Heparan Sulfate Proteoglycans Biosynthesis and Post Synthesis Mechanisms Combine Few Enzymes and Few Core Proteins to Generate Extensive Structural and Functional Diversity
Abstract
1. Introduction
2. Heparan Sulfate Proteoglycans
2.1. Pericellular and Extracellular Matrix HSPGs
2.2. Cell-Surface HSPGs
2.3. A Unique Intracellular PG: Serglycin
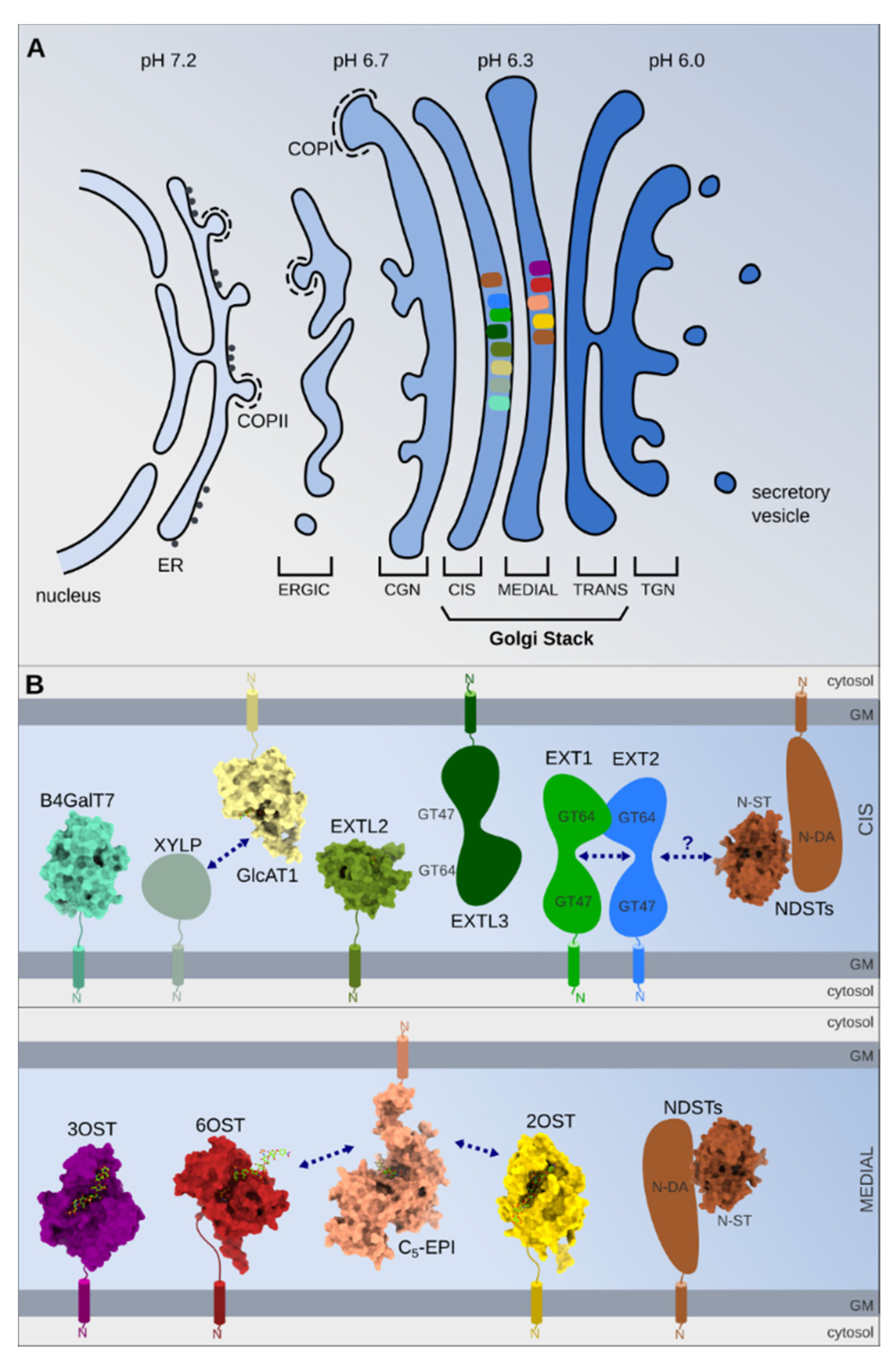
3. The Gagosylation Process Initiation and the Golgi Apparatus
4. Heparan Sulfate Elongation
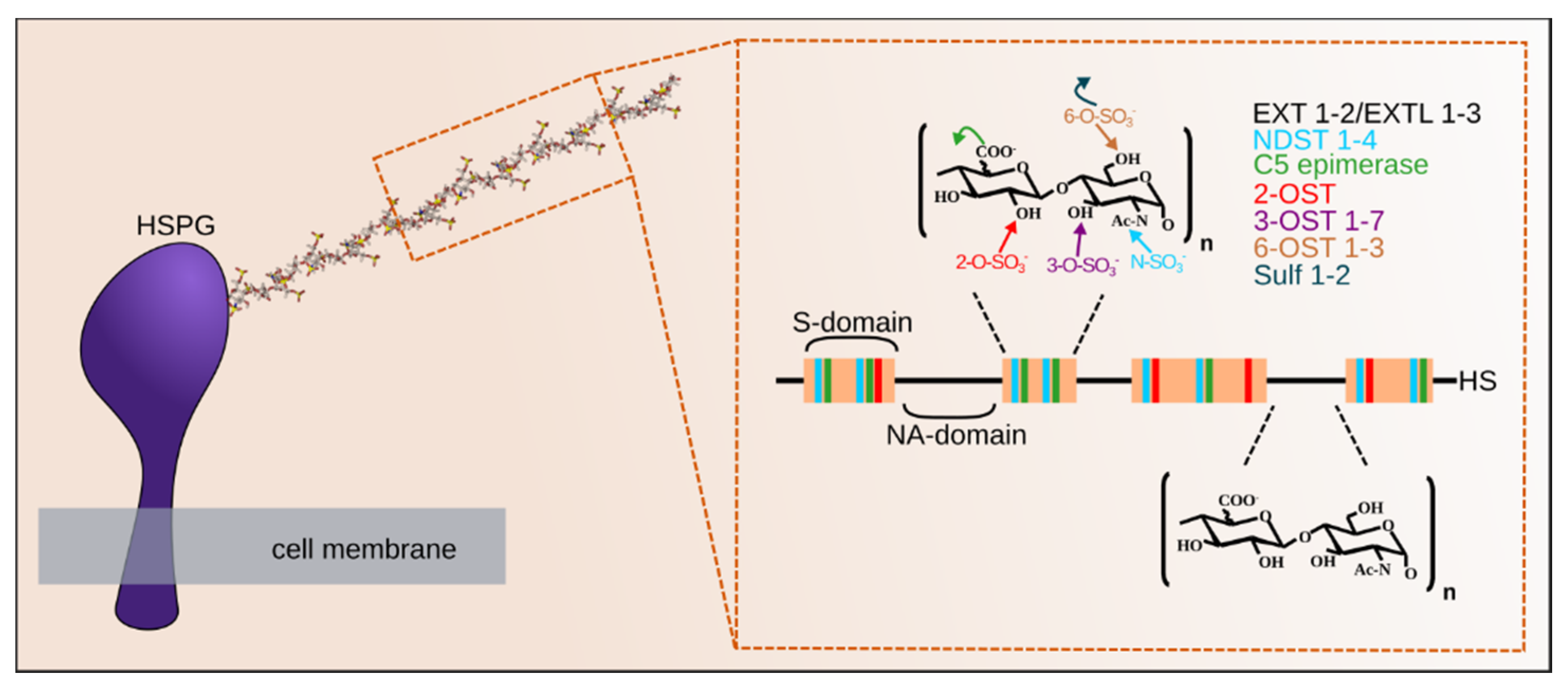
5. Heparan Sulfate Maturation
5.1. GlcNAc De-N-Acetylation/Re-N-Sulfation
5.2. GlcA C5 Epimerization
5.3. GlcA/IdoA and GlcN O-Sulfation
6. Post Synthetic Mechanism Regulating HS Structure
6.1. Heparanase
6.2. Sheddase
6.3. The Sulfs
7. Conclusion and Future Directions
Funding
Acknowledgments
Conflicts of Interest
References
- Varki, A.; Gagneux, P. Biological functions of glycans. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Iozzo, R.V.; Schaefer, L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015, 42, 11–55. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Olsen, S.K.; Goetz, R. A protein canyon in the FGF-FGF receptor dimer selects from an à la carte menu of heparan sulfate motifs. Curr. Opin. Struct. Biol. 2005, 15, 506–516. [Google Scholar] [CrossRef]
- Connell, B.J.; Sadir, R.; Baleux, F.; Laguri, C.; Kleman, J.-P.; Luo, L.; Arenzana-Seisdedos, F.; Lortat-Jacob, H. Heparan sulfate differentially controls CXCL12α-and CXCL12γ-mediated cell migration through differential presentation to their receptor CXCR4. Sci. Signal. 2016, 9, ra107. [Google Scholar] [CrossRef]
- Lortat-Jacob, H.; Grimaud, J.A. Interferon-gamma binds to heparan sulfate by a cluster of amino acids located in the C-terminal part of the molecule. FEBS Lett. 1991, 280, 152–154. [Google Scholar] [CrossRef]
- Monneau, Y.; Arenzana-Seisdedos, F.; Lortat-Jacob, H. The sweet spot: How GAGs help chemokines guide migrating cells. J. Leukoc. Biol. 2016, 99, 935–953. [Google Scholar] [CrossRef]
- Björk, I.; Lindahl, U. Mechanism of the anticoagulant action of heparin. Mol. Cell. Biochem. 1982, 48, 161–182. [Google Scholar] [CrossRef]
- Connell, B.J.; Lortat-Jacob, H. Human immunodeficiency virus and heparan sulfate: From attachment to entry inhibition. Front. Immunol. 2013, 4, 385. [Google Scholar] [CrossRef]
- Bishop, J.R.; Schuksz, M.; Esko, J.D. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 2007, 446, 1030–1037. [Google Scholar] [CrossRef]
- Ori, A.; Wilkinson, M.C.; Fernig, D.G. The heparanome and regulation of cell function: Structures, functions and challenges. Front. Biosci. 2008, 13, 4309–4338. [Google Scholar] [CrossRef]
- Busse-Wicher, M.; Wicher, K.B.; Kusche-Gullberg, M. The exostosin family: Proteins with many functions. Matrix Biol. 2014, 35, 25–33. [Google Scholar] [CrossRef]
- Kreuger, J.; Kjellen, L. Heparan sulfate biosynthesis: Regulation and variability. J. Histochem. Cytochem. 2012, 60, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-P.; Kusche-Gullberg, M. Heparan sulfate: Biosynthesis, structure, and function. Int. Rev. Cell Mol. Biol. 2016, 325, 215–273. [Google Scholar] [CrossRef] [PubMed]
- Prydz, K. Determinants of Glycosaminoglycan (GAG) Structure. Biomolecules 2015, 5, 2003–2022. [Google Scholar] [CrossRef]
- Vives, R.R.; Seffouh, A.; Lortat-Jacob, H. Post-Synthetic Regulation of HS Structure: The Yin and Yang of the Sulfs in Cancer. Front. Oncol. 2014, 3, 331. [Google Scholar] [CrossRef]
- Thompson, S.M.; Fernig, D.G.; Jesudason, E.C.; Losty, P.D.; van de Westerlo, E.M.A.; van Kuppevelt, T.H.; Turnbull, J.E. Heparan sulfate phage display antibodies identify distinct epitopes with complex binding characteristics: Insights into protein binding specificities. J. Biol. Chem. 2009, 284, 35621–35631. [Google Scholar] [CrossRef]
- Van den Born, J.; Salmivirta, K.; Henttinen, T.; Ostman, N.; Ishimaru, T.; Miyaura, S.; Yoshida, K.; Salmivirta, M. Novel heparan sulfate structures revealed by monoclonal antibodies. J. Biol. Chem. 2005, 280, 20516–20523. [Google Scholar] [CrossRef] [PubMed]
- Ledin, J.; Staatz, W.; Li, J.-P.; Götte, M.; Selleck, S.; Kjellén, L.; Spillmann, D. Heparan sulfate structure in mice with genetically modified heparan sulfate production. J. Biol. Chem. 2004, 279, 42732–42741. [Google Scholar] [CrossRef] [PubMed]
- Leymarie, N.; McComb, M.E.; Naimy, H.; Staples, G.O.; Zaia, J. Differential characterization and classification of tissue specific glycosaminoglycans by tandem mass spectrometry and statistical methods. Int. J. Mass Spectrom. 2012, 312, 144–154. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Feyzi, E.; Saldeen, T.; Larsson, E.; Lindahl, U.; Salmivirta, M. Age-dependent modulation of heparan sulfate structure and function. J. Biol. Chem. 1998, 273, 13395–13398. [Google Scholar] [CrossRef] [PubMed]
- Suhovskih, A.V.; Domanitskaya, N.V.; Tsidulko, A.Y.; Prudnikova, T.Y.; Kashuba, V.I.; Grigorieva, E.V. Tissue-specificity of heparan sulfate biosynthetic machinery in cancer. Cell Adhes. Migr. 2015, 9, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Melrose, J.; Smith, S.; Whitelock, J. Perlecan immunolocalizes to perichondrial vessels and canals in human fetal cartilaginous primordia in early vascular and matrix remodeling events associated with diarthrodial joint development. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2004, 52, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Farach-Carson, M.C.; Warren, C.R.; Harrington, D.A.; Carson, D.D. Border patrol: Insights into the unique role of perlecan/heparan sulfate proteoglycan 2 at cell and tissue borders. Matrix Biol. J. Int. Soc. Matrix Biol. 2014, 34, 64–79. [Google Scholar] [CrossRef] [PubMed]
- Mongiat, M.; Sweeney, S.M.; San Antonio, J.D.; Fu, J.; Iozzo, R.V. Endorepellin, a novel inhibitor of angiogenesis derived from the C terminus of perlecan. J. Biol. Chem. 2003, 278, 4238–4249. [Google Scholar] [CrossRef]
- Costell, M.; Gustafsson, E.; Aszódi, A.; Mörgelin, M.; Bloch, W.; Hunziker, E.; Addicks, K.; Timpl, R.; Fässler, R. Perlecan maintains the integrity of cartilage and some basement membranes. J. Cell Biol. 1999, 147, 1109–1122. [Google Scholar] [CrossRef]
- Arikawa-Hirasawa, E.; Wilcox, W.R.; Le, A.H.; Silverman, N.; Govindraj, P.; Hassell, J.R.; Yamada, Y. Dyssegmental dysplasia, Silverman-Handmaker type, is caused by functional null mutations of the perlecan gene. Nat. Genet. 2001, 27, 431–434. [Google Scholar] [CrossRef]
- Arikawa-Hirasawa, E.; Le, A.H.; Nishino, I.; Nonaka, I.; Ho, N.C.; Francomano, C.A.; Govindraj, P.; Hassell, J.R.; Devaney, J.M.; Spranger, J.; et al. Structural and functional mutations of the perlecan gene cause Schwartz-Jampel syndrome, with myotonic myopathy and chondrodysplasia. Am. J. Hum. Gen. 2002, 70, 1368–1375. [Google Scholar] [CrossRef]
- Denzer, A.J.; Schulthess, T.; Fauser, C.; Schumacher, B.; Kammerer, R.A.; Engel, J.; Ruegg, M.A. Electron microscopic structure of agrin and mapping of its binding site in laminin-1. EMBO J. 1998, 17, 335–343. [Google Scholar] [CrossRef]
