [CrossRef] [Green Version]
- Wessjohann, L.A.; Brandt, W.; Thieman, T. Biosynthesis and metabolism of cyclopropane rings in natural compounds. Chemcial Rev. 2003, 103, 1625–1648. [Google Scholar] [CrossRef]
- Sambrook, J.; Russel, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory: New York, NY, USA, 2001. [Google Scholar]
- Khang, C.H.; Park, S.-Y.; Rho, H.-S.; Lee, Y.-H.; Kang, S. Filamentous fungi (Magnaporthe grisea and Fusarium oxysporum). Methods Mol. Biol. 2006, 344, 403–420. [Google Scholar] [PubMed]
- Guo, Z.; Hao, T.; Wang, Y.; Pan, Y.; Ren, F.; Liu, X.; Che, Y.; Liu, G. Verz, a Zn(II)2Cys6 DNA-binding protein, regulates the biosynthesis of verticillin in Clonostachys rogersoniana. Microbiology 2017, 163, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pan, Y.; Liu, G. Disruption of the nitrogen regulatory gene Acarea in Acremonium chrysogenum leads to reduction of cephalosporin production and repression of nitrogen metabolism. Fungal Genet. Biol. 2013, 61, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Yanling, W.; Pengjie, H.; Erwei, L.; Xingzhong, L.; Yongsheng, C.; Gang, L. Genetic transformation of the fungus Gliocladium sp. mediated by Agrobacterium tumefaciens. Acta Microbiol. Sin. 2013, 53, 1233–1239. [Google Scholar]
- SchrödingerLlc. Macromodel, Version 9.7; SchrödingerLlc: New York, NY, USA, 2009. [Google Scholar]
- Frisch, M.J.; Trucks, G.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09 Revision A.1; Gaussian inc: Wallingford, CT, USA, 2009. [Google Scholar]