- Jury, E.C.; Kabouridis, P.S. New role for Agrin in T cells and its potential importance in immune system regulation. Arthritis Res. Ther. 2010, 12, 205. [Google Scholar] [CrossRef]
- Burgess, R.W.; Skarnes, W.C.; Sanes, J. Agrin isoforms with distinct amino termini: Differential expression, localization, and function. J. Cell Biol. 2000, 151, 41–52. [Google Scholar] [CrossRef]
- Gautam, M.; Noakes, P.G.; Moscoso, L.; Rupp, F.; Scheller, R.H.; Merlie, J.P.; Sanes, J. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell 1996, 85, 525–535. [Google Scholar] [CrossRef]
- Khan, A.A.; Bose, C.; Yam, L.S.; Soloski, M.J.; Rupp, F. Physiological regulation of the immunological synapse by agrin. Science 2001, 292, 1681–1686. [Google Scholar] [CrossRef] [PubMed]
- Alfsen, A.; Yu, H.; Magérus-Chatinet, A.; Schmitt, A.; Bomsel, M. HIV-1-infected blood mononuclear cells form an integrin- and agrin-dependent viral synapse to induce efficient HIV-1 transcytosis across epithelial cell monolayer. Mol. Biol. Cell 2005, 16, 4267–4279. [Google Scholar] [CrossRef]
- Seppinen, L.; Sormunen, R.; Soini, Y.; Elamaa, H.; Heljasvaara, R.; Pihlajaniemi, T. Lack of collagen XVIII accelerates cutaneous wound healing, while overexpression of its endostatin domain leads to delayed healing. Matrix Biol. J. Int. Soc. Matrix Biol. 2008, 27, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Moulton, K.S.; Olsen, B.R.; Sonn, S.; Fukai, N.; Zurakowski, D.; Zeng, X. Loss of collagen XVIII enhances neovascularization and vascular permeability in atherosclerosis. Circulation 2004, 110, 1330–1336. [Google Scholar] [CrossRef] [PubMed]
- Zaferani, A.; Talsma, D.T.; Yazdani, S.; Celie, J.W.A.M.; Aikio, M.; Heljasvaara, R.; Navis, G.J.; Pihlajaniemi, T.; van den Born, J. Basement membrane zone collagens XV and XVIII/proteoglycans mediate leukocyte influx in renal ischemia/reperfusion. PLoS ONE 2014, 9, e106732. [Google Scholar] [CrossRef]
- Van Horssen, J.; Wilhelmus, M.M.M.; Heljasvaara, R.; Pihlajaniemi, T.; Wesseling, P.; de Waal, R.M.W.; Verbeek, M.M. Collagen XVIII: A novel heparan sulfate proteoglycan associated with vascular amyloid depositions and senile plaques in Alzheimer’s disease brains. Brain Pathol. 2002, 12, 456–462. [Google Scholar] [CrossRef]
- Suzuki, O.T.; Sertié, A.L.; Der Kaloustian, V.M.; Kok, F.; Carpenter, M.; Murray, J.; Czeizel, A.E.; Kliemann, S.E.; Rosemberg, S.; Monteiro, M.; et al. Molecular analysis of collagen XVIII reveals novel mutations, presence of a third isoform, and possible genetic heterogeneity in Knobloch syndrome. Am. J. Hum. Genet. 2002, 71, 1320–1329. [Google Scholar] [CrossRef]
- Bishop, J.R.; Passos-Bueno, M.R.; Fong, L.; Stanford, K.I.; Gonzales, J.C.; Yeh, E.; Young, S.G.; Bensadoun, A.; Witztum, J.L.; Esko, J.D.; et al. Deletion of the basement membrane heparan sulfate proteoglycan type XVIII collagen causes hypertriglyceridemia in mice and humans. PLoS ONE 2010, 5, e13919. [Google Scholar] [CrossRef]
- Bonnet, F.; Périn, J.P.; Charbonnier, F.; Camuzat, A.; Roussel, G.; Nussbaum, J.L.; Alliel, P.M. Structure and cellular distribution of mouse brain testican. Association with the postsynaptic area of hippocampus pyramidal cells. J. Biol. Chem. 1996, 271, 4373–4380. [Google Scholar] [CrossRef]
- Vannahme, C.; Schübel, S.; Herud, M.; Gösling, S.; Hülsmann, H.; Paulsson, M.; Hartmann, U.; Maurer, P. Molecular cloning of testican-2: Defining a novel calcium-binding proteoglycan family expressed in brain. J. Neurochem. 1999, 73, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, U.; Hülsmann, H.; Seul, J.; Röll, S.; Midani, H.; Breloy, I.; Hechler, D.; Müller, R.; Paulsson, M. Testican-3: A brain-specific proteoglycan member of the BM-40/SPARC/osteonectin family. J. Neurochem. 2013, 125, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Dhamija, R.; Graham, J.M.; Smaoui, N.; Thorland, E.; Kirmani, S. Novel de novo SPOCK1 mutation in a proband with developmental delay, microcephaly and agenesis of corpus callosum. Eur. J. Med. Genet. 2014, 57, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, A.; Uchiyama, K.; Nara, T.; Nishimura, N.; Hayasaka, M.; Hanaoka, K.; Yamamoto, T. Structural abnormalities of corpus callosum and cortical axonal tracts accompanied by decreased anxiety-like behavior and lowered sociability in spock3-mutant mice. Dev. Neurosci. 2014, 36, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Bernfield, M.; Kokenyesi, R.; Kato, M.; Hinkes, M.T.; Spring, J.; Gallo, R.L.; Lose, E.J. Biology of the syndecans: A family of transmembrane heparan sulfate proteoglycans. Annu. Rev. Cell Biol. 1992, 8, 365–393. [Google Scholar] [CrossRef] [PubMed]
- Manon-Jensen, T.; Itoh, Y.; Couchman, J.R. Proteoglycans in health and disease: The multiple roles of syndecan shedding. FEBS J. 2010, 277, 3876–3889. [Google Scholar] [CrossRef]
- Dews, I.C.; Mackenzie, K.R. Transmembrane domains of the syndecan family of growth factor coreceptors display a hierarchy of homotypic and heterotypic interactions. Proc. Natl. Acad. Sci. USA 2007, 104, 20782–20787. [Google Scholar] [CrossRef]
- Longley, R.L.; Woods, A.; Fleetwood, A.; Cowling, G.J.; Gallagher, J.T.; Couchman, J.R. Control of morphology, cytoskeleton and migration by syndecan-4. J. Cell Sci. 1999, 112, 3421–3431. [Google Scholar]
- Woods, A.; Couchman, J.R. Syndecan 4 heparan sulfate proteoglycan is a selectively enriched and widespread focal adhesion component. Mol. Biol. Cell 1994, 5, 183–192. [Google Scholar] [CrossRef]
- Kinnunen, T.; Kaksonen, M.; Saarinen, J.; Kalkkinen, N.; Peng, H.B.; Rauvala, H. Cortactin-Src kinase signaling pathway is involved in N-syndecan-dependent neurite outgrowth. J. Biol. Chem. 1998, 273, 10702–10708. [Google Scholar] [CrossRef]
- Grootjans, J.J.; Zimmermann, P.; Reekmans, G.; Smets, A.; Degeest, G.; Durr, J.; David, G. Syntenin, a PDZ protein that binds syndecan cytoplasmic domains. Proc. Natl. Acad. Sci. USA 1997, 94, 13683–13688. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.R.; Woods, D.F.; Marfatia, S.M.; Walther, Z.; Chishti, A.H.; Anderson, J.M.; Wood, D.F. Human CASK/LIN-2 binds syndecan-2 and protein 4.1 and localizes to the basolateral membrane of epithelial cells. J. Cell Biol. 1998, 142, 129–138. [Google Scholar] [CrossRef]
- Kim, C.W.; Goldberger, O.A.; Gallo, R.L.; Bernfield, M. Members of the syndecan family of heparan sulfate proteoglycans are expressed in distinct cell-, tissue-, and development-specific patterns. Mol. Biol. Cell 1994, 5, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Salmivirta, M.; Jalkanen, M. Syndecan family of cell surface proteoglycans: Developmentally regulated receptors for extracellular effector molecules. Experientia 1995, 51, 863–872. [Google Scholar] [CrossRef] [PubMed]
- David, G.; Bai, X.M.; Van der Schueren, B.; Marynen, P.; Cassiman, J.J.; Van den Berghe, H. Spatial and temporal changes in the expression of fibroglycan (syndecan-2) during mouse embryonic development. Development 1993, 119, 841–854. [Google Scholar]