- Meerloo, J.V.; Kaspers, G.J.L.; Cloos, J. Cell sensitivity assays: The MTT assay. Methods Mol. Biol. 2011, 731, 237–245. [Google Scholar]
- Lyu, H.; Liu, H.; Keller, N.P.; Yin, W. Harnessing diverse transcriptional regulators for natural product discovery in fungi. Nat. Prod. Rep. 2020, 37, 6–16. [Google Scholar] [CrossRef]
- Knestrick, M.A.; Tawfik, R.; Shaw, L.N.; Baker, B.J. Chromatographic editing enhances natural product discovery. J. Pharm. Biomed. Anal. 2019, 176, 112831–112837. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 1–4 are available from the authors. |


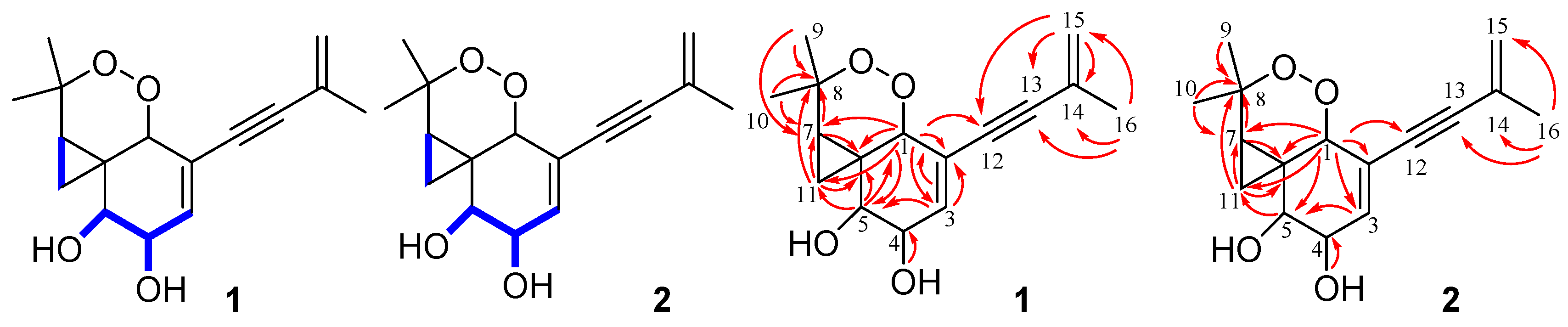
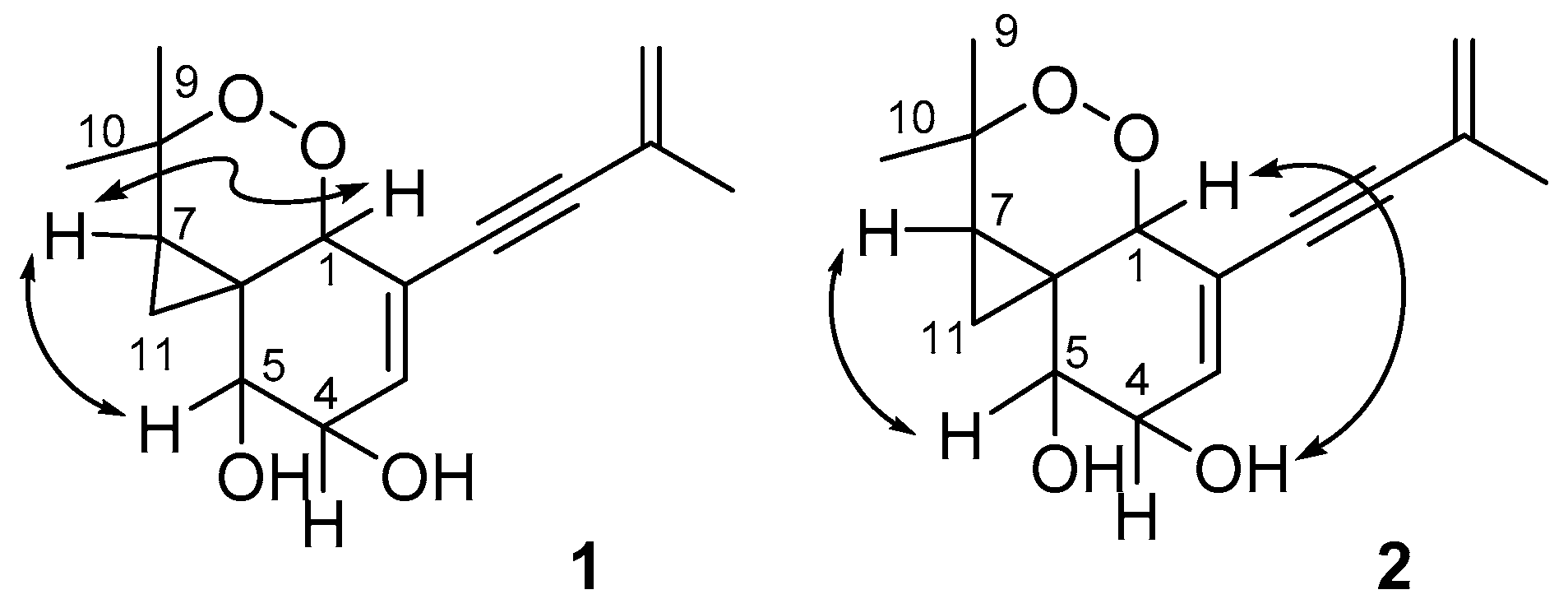
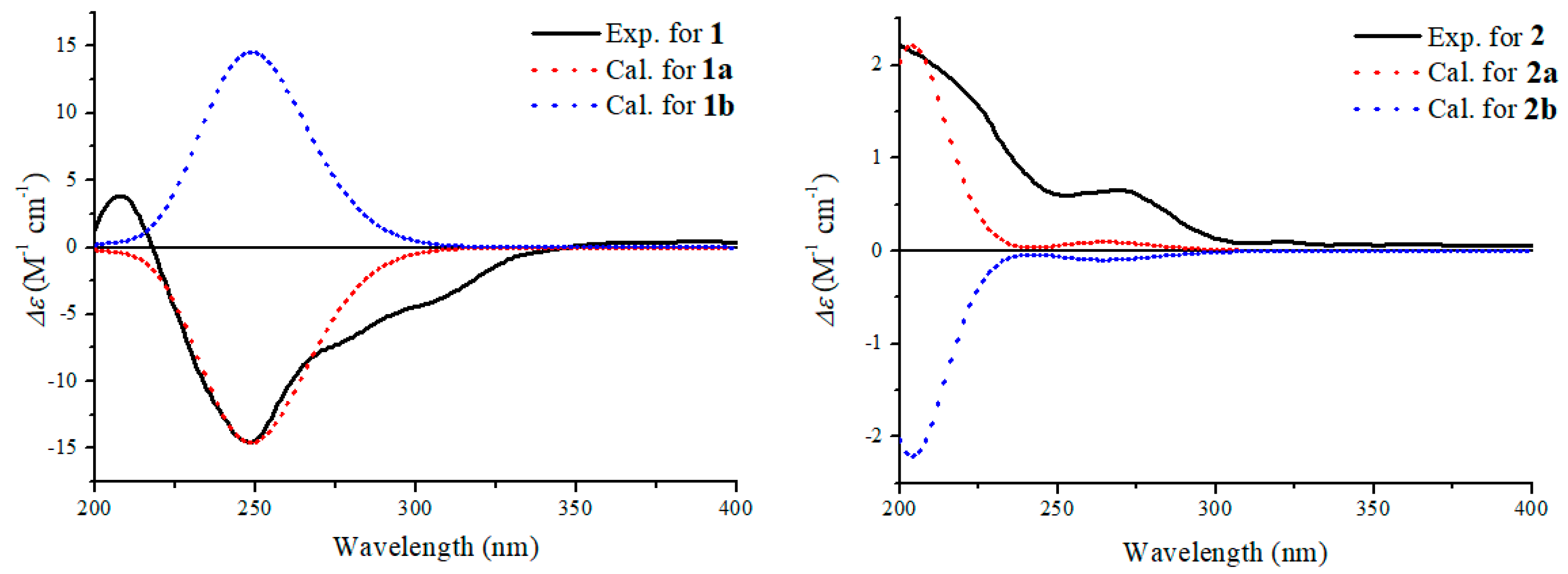
| Pos. | 1 | 2 | ||||
|---|---|---|---|---|---|---|
| δCa,mult. | δHb(J in Hz) | HMBC | δCa,mult. | δHb(J in Hz) | HMBC | |
| 1 | 76.8, CH | 4.41, dd (3.0, 2.4) | 2, 3, 4, 5, 6, 7, 11, 12 | 77.3, CH | 4.19, dd (2.4,1.8) | 2, 3, 4, 5, 7, 11, 12 |
| 2 | 124.1, qC | 128.8, qC | ||||
| 3 | 139.5, CH | 5.85, dd (3.0, 2.4) | 1, 2, 5, 6 | 135.0, CH | 6.11, ddd (5.4, 2.4, 1.2) | 5 |
| 4 | 77.0, CH | 4.15, m | 69.1, CH | 4.12, m | ||
| 5 | 72.7, CH | 4.10, dd (7.2, 3.0) | 1, 6, 11 | 75.0, CH | 3.48, ddd (5.4, 3.6, 1.8) | |
| 6 | 36.5, qC | 32.4, qC | ||||
| 7 | 29.1, CH | 1.64, dd (7.8, 3.0) | 1, 5, 6, 8, 9, 10, 11 | 33.2, CH | 1.64, dd (7.8, 4.2) | 1, 5, 6, 8, 9 |
| 8 | 82.6, qC | 82.3, qC | ||||
| 9 | 24.8, CH3 | 1.23, s | 7, 8, 10 | 28.4, CH3 | 1.33, s | |
| 10 | 24.7, CH3 | 1.17, s | 7, 8, 9 | 26.2, CH3 | 1.12, s | |
| 11 | 3.9, CH2 | 0.60, dd (7.8, 3.6) | 1, 5, 6, 7, 8 | 12.0, CH2 | 0.64, dd (7.8, 4.8) | 1, 5, 6, 7, 8 |
| 0.28, dd (3.6, 3.0) | 1, 5, 6, 7, 8, 9, 10 | 0.58, dd (4.8, 4.2) | 1, 5, 6, 7, 8 | |||
| 12 | 86.7, qC | 89.0, qC | ||||
| 13 | 92.7, qC | 91.4, qC | ||||
| 14 | 128.0, qC | 128.0, qC | ||||
| 15 | 122.0, CH2 | 5.27, m; 5.24, m | 13, 14 | 121.8, CH2 | 5.26, m; 5.24, m | |
| 16 | 23.6, CH3 | 1.87, br. s | 13, 14, 15 | 23.6, CH3 | 1.88, br. s | 13, 14, 15 |
| 4-OH | 4.26, d (6.0) | 4.41, d (4.2) | 3, 4, 5, | |||
| 5-OH | 4.32, d (3.0) | 4.42, d (3.6) | 4, 5, 6 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Z.; Zou, Z.-M. Discovery of New Secondary Metabolites by Epigenetic Regulation and NMR Comparison from the Plant Endophytic Fungus Monosporascus eutypoides. Molecules 2020, 25, 4192. https://doi.org/10.3390/molecules25184192
Guo Z, Zou Z-M. Discovery of New Secondary Metabolites by Epigenetic Regulation and NMR Comparison from the Plant Endophytic Fungus Monosporascus eutypoides. Molecules. 2020; 25(18):4192. https://doi.org/10.3390/molecules25184192
Chicago/Turabian StyleGuo, Zhe, and Zhong-Mei Zou. 2020. "Discovery of New Secondary Metabolites by Epigenetic Regulation and NMR Comparison from the Plant Endophytic Fungus Monosporascus eutypoides" Molecules 25, no. 18: 4192. https://doi.org/10.3390/molecules25184192