- Pierce, A.; Lyon, M.; Hampson, I.N.; Cowling, G.J.; Gallagher, J.T. Molecular cloning of the major cell surface heparan sulfate proteoglycan from rat liver. J. Biol. Chem. 1992, 267, 3894–3900. [Google Scholar]
- Carey, D.J.; Evans, D.M.; Stahl, R.C.; Asundi, V.K.; Conner, K.J.; Garbes, P.; Cizmeci Smith, G. Molecular cloning and characterization of N-syndecan, a novel transmembrane heparan sulfate proteoglycan. J. Cell Biol. 1992, 117, 191–201. [Google Scholar] [CrossRef]
- Gould, S.E.; Upholt, W.B.; Kosher, R.A. Syndecan 3: A member of the syndecan family of membrane-intercalated proteoglycans that is expressed in high amounts at the onset of chicken limb cartilage differentiation. Proc. Natl. Acad. Sci. USA 1992, 89, 3271–3275. [Google Scholar] [CrossRef]
- David, G.; van der Schueren, B.; Marynen, P.; Cassiman, J.J.; van den Berghe, H. Molecular cloning of amphiglycan, a novel integral membrane heparan sulfate proteoglycan expressed by epithelial and fibroblastic cells. J. Cell Biol. 1992, 118, 961–969. [Google Scholar] [CrossRef]
- David, G.; Lories, V.; Decock, B.; Marynen, P.; Cassiman, J.J.; Van den Berghe, H. Molecular cloning of a phosphatidylinositol-anchored membrane heparan sulfate proteoglycan from human lung fibroblasts. J. Cell. Biol. 1990, 111, 3165–3176. [Google Scholar] [CrossRef] [PubMed]
- Stipp, C.S.; Litwack, E.D.; Lander, A.D. Cerebroglycan: An integral membrane heparan sulfate proteoglycan that is unique to the developing nervous system and expressed specifically during neuronal differentiation. J. Cell Biol. 1994, 124, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Filmus, J.; Shi, W.; Wong, Z.M.; Wong, M.J. Identification of a new membrane-bound heparan sulphate proteoglycan. Biochem. J. 1995, 311, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Yamada, H.; Yamaguchi, Y. K-glypican: A novel GPI-anchored heparan sulfate proteoglycan that is highly expressed in developing brain and kidney. J. Cell Biol. 1995, 130, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Veugelers, M.; Vermeesch, J.; Reekmans, G.; Steinfeld, R.; Marynen, P.; David, G. Characterization of glypican-5 and chromosomal localization of human GPC5, a new member of the glypican gene family. Genomics 1997, 40, 24–30. [Google Scholar] [CrossRef]
- Veugelers, M.; De Cat, B.; Ceulemans, H.; Bruystens, A.M.; Coomans, C.; Dürr, J.; Vermeesch, J.; Marynen, P.; David, G. Glypican-6, a new member of the glypican family of cell surface heparan sulfate proteoglycans. J. Biol. Chem. 1999, 274, 26968–26977. [Google Scholar] [CrossRef]
- Paine Saunders, S.; Viviano, B.L.; Saunders, S. GPC6, a novel member of the glypican gene family, encodes a product structurally related to GPC4 and is colocalized with GPC5 on human chromosome 13. Genomics 1999, 57, 455–458. [Google Scholar] [CrossRef]
- Saunders, S.; Paine-Saunders, S.; Lander, A.D. Expression of the cell surface proteoglycan glypican-5 is developmentally regulated in kidney, limb, and brain. Dev. Biol. 1997, 190, 78–93. [Google Scholar] [CrossRef]
- Filmus, J.; Capurro, M.; Rast, J. Glypicans. Genome Biol. 2008, 9, 224. [Google Scholar] [CrossRef]
- Fico, A.; Maina, F.; Dono, R. Fine-tuning of cell signalling by glypicans. Cell Mol. Life Sci. 2007, 68, 23–29. [Google Scholar] [CrossRef]
- Yan, D.; Lin, X. Opposing roles for glypicans in Hedgehog signalling. Nat. Cell Biol. 2008, 10, 761–763. [Google Scholar] [CrossRef]
- Gutierrez, J.; Brandan, E. A novel mechanism of sequestering fibroblast growth factor 2 by glypican in lipid rafts, allowing skeletal muscle differentiation. Mol. Cell Biol. 2010, 30, 1634–1649. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Toomre, D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000, 1, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.R.; Whitehouse, I.J.; Hooper, N.M. Glypican-1 mediates both prion protein lipid raft association and disease isoform formation. PLoS Pathog. 2009, 5, e1000666. [Google Scholar] [CrossRef] [PubMed]
- Kreuger, J.; Perez, L.; Giraldez, A.J.; Cohen, S.M. Opposing Activities of Dally-like Glypican at High and Low Levels of Wingless Morphogen Activity. Dev. Cell 2004, 7, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Traister, A.; Shi, W.; Filmus, J. Mammalian Notum induces the release of glypicans and other GPI-anchored proteins from the cell surface. BioChem. J. 2008, 410, 503–511. [Google Scholar] [CrossRef]
- Fransson, L.-A.; Belting, M.; Cheng, F.; Jönsson, M.; Mani, K.; Sandgren, S. Novel aspects of glypican glycobiology. Cell. Mol. Life Sci. 2004, 61, 1016–1024. [Google Scholar] [CrossRef]
- Filmus, J. Glypicans in growth control and cancer. Glycobiology 2001, 11, 19R–23R. [Google Scholar] [CrossRef]
- Kaur, S.P.; Cummings, B.S. Role of glypicans in regulation of the tumor microenvironment and cancer progression. Biochem. Pharmacol. 2019, 168, 108–118. [Google Scholar] [CrossRef]
- Li, N.; Gao, W.; Zhang, Y.-F.; Ho, M. Glypicans as cancer therapeutic targets. Trends Cancer 2018, 4, 741–754. [Google Scholar] [CrossRef]
- Cano Gauci, D.F.; Song, H.H.; Yang, H.; McKerlie, C.; Choo, B.; Shi, W.; Pullano, R.; Piscione, T.D.; Grisaru, S.; Soon, S.; et al. Glypican-3-deficient mice exhibit developmental overgrowth and some of the abnormalities typical of Simpson-Golabi-Behmel syndrome. J. Cell Biol. 1999, 146, 255–264. [Google Scholar] [CrossRef]
- DeBaun, M.R.; Ess, J.; Saunders, S. Simpson Golabi Behmel syndrome: Progress toward understanding the molecular basis for overgrowth, malformation, and cancer predisposition. Mol. Genet. Metab. 2001, 72, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Veugelers, M.; Vermeesch, J.; Watanabe, K.; Yamaguchi, Y.; Marynen, P.; David, G. GPC4, the gene for human K-glypican, flanks GPC3 on xq26: Deletion of the GPC3-GPC4 gene cluster in one family with Simpson-Golabi-Behmel syndrome. Genomics 1998, 53, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bilandzic, M.; Stenvers, K.L. Betaglycan: A multifunctional accessory. Mol. Cell. Endocrinol. 2011, 339, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Massague, J. Receptors for the TGF-beta family. Cell 1992, 69, 1067–1070. [Google Scholar] [CrossRef]
- Andres, J.L.; DeFalcis, D.; Noda, M.; Massague, J. Binding of two growth factor families to separate domains of the proteoglycan betaglycan. J. Biol. Chem. 1992, 267, 5927–5930. [Google Scholar]
- Boivin, W.A.; Shackleford, M.; Vanden Hoek, A.; Zhao, H.; Hackett, T.L.; Knight, D.A.; Granville, D.J. Granzyme B cleaves decorin, biglycan and soluble betaglycan, releasing active transforming growth factor-β1. PLoS ONE 2012, 7, e33163. [Google Scholar] [CrossRef]
- Brown, C.B.; Boyer, A.S.; Runyan, R.B.; Barnett, J.V. Requirement of type III TGF-beta receptor for endocardial cell transformation in the heart. Science 1999, 283, 2080–2082. [Google Scholar] [CrossRef]
- Bernabeu, C.; Lopez-Novoa, J.M.; Quintanilla, M. The emerging role of TGF-beta superfamily coreceptors in cancer. Biochim. Biophys. Acta 2009, 1792, 954–973. [Google Scholar] [CrossRef]
- Haggerty, J.G.; Bretton, R.H.; Milstone, L.M. Identification and characterization of a cell surface proteoglycan on keratinocytes. J. Investig. Dermatol. 1992, 99, 374–380. [Google Scholar] [CrossRef]
- Jones, M.; Tussey, L.; Athanasou, N.; Jackson, D.G. Heparan sulfate proteoglycan isoforms of the CD44 hyaluronan receptor induced in human inflammatory macrophages can function as paracrine regulators of fibroblast growth factor action. J. Biol. Chem. 2000, 275, 7964–7974. [Google Scholar] [CrossRef]
- Tanaka, Y.; Adams, D.H.; Hubscher, S.; Hirano, H.; Siebenlist, U.; Shaw, S. T-cell adhesion induced by proteoglycan-immobilized cytokine MIP-lβ. Nature 1993, 361, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Milstone, L.M.; Hough Monroe, L.; Kugelman, L.C.; Bender, J.R.; Haggerty, J.G. Epican, a heparan/chondroitin sulfate proteoglycan form of CD44, mediates cell-cell adhesion. J. Cell Sci. 1994, 107, 3183–3190. [Google Scholar] [PubMed]
- Wielenga, V.J.M.; van der Voort, R.; Taher, T.E.I.; Smit, L.; Beuling, E.A.; van Krimpen, C.; Spaargaren, M.; Pals, S.T. Expression of c-Met and heparan-sulfate proteoglycan forms of CD44 in colorectal cancer. Am. J. Pathol. 2000, 157, 1563–1573. [Google Scholar] [CrossRef]
- Taylor, K.R.; Gallo, R.L. Glycosaminoglycans and their proteoglycans: Host-associated molecular patterns for initiation and modulation of inflammation. FASEB J. 2006, 20, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Shintani, Y.; Takashima, S.; Asano, Y.; Kato, H.; Liao, Y.; Yamazaki, S.; Tsukamoto, O.; Seguchi, O.; Yamamoto, H.; Fukushima, T.; et al. Glycosaminoglycan modification of neuropilin-1 modulates VEGFR2 signaling. EMBO J. 2006, 25, 3045–3055. [Google Scholar] [CrossRef]
- Sakaguchi, K.; Lorenzi, M.V.; Bottaro, D.P.; Miki, T. The acidic domain and first immunoglobulin-like loop of fibroblast growth factor receptor 2 modulate downstream signaling through glycosaminoglycan modification. Mol. Cell Biol. 1999, 19, 6754–6764. [Google Scholar] [CrossRef][Green Version]
- Takagi, Y.; Shrivastav, S.; Miki, T.; Sakaguchi, K. Molecular cloning and expression of the acidic fibroblast growth factor receptors in a rat parathyroid cell line (PT-r). Parathyroid cell-specific calcium-dependent change of ligand accessibility and covalent attachment of heparan sulfate glycosaminoglycan to the receptors. J. Biol. Chem. 1994, 269, 23743–23749. [Google Scholar]
- Tantravahi, R.V.; Stevens, R.L.; Austen, K.F.; Weis, J.H. A single gene in mast cells encodes the core peptides of heparin and chondroitin sulfate proteoglycans. PNAS 1986, 83, 9207–9210. [Google Scholar] [CrossRef]
- Kolset, S.O.; Tveit, H. Serglycin--structure and biology. Cell Mol. Life Sci. 2008, 65, 1073–1085. [Google Scholar] [CrossRef]
- Schick, B.P.; Gradowski, J.F.; San Antonio, J.D. Synthesis, secretion, and subcellular localization of serglycin proteoglycan in human endothelial cells. Blood 2001, 97, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Schick, B.P.; Ho, H.C.; Brodbeck, K.C.; Wrigley, C.W.; Klimas, J. Serglycin proteoglycan expression and synthesis in embryonic stem cells. Biochim. Biophys. Acta 2003, 1593, 259–267. [Google Scholar] [CrossRef]
- Kolset, S.O.; Pejler, G. Serglycin: A structural and functional chameleon with wide impact on immune cells. J. Immunol. 2011, 1950, 4927–4933. [Google Scholar] [CrossRef] [PubMed]
- Ringvall, M.; Rönnberg, E.; Wernersson, S.; Duelli, A.; Henningsson, F.; Abrink, M.; García-Faroldi, G.; Fajardo, I.; Pejler, G. Serotonin and histamine storage in mast cell secretory granules is dependent on serglycin proteoglycan. J. Allergy Clin. Immunol. 2008, 121, 1020–1026. [Google Scholar] [CrossRef] [PubMed]
- Braga, T.; Grujic, M.; Lukinius, A.; Hellman, L.; Åbrink, M.; Pejler, G. Serglycin proteoglycan is required for secretory granule integrity in mucosal mast cells. BioChem. J. 2007, 403, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Åbrink, M.; Grujic, M.; Pejler, G. Serglycin is essential for maturation of mast cell secretory granule. J. Biol. Chem. 2004, 279, 40897–40905. [Google Scholar] [CrossRef]
- Forsberg, E.; Pejler, G.; Ringvall, M.; Lunderius, C.; Tomasini-Johansson, B.; Kusche-Gullberg, M.; Eriksson, I.; Ledin, J.; Hellman, L.; Kjellen, L. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature 1999, 400, 773–776. [Google Scholar] [CrossRef]
- Sutton, V.R.; Brennan, A.J.; Ellis, S.; Danne, J.; Thia, K.; Jenkins, M.R.; Voskoboinik, I.; Pejler, G.; Johnstone, R.W.; Andrews, D.M.; et al. Serglycin determines secretory granule repertoire and regulates natural killer cell and cytotoxic T lymphocyte cytotoxicity. FEBS J. 2016, 283, 947–961. [Google Scholar] [CrossRef]
- Meen, A.J.; Øynebråten, I.; Reine, T.M.; Duelli, A.; Svennevig, K.; Pejler, G.; Jenssen, T.; Kolset, S.O. Serglycin is a major proteoglycan in polarized human endothelial cells and is implicated in the secretion of the chemokine GROα/CXCL1. J. Biol. Chem. 2011, 286, 2636–2647. [Google Scholar] [CrossRef]
- Woulfe, D.S.; Lilliendahl, J.K.; August, S.; Rauova, L.; Kowalska, M.A.; Abrink, M.; Pejler, G.; White, J.G.; Schick, B.P. Serglycin proteoglycan deletion induces defects in platelet aggregation and thrombus formation in mice. Blood 2008, 111, 3458–3467. [Google Scholar] [CrossRef]
- Scully, O.J.; Chua, P.-J.; Harve, K.S.; Bay, B.-H.; Yip, G.W. Serglycin in health and diseases. Anat. Rec. (Hoboken) 2012, 295, 1415–1420. [Google Scholar] [CrossRef]
- Korpetinou, A.; Skandalis, S.S.; Moustakas, A.; Happonen, K.E.; Tveit, H.; Prydz, K.; Labropoulou, V.T.; Giannopoulou, E.; Kalofonos, H.P.; Blom, A.M.; et al. Serglycin is implicated in the promotion of aggressive phenotype of breast cancer cells. PLoS ONE 2013, 8, e78157. [Google Scholar] [CrossRef] [PubMed]
- Dalton, A.J.; Felix, M.D. Studies on the Golgi substance of the epithelial cells of the epididymis and duodenum of the mouse. Am. J. Anat. 1953, 92, 277–305. [Google Scholar] [CrossRef]
- Rambourg, A.; Clermont, Y.; Hermo, L.; Segretain, D. Tridimensional architecture of the Golgi apparatus and its components in mucous cells of Brunner’s glands of the mouse. Am. J. Anat. 1987, 179, 95–107. [Google Scholar] [CrossRef]
- Klumperman, J. Architecture of the mammalian Golgi. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef]
- Boncompain, G.; Perez, F. The many routes of Golgi-dependent trafficking. Histochem. Cell Biol. 2013, 140, 251–260. [Google Scholar] [CrossRef]
- Arakel, E.C.; Schwappach, B. Formation of COPI-coated vesicles at a glance. J. Cell. Sci. 2018, 131. [Google Scholar] [CrossRef]
- Emr, S.; Glick, B.S.; Linstedt, A.D.; Lippincott-Schwartz, J.; Luini, A.; Malhotra, V.; Marsh, B.J.; Nakano, A.; Pfeffer, S.R.; Rabouille, C.; et al. Journeys through the Golgi--taking stock in a new era. J. Cell Biol. 2009, 187, 449–453. [Google Scholar] [CrossRef]
- Jensen, D.; Schekman, R. COPII-mediated vesicle formation at a glance. J. Cell. Sci. 2011, 124, 1–4. [Google Scholar] [CrossRef]
- Götting, C.; Kuhn, J.; Zahn, R.; Brinkmann, T.; Kleesiek, K. Molecular cloning and expression of human UDP-d-Xylose:proteoglycan core protein beta-d-xylosyltransferase and its first isoform XT-II. J. Mol. Biol. 2000, 304, 517–528. [Google Scholar] [CrossRef]
- Almeida, R.; Levery, S.B.; Mandel, U.; Kresse, H.; Schwientek, T.; Bennett, E.P.; Clausen, H. Cloning and expression of a proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I. A seventh member of the human beta4-galactosyltransferase gene family. J. Biol. Chem. 1999, 274, 26165–26171. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, H.; Taoka, M.; Tone, Y.; Sugahara, K. Human glycosaminoglycan glucuronyltransferase I gene and a related processed pseudogene: Genomic structure, chromosomal mapping and characterization. Biochem. J. 2001, 358, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Pinhal, M.A.; Smith, B.; Olson, S.; Aikawa, J.; Kimata, K.; Esko, J.D. Enzyme interactions in heparan sulfate biosynthesis: Uronosyl 5-epimerase and 2-O-sulfotransferase interact in vivo. Proc. Natl. Acad. Sci. USA 2001, 98, 12984–12989. [Google Scholar] [CrossRef] [PubMed]
- Schön, S.; Prante, C.; Bahr, C.; Kuhn, J.; Kleesiek, K.; Götting, C. Cloning and recombinant expression of active full-length xylosyltransferase I (XT-I) and characterization of subcellular localization of XT-I and XT-II. J. Biol. Chem. 2006, 281, 14224–14231. [Google Scholar] [CrossRef]
- Lind, T.; Tufaro, F.; McCormick, C.; Lindahl, U.; Lidholt, K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J. Biol. Chem. 1998, 273, 26265–26268. [Google Scholar] [CrossRef]
- Senay, C.; Lind, T.; Muguruma, K.; Tone, Y.; Kitagawa, H.; Sugahara, K.; Lidholt, K.; Lindahl, U.; Kusche-Gullberg, M. The EXT1/EXT2 tumor suppressors: Catalytic activities and role in heparan sulfate biosynthesis. EMBO Rep. 2000, 1, 282–286. [Google Scholar] [CrossRef]
- Merry, C.L.; Bullock, S.L.; Swan, D.C.; Backen, A.C.; Lyon, M.; Beddington, R.S.; Wilson, V.A.; Gallagher, J.T. The molecular phenotype of heparan sulfate in the Hs2st-/-mutant mouse. J. Biol. Chem. 2001, 276, 35429–35434. [Google Scholar] [CrossRef] [PubMed]
- Delos, M.; Foulquier, F.; Hellec, C.; Vicogne, D.; Fifre, A.; Carpentier, M.; Papy-Garcia, D.; Allain, F.; Denys, A. Heparan sulfate 3-O-sulfotransferase 2 (HS3ST2) displays an unexpected subcellular localization in the plasma membrane. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 1644–1655. [Google Scholar] [CrossRef]
- Park, P.W.; Foster, T.J.; Nishi, E.; Duncan, S.J.; Klagsbrun, M.; Chen, Y. Activation of syndecan-1 ectodomain shedding by Staphylococcus aureus alpha-toxin and beta-toxin. J. Biol. Chem. 2004, 279, 251–258. [Google Scholar] [CrossRef]
- Kitagawa, H.; Shimakawa, H.; Sugahara, K. The tumor suppressor EXT-like gene EXTL2 encodes an alpha1, 4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region. The key enzyme for the chain initiation of heparan sulfate. J. Biol. Chem. 1999, 274, 13933–13937. [Google Scholar] [CrossRef]
- Katta, K.; Imran, T.; Busse-Wicher, M.; Grønning, M.; Czajkowski, S.; Kusche-Gullberg, M. Reduced expression of EXTL2, a member of the exostosin (EXT) family of glycosyltransferases, in human embryonic kidney 293 cells results in longer heparan sulfate chains. J. Biol. Chem. 2015, 290, 13168–13177. [Google Scholar] [CrossRef]
- Wen, J.; Xiao, J.; Rahdar, M.; Choudhury, B.P.; Cui, J.; Taylor, G.S.; Esko, J.D.; Dixon, J.E. Xylose phosphorylation functions as a molecular switch to regulate proteoglycan biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 15723–15728. [Google Scholar] [CrossRef] [PubMed]
- Koike, T.; Izumikawa, T.; Tamura, J.-I.; Kitagawa, H. FAM20B is a kinase that phosphorylates xylose in the glycosaminoglycan-protein linkage region. Biochem. J. 2009, 421, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Nadanaka, S.; Zhou, S.; Kagiyama, S.; Shoji, N.; Sugahara, K.; Sugihara, K.; Asano, M.; Kitagawa, H. EXTL2, a member of the EXT family of tumor suppressors, controls glycosaminoglycan biosynthesis in a xylose kinase-dependent manner. J. Biol. Chem. 2013, 288, 9321–9333. [Google Scholar] [CrossRef] [PubMed]
- Tone, Y.; Pedersen, L.C.; Yamamoto, T.; Izumikawa, T.; Kitagawa, H.; Nishihara, J.; Tamura, J.-I.; Negishi, M.; Sugahara, K. 2-o-phosphorylation of xylose and 6-o-sulfation of galactose in the protein linkage region of glycosaminoglycans influence the glucuronyltransferase-I activity involved in the linkage region synthesis. J. Biol. Chem. 2008, 283, 16801–16807. [Google Scholar] [CrossRef] [PubMed]
- Koike, T.; Izumikawa, T.; Sato, B.; Kitagawa, H. Identification of phosphatase that dephosphorylates xylose in the glycosaminoglycan-protein linkage region of proteoglycans. J. Biol. Chem. 2014, 289, 6695–6708. [Google Scholar] [CrossRef]
- Gulberti, S.; Lattard, V.; Fondeur, M.; Jacquinet, J.-C.; Mulliert, G.; Netter, P.; Magdalou, J.; Ouzzine, M.; Fournel-Gigleux, S. Phosphorylation and sulfation of oligosaccharide substrates critically influence the activity of human beta1,4-galactosyltransferase 7 (GalT-I) and beta1,3-glucuronosyltransferase I (GlcAT-I) involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. J. Biol. Chem. 2005, 280, 1417–1425. [Google Scholar] [CrossRef]
- Kim, B.-T.; Kitagawa, H.; Tanaka, J.; Tamura, J.; Sugahara, K. In vitro heparan sulfate polymerization: Crucial roles of core protein moieties of primer substrates in addition to the EXT1-EXT2 interaction. J. Biol. Chem. 2003, 278, 41618–41623. [Google Scholar] [CrossRef]
- Busse, M.; Kusche-Gullberg, M. In vitro polymerization of heparan sulfate backbone by the EXT proteins. J. Biol. Chem. 2003, 278, 41333–41337. [Google Scholar] [CrossRef]
- McCormick, C.; Duncan, G.; Goutsos, K.T.; Tufaro, F. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc. Natl. Acad. Sci. USA 2000, 97, 668–673. [Google Scholar] [CrossRef]
- Busse, M.; Feta, A.; Presto, J.; Wilén, M.; Grønning, M.; Kjellén, L.; Kusche-Gullberg, M. Contribution of EXT1, EXT2, and EXTL3 to heparan sulfate chain elongation. J. Biol. Chem. 2007, 282, 32802–32810. [Google Scholar] [CrossRef]
- Kobayashi, S.; Morimoto, K.; Shimizu, T.; Takahashi, M.; Kurosawa, H.; Shirasawa, T. Association of EXT1 and EXT2, hereditary multiple exostoses gene products, in Golgi apparatus. BioChem. Biophys. Res. Commun. 2000, 268, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.-L.; Chang, C.-W.; Chang, Y.-Y.; Sung, H.-H.; Lin, M.-D.; Chang, S.-C.; Chen, C.-H.; Huang, C.-W.; Tung, K.-S.; Chou, T.-B. The Drosophila GOLPH3 homolog regulates the biosynthesis of heparan sulfate proteoglycans by modulating the retrograde trafficking of exostosins. Dev. Camb. Engl. 2013, 140, 2798–2807. [Google Scholar] [CrossRef]
- Ropero, S.; Setien, F.; Espada, J.; Fraga, M.F.; Herranz, M.; Asp, J.; Benassi, M.S.; Franchi, A.; Patiño, A.; Ward, L.S.; et al. Epigenetic loss of the familial tumor-suppressor gene exostosin-1 (EXT1) disrupts heparan sulfate synthesis in cancer cells. Hum. Mol. Genet. 2004, 13, 2753–2765. [Google Scholar] [CrossRef]
- Liu, N.-W.; Huang, X.; Liu, S.; Lu, Y. EXT1, Regulated by MiR-665, Promotes cell apoptosis via ERK1/2 signaling pathway in acute lymphoblastic leukemia. Med. Sci. Monit. 2019, 25, 6491–6503. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, P.; Presto, J.; Spillmann, D.; Lindahl, U.; Kjellén, L. Heparin/heparan sulfate biosynthesis: Processive formation of N-sulfated domains. J. Biol. Chem. 2008, 283, 20008–20014. [Google Scholar] [CrossRef]
- Pikas, D.S.; Eriksson, I.; Kjellén, L. Overexpression of different isoforms of glucosaminyl N-deacetylase/N-sulfotransferase results in distinct heparan sulfate N-sulfation patterns. Biochemistry 2000, 39, 4552–4558. [Google Scholar] [CrossRef]
- Maccarana, M.; Sakura, Y.; Tawada, A.; Yoshida, K.; Lindahl, U. Domain structure of heparan sulfates from bovine organs. J. Biol. Chem. 1996, 271, 17804–17810. [Google Scholar] [CrossRef]
- Wei, Z.; Deakin, J.A.; Blaum, B.S.; Uhrín, D.; Gallagher, J.T.; Lyon, M. Preparation of heparin/heparan sulfate oligosaccharides with internal N-unsubstituted glucosamine residues for functional studies. Glycoconj. J. 2011, 28, 525–535. [Google Scholar] [CrossRef]
- Sheng, J.; Liu, R.; Xu, Y.; Liu, J. The dominating role of N-deacetylase/N-sulfotransferase 1 in forming domain structures in heparan sulfate. J. Biol. Chem. 2011, 286, 19768–19776. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Orellana, A.; Gil, G.; Hirschberg, C.B. Molecular cloning and expression of rat liver N-heparan sulfate sulfotransferase. J. Biol. Chem. 1992, 267, 15744–15750. [Google Scholar] [PubMed]
- Orellana, A.; Hirschberg, C.B.; Wei, Z.; Swiedler, S.J.; Ishihara, M. Molecular cloning and expression of a glycosaminoglycan N-acetylglucosaminyl N-deacetylase/N-sulfotransferase from a heparin-producing cell line. J. Biol. Chem. 1994, 269, 2270–2276. [Google Scholar] [PubMed]
- Eriksson, I.; Sandbäck, D.; Ek, B.; Lindahl, U.; Kjellén, L. cDNA cloning and sequencing of mouse mastocytoma glucosaminyl N-deacetylase/N-sulfotransferase, an enzyme involved in the biosynthesis of heparin. J. Biol. Chem. 1994, 269, 10438–10443. [Google Scholar] [PubMed]
- Aikawa, J.; Esko, J.D. Molecular cloning and expression of a third member of the heparan sulfate/heparin GlcNAc N-deacetylase/ N-sulfotransferase family. J. Biol. Chem. 1999, 274, 2690–2695. [Google Scholar] [CrossRef] [PubMed]
- Aikawa, J.; Grobe, K.; Tsujimoto, M.; Esko, J.D. Multiple isozymes of heparan sulfate/heparin GlcNAc N-deacetylase/GlcN N-sulfotransferase. Structure and activity of the fourth member, NDST4. J. Biol. Chem. 2001, 276, 5876–5882. [Google Scholar] [CrossRef]
- Fan, G.; Xiao, L.; Cheng, L.; Wang, X.; Sun, B.; Hu, G. Targeted disruption of NDST-1 gene leads to pulmonary hypoplasia and neonatal respiratory distress in mice. FEBS Lett. 2000, 467, 7–11. [Google Scholar] [CrossRef]
- Ringvall, M.; Ledin, J.; Holmborn, K.; van Kuppevelt, T.; Ellin, F.; Eriksson, I.; Olofsson, A.M.; Kjellen, L.; Forsberg, E. Defective heparan sulfate biosynthesis and neonatal lethality in mice lacking N-deacetylase/N-sulfotransferase-1. J. Biol. Chem. 2000, 275, 25926–25930. [Google Scholar] [CrossRef]
- Deligny, A.; Dierker, T.; Dagälv, A.; Lundequist, A.; Eriksson, I.; Nairn, A.V.; Moremen, K.W.; Merry, C.L.R.; Kjellén, L. NDST2 (N-Deacetylase/N-Sulfotransferase-2) Enzyme regulates heparan sulfate chain length. J. Biol. Chem. 2016, 291, 18600–18607. [Google Scholar] [CrossRef]
- Pallerla, S.R.; Lawrence, R.; Lewejohann, L.; Pan, Y.; Fischer, T.; Schlomann, U.; Zhang, X.; Esko, J.D.; Grobe, K. Altered heparan sulfate structure in mice with deleted NDST3 gene function. J. Biol. Chem. 2008, 283, 16885–16894. [Google Scholar] [CrossRef]
- Jao, T.-M.; Li, Y.-L.; Lin, S.-W.; Tzeng, S.-T.; Yu, I.-S.; Yen, S.-J.; Tsai, M.-H.; Yang, Y.-C. Alteration of colonic epithelial cell differentiation in mice deficient for glucosaminyl N-deacetylase/N-sulfotransferase 4. Oncotarget 2016, 7, 84938–84950. [Google Scholar] [CrossRef]
- Kusche-Gullberg, M.; Eriksson, I.; Pikas, D.S.; Kjellén, L. Identification and expression in mouse of two heparan sulfate glucosaminyl N-deacetylase/N-sulfotransferase genes. J. Biol. Chem. 1998, 273, 11902–11907. [Google Scholar] [CrossRef]
- Duncan, M.B.; Liu, M.; Fox, C.; Liu, J. Characterization of the N-deacetylase domain from the heparan sulfate N-deacetylase/N-sulfotransferase 2. Biochem. Biophys. Res. Commun. 2006, 339, 1232–1237. [Google Scholar] [CrossRef]
- Dou, W.; Xu, Y.; Pagadala, V.; Pedersen, L.C.; Liu, J. Role of deacetylase activity of N-deacetylase/N-sulfotransferase 1 in forming N-sulfated domain in heparan sulfate. J. Biol. Chem. 2015, 290, 20427–20437. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-J.; Yin, F.-X.; Zhang, X.-K.; Yu, J.; Zheng, S.; Song, X.-L.; Wang, F.-S.; Sheng, J.-Z. Characterization of heparan sulfate N-deacetylase/N-sulfotransferase isoform 4 using synthetic oligosaccharide substrates. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Norgard-Sumnicht, K.; Varki, A. Endothelial heparan sulfate proteoglycans that bind to L-selectin have glucosamine residues with unsubstituted amino groups. J. Biol. Chem. 1995, 270, 12012–12024. [Google Scholar] [CrossRef] [PubMed]
- Vanpouille, C.; Deligny, A.; Delehedde, M.; Denys, A.; Melchior, A.; Liénard, X.; Lyon, M.; Mazurier, J.; Fernig, D.G.; Allain, F. The heparin/heparan sulfate sequence that interacts with cyclophilin B contains a 3-O-sulfated N-unsubstituted glucosamine residue. J. Biol. Chem. 2007, 282, 24416–24429. [Google Scholar] [CrossRef]
- Nadanaka, S.; Purunomo, E.; Takeda, N.; Tamura, J.; Kitagawa, H. Heparan sulfate containing unsubstituted glucosamine residues: Biosynthesis and heparanase-inhibitory activity. J. Biol. Chem. 2014, 289, 15231–15243. [Google Scholar] [CrossRef]
- Kakuta, Y.; Sueyoshi, T.; Negishi, M.; Pedersen, L.C. Crystal structure of the sulfotransferase domain of human heparan sulfate N-deacetylase/ N-sulfotransferase 1. J. Biol. Chem. 1999, 274, 10673–10676. [Google Scholar] [CrossRef]
- Presto, J.; Thuveson, M.; Carlsson, P.; Busse, M.; Wilén, M.; Eriksson, I.; Kusche-Gullberg, M.; Kjellén, L. Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc. Natl. Acad. Sci. USA 2008, 105, 4751–4756. [Google Scholar] [CrossRef]
- Lindahl, U.; Bäckström, G.; Malmström, A.; Fransson, L.A. Biosynthesis of L-iduronic acid in heparin: Epimerization of D-glucuronic acid on the polymer level. Biochem. Biophys. Res. Commun. 1972, 46, 985–991. [Google Scholar] [CrossRef]
- Hagner-Mcwhirter, A.; Lindahl, U.; Li, J. p Biosynthesis of heparin/heparan sulphate: Mechanism of epimerization of glucuronyl C-5. Biochem. J. 2000, 347, 69–75. [Google Scholar] [CrossRef]
- Sheng, J.; Xu, Y.; Dulaney, S.B.; Huang, X.; Liu, J. Uncovering biphasic catalytic mode of C5-epimerase in heparan sulfate biosynthesis. J. Biol. Chem. 2012, 287, 20996–21002. [Google Scholar] [CrossRef] [PubMed]
- Jacobsson, I.; Lindahl, U.; Jensen, J.W.; Rodén, L.; Prihar, H.; Feingold, D.S. Biosynthesis of heparin. Substrate specificity of heparosan N-sulfate D-glucuronosyl 5-epimerase. J. Biol. Chem. 1984, 259, 1056–1063. [Google Scholar] [PubMed]
- Debarnot, C.; Monneau, Y.R.; Roig-Zamboni, V.; Delauzun, V.; Le Narvor, C.; Richard, E.; Hénault, J.; Goulet, A.; Fadel, F.; Vivès, R.R.; et al. Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase. Proc. Natl. Acad. Sci. USA 2019, 116, 6760–6765. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Ke, J.; Gu, X.; Fang, J.; Wang, W.; Cong, Q.; Li, J.; Tan, J.; Brunzelle, J.S.; Zhang, C.; et al. Structural and functional study of D-glucuronyl C5-epimerase. J. Biol. Chem. 2015, 290, 4620–4630. [Google Scholar] [CrossRef]
- Hagner-McWhirter, A.; Li, J.P.; Oscarson, S.; Lindahl, U. Irreversible glucuronyl C5-epimerization in the biosynthesis of heparan sulfate. J. Biol. Chem. 2004, 279, 14631–14638. [Google Scholar] [CrossRef]
- Préchoux, A.; Halimi, C.; Simorre, J.-P.; Lortat-Jacob, H.; Laguri, C. C5-epimerase and 2-O-sulfotransferase associate in vitro to generate contiguous epimerized and 2-O-sulfated heparan sulfate domains. ACS Chem. Biol. 2015, 10, 1064–1071. [Google Scholar] [CrossRef]
- Lindahl, U.; Li, J.P. Interactions between heparan sulfate and proteins-design and functional implications. Int. Rev. Cell Mol. Biol. 2009, 276, 105–159. [Google Scholar] [CrossRef]
- Li, J.-P.; Gong, F.; Hagner-McWhirter, A.; Forsberg, E.; Abrink, M.; Kisilevsky, R.; Zhang, X.; Lindahl, U. Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking L-iduronic acid and in neonatal lethality. J. Biol. Chem. 2003, 278, 28363–28366. [Google Scholar] [CrossRef]
- Mulloy, B.; Forster, M.J. Conformation and dynamics of heparin and heparan sulfate. Glycobiology 2000, 10, 1147–1156. [Google Scholar] [CrossRef]
- Jia, J.; Maccarana, M.; Zhang, X.; Bespalov, M.; Lindahl, U.; Li, J.-P. Lack of L-iduronic acid in heparan sulfate affects interaction with growth factors and cell signaling. J. Biol. Chem. 2009, 284, 15942–15950. [Google Scholar] [CrossRef]
- Feyerabend, T.B.; Li, J.-P.; Lindahl, U.; Rodewald, H.-R. Heparan sulfate C5-epimerase is essential for heparin biosynthesis in mast cells. Nat. Chem. Biol. 2006, 2, 195–196. [Google Scholar] [CrossRef] [PubMed]
- Rong, J.; Habuchi, H.; Kimata, K.; Lindahl, U.; Kusche-Gullberg, M. Substrate specificity of the heparan sulfate hexuronic acid 2-O-sulfotransferase. Biochemistry 2001, 40, 5548–5555. [Google Scholar] [CrossRef] [PubMed]
- Wilson, V.A.; Gallagher, J.T.; Merry, C.L.R. Heparan sulfate 2-O-sulfotransferase (Hs2st) and mouse development. Glycoconj. J. 2002, 19, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, B.; Eriksson, L.; Lindahl, U. Structure of heparan sulphate from human brain, with special regard to Alzheimer’s disease. Biochem. J. 1995, 306((Pt. 1)), 177–184. [Google Scholar] [CrossRef]
- Fedarko, N.S.; Conrad, H.E. A unique heparan sulfate in the nuclei of hepatocytes: Structural changes with the growth state of the cells. J. Cell Biol. 1986, 102, 587–599. [Google Scholar] [CrossRef]
- Liu, C.; Sheng, J.; Krahn, J.M.; Perera, L.; Xu, Y.; Hsieh, P.-H.; Dou, W.; Liu, J.; Pedersen, L.C. Molecular mechanism of substrate specificity for heparan sulfate 2-O-sulfotransferase. J. Biol. Chem. 2014, 289, 13407–13418. [Google Scholar] [CrossRef]
- Thieker, D.F.; Xu, Y.; Chapla, D.; Nora, C.; Qiu, H.; Felix, T.; Wang, L.; Moremen, K.W.; Liu, J.; Esko, J.D.; et al. Downstream products are potent inhibitors of the heparan sulfate 2-O-sulfotransferase. Sci. Rep. 2018, 8, 11832. [Google Scholar] [CrossRef]
- Jemth, P.; Smeds, E.; Do, A.-T.; Habuchi, H.; Kimata, K.; Lindahl, U.; Kusche-Gullberg, M. Oligosaccharide library-based assessment of heparan sulfate 6-O-sulfotransferase substrate specificity. J. Biol. Chem. 2003, 278, 24371–24376. [Google Scholar] [CrossRef]
- Smeds, E.; Habuchi, H.; Do, A.-T.; Hjertson, E.; Grundberg, H.; Kimata, K.; Lindahl, U.; Kusche-Gullberg, M. Substrate specificities of mouse heparan sulphate glucosaminyl 6-O-sulphotransferases. Biochem. J. 2003, 372, 371–380. [Google Scholar] [CrossRef]
- Sedita, J.; Izvolsky, K.; Cardoso, W.V. Differential expression of heparan sulfate 6-O-sulfotransferase isoforms in the mouse embryo suggests distinctive roles during organogenesis. Dev. Dyn. 2004, 231, 782–794. [Google Scholar] [CrossRef]
- Habuchi, H.; Tanaka, M.; Habuchi, O.; Yoshida, K.; Suzuki, H.; Ban, K.; Kimata, K. The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for hexuronic acid adjacent to the targeted N-sulfoglucosamine. J. Biol. Chem. 2000, 275, 2859–2868. [Google Scholar] [CrossRef] [PubMed]
- Habuchi, H.; Miyake, G.; Nogami, K.; Kuroiwa, A.; Matsuda, Y.; Kusche-Gullberg, M.; Habuchi, O.; Tanaka, M.; Kimata, K. Biosynthesis of heparan sulphate with diverse structures and functions: Two alternatively spliced forms of human heparan sulphate 6-O-sulphotransferase-2 having different expression patterns and properties. Biochem. J. 2003, 371, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Thacker, B.E.; Xu, D.; Lawrence, R.; Esko, J.D. Heparan sulfate 3-O-sulfation: A rare modification in search of a function. Matrix Biol. 2014, 35, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Marcum, J.A.; Atha, D.H.; Fritze, L.M.; Nawroth, P.; Stern, D.; Rosenberg, R.D. Cloned bovine aortic endothelial cells synthesize anticoagulantly active heparan sulfate proteoglycan. J. Biol. Chem. 1986, 261, 7507–7517. [Google Scholar]
- Pejler, G.; Bäckström, G.; Lindahl, U.; Paulsson, M.; Dziadek, M.; Fujiwara, S.; Timpl, R. Structure and affinity for antithrombin of heparan sulfate chains derived from basement membrane proteoglycans. J. Biol. Chem. 1987, 262, 5036–5043. [Google Scholar]
- De Agostini, A.I.; Dong, J.-C.; de Vantéry Arrighi, C.; Ramus, M.-A.; Dentand-Quadri, I.; Thalmann, S.; Ventura, P.; Ibecheole, V.; Monge, F.; Fischer, A.-M.; et al. Human follicular fluid heparan sulfate contains abundant 3-O-sulfated chains with anticoagulant activity. J. Biol. Chem. 2008, 283, 28115–28124. [Google Scholar] [CrossRef]
- Shukla, D.; Spear, P.G. Herpesviruses and heparan sulfate: An intimate relationship in aid of viral entry. J. Clin. Investig. 2001, 108, 503–510. [Google Scholar] [CrossRef]
- Xia, G.; Chen, J.; Tiwari, V.; Ju, W.; Li, J.-P.; Malmstrom, A.; Shukla, D.; Liu, J. Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1. J. Biol. Chem. 2002, 277, 37912–37919. [Google Scholar] [CrossRef]
- Edavettal, S.C.; Lee, K.A.; Negishi, M.; Linhardt, R.J.; Liu, J.; Pedersen, L.C. Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1. J. Biol. Chem. 2004, 279, 25789–25797. [Google Scholar] [CrossRef]
- Moon, A.F.; Edavettal, S.C.; Krahn, J.M.; Munoz, E.M.; Negishi, M.; Linhardt, R.J.; Liu, J.; Pedersen, L.C. Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1. J. Biol. Chem. 2004, 279, 45185–45193. [Google Scholar] [CrossRef]
- Xu, D.; Moon, A.F.; Song, D.; Pedersen, L.C.; Liu, J. Engineering sulfotransferases to modify heparan sulfate. Nat. Chem. Biol. 2008, 4, 200–202. [Google Scholar] [CrossRef] [PubMed]
- Sousa, R.P.; Fernandes, P.A.; Ramos, M.J.; Brás, N.F. Insights into the reaction mechanism of 3-O-sulfotransferase through QM/MM calculations. Phys. Chem. Chem. Phys. 2016, 18, 11488–11496. [Google Scholar] [CrossRef] [PubMed]
- Denys, A.; Allain, F. The emerging roles of heparan sulfate 3-O-sulfotransferases in cancer. Front. Oncol. 2019, 9, 507. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.; Best, M.D.; Hanson, S.R.; Wong, C.-H. Sulfotransferases: Structure, mechanism, biological activity, inhibition, and synthetic utility. Angew. Chem. Int. Ed. Engl. 2004, 43, 3526–3548. [Google Scholar] [CrossRef] [PubMed]
- Byrne, D.P.; Li, Y.; Ramakrishnan, K.; Barsukov, I.L.; Yates, E.A.; Eyers, C.E.; Papy-Garcia, D.; Chantepie, S.; Pagadala, V.; Liu, J.; et al. New tools for carbohydrate sulfation analysis: Heparan sulfate 2-O-sulfotransferase (HS2ST) is a target for small-molecule protein kinase inhibitors. Biochem. J. 2018, 475, 2417–2433. [Google Scholar] [CrossRef]
- Wu, L.; Viola, C.M.; Brzozowski, A.M.; Davies, G.J. Structural characterization of human heparanase reveals insights into substrate recognition. Nat. Struct. Mol. Biol. 2015, 22, 1016–1022. [Google Scholar] [CrossRef]
- Vlodavsky, I.; Gross-Cohen, M.; Weissmann, M.; Ilan, N.; Sanderson, R.D. Opposing functions of heparanase-1 and heparanase-2 in cancer progression. Trends Biochem. Sci. 2018, 43, 18–31. [Google Scholar] [CrossRef]
- He, Y.Q.; Sutcliffe, E.L.; Bunting, K.L.; Li, J.; Goodall, K.J.; Poon, I.K.A.; Hulett, M.D.; Freeman, C.; Zafar, A.; McInnes, R.L.; et al. The endoglycosidase heparanase enters the nucleus of T lymphocytes and modulates H3 methylation at actively transcribed genes via the interplay with key chromatin modifying enzymes. Transcription 2012, 3, 130–145. [Google Scholar] [CrossRef]
- Masola, V.; Bellin, G.; Gambaro, G.; Onisto, M. Heparanase: A multitasking protein involved in extracellular matrix (ECM) remodeling and intracellular events. Cells 2018, 7. [Google Scholar] [CrossRef]
- Koganti, R.; Suryawanshi, R.; Shukla, D. Heparanase, cell signaling, and viral infections. Cell. Mol. Life Sci. 2020. [Google Scholar] [CrossRef]
- Vlodavsky, I.; Singh, P.; Boyango, I.; Gutter-Kapon, L.; Elkin, M.; Sanderson, R.D.; Ilan, N. Heparanase: From basic research to therapeutic applications in cancer and inflammation. Drug Resist. Updat. 2016, 29, 54–75. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, E.; Tyson, K.; Stamps, A.; Smith, P.; Turner, P.; Barry, R.; Hircock, M.; Patel, S.; Barry, E.; Stubberfield, C.; et al. Cloning and expression profiling of Hpa2, a novel mammalian heparanase family member. BioChem. Biophys. Res. Commun. 2000, 276, 1170–1177. [Google Scholar] [CrossRef] [PubMed]
- Manon-Jensen, T.; Multhaupt, H.A.B.; Couchman, J.R. Mapping of matrix metalloproteinase cleavage sites on syndecan-1 and syndecan-4 ectodomains. FEBS J. 2013, 280, 2320–2331. [Google Scholar] [CrossRef]
- Gopal, S. Syndecans in Inflammation at a Glance. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Akbarshahi, H.; Axelsson, J.B.F.; Said, K.; Malmström, A.; Fischer, H.; Andersson, R. TLR4 dependent heparan sulphate-induced pancreatic inflammatory response is IRF3-mediated. J. Transl. Med. 2011, 9, 219. [Google Scholar] [CrossRef]
- Li, Q.; Park, P.W.; Wilson, C.L.; Parks, W.C. Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell 2002, 111, 635–646. [Google Scholar] [CrossRef]
- Gill, S.E.; Nadler, S.T.; Li, Q.; Frevert, C.W.; Park, P.W.; Chen, P.; Parks, W.C. Shedding of syndecan-1/CXCL1 complexes by matrix metalloproteinase 7 functions as an epithelial checkpoint of neutrophil activation. Am. J. Respir. Cell Mol. Biol. 2016, 55, 243–251. [Google Scholar] [CrossRef]
- Zhang, C.; Guo, F.; Chang, M.; Zhou, Z.; Yi, L.; Gao, C.; Huang, X.; Huan, J. Exosome-delivered syndecan-1 rescues acute lung injury via a FAK/p190RhoGAP/RhoA/ROCK/NF-κB signaling axis and glycocalyx enhancement. Exp. Cell Res. 2019, 384, 111596. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, K.; Parks, W.C.; Park, P.W. Syndecan-1 shedding facilitates the resolution of neutrophilic inflammation by removing sequestered CXC chemokines. Blood 2009, 114, 3033–3043. [Google Scholar] [CrossRef] [PubMed]
- Eustace, A.D.; McNaughton, E.F.; King, S.; Kehoe, O.; Kungl, A.; Mattey, D.; Nobbs, A.H.; Williams, N.; Middleton, J. Soluble syndecan-3 binds chemokines, reduces leukocyte migration in vitro and ameliorates disease severity in models of rheumatoid arthritis. Arthritis Res. Ther. 2019, 21, 172. [Google Scholar] [CrossRef]
- Xu, J.; Park, P.W.; Kheradmand, F.; Corry, D.B. Endogenous attenuation of allergic lung inflammation by syndecan-1. J. Immunol. 2005, 174, 5758–5765. [Google Scholar] [CrossRef]
- Yang, Y.; Yaccoby, S.; Liu, W.; Langford, J.K.; Pumphrey, C.Y.; Theus, A.; Epstein, J.; Sanderson, R.D. Soluble syndecan-1 promotes growth of myeloma tumors in vivo. Blood 2002, 100, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Endo, K.; Takino, T.; Miyamori, H.; Kinsen, H.; Yoshizaki, T.; Furukawa, M.; Sato, H. Cleavage of syndecan-1 by membrane type matrix metalloproteinase-1 stimulates cell migration. J. Biol. Chem. 2003, 278, 40764–40770. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Blaine, S.A.; Qiao, D.; Friedl, A. Membrane type 1 matrix metalloproteinase-mediated stromal syndecan-1 shedding stimulates breast carcinoma cell proliferation. Cancer Res. 2008, 68, 9558–9565. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, V.; Koo, C.Y.; Ibrahim, S.A.; Wang, Z.; Spillmann, D.; Dreier, R.; Kelsch, R.; Fischgrabe, J.; Smollich, M.; Rossi, L.H.; et al. Differential roles for membrane-bound and soluble syndecan-1 (CD138) in breast cancer progression. Carcinogenesis 2009, 30, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Macleod, V.; Miao, H.-Q.; Theus, A.; Zhan, F.; Shaughnessy, J.D.; Sawyer, J.; Li, J.-P.; Zcharia, E.; Vlodavsky, I.; et al. Heparanase enhances syndecan-1 shedding: A novel mechanism for stimulation of tumor growth and metastasis. J. Biol. Chem. 2007, 282, 13326–13333. [Google Scholar] [CrossRef] [PubMed]
- Aquino, R.S.; Teng, Y.H.-F.; Park, P.W. Glycobiology of syndecan-1 in bacterial infections. Biochem. Soc. Trans. 2018, 46, 371–377. [Google Scholar] [CrossRef]
- Park, P.W.; Pier, G.B.; Hinkes, M.T.; Bernfield, M. Exploitation of syndecan-1 shedding by Pseudomonas aeruginosa enhances virulence. Nature 2001, 411, 98–102. [Google Scholar] [CrossRef]
- Aquino, R.S.; Hayashida, A.; Park, P.W. Host syndecan-1 promotes listeriosis by inhibiting intravascular neutrophil extracellular traps. PLoS Pathog. 2020, 16, e1008497. [Google Scholar] [CrossRef]
- Dhoot, G.K.; Gustafsson, M.K.; Ai, X.; Sun, W.; Standiford, D.M.; CP, E., Jr. Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science 2001, 293, 1663–1666. [Google Scholar] [CrossRef]
- Morimoto-Tomita, M.; Uchimura, K.; Werb, Z.; Hemmerich, S.; Rosen, S.D. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J. Biol. Chem. 2002, 277, 49175–49185. [Google Scholar] [CrossRef] [PubMed]
- Frese, M.A.; Milz, F.; Dick, M.; Lamanna, W.C.; Dierks, T. Characterization of the human sulfatase Sulf1 and its high affinity heparin/heparan sulfate interaction domain. J. Biol. Chem. 2009, 284, 28033–28044. [Google Scholar] [CrossRef]
- Tang, R.; Rosen, S.D. Functional consequences of the subdomain organization of the sulfs. J. Biol. Chem. 2009, 284, 21505–21514. [Google Scholar] [CrossRef] [PubMed]
- Ai, X.; Do, A.T.; Kusche-Gullberg, M.; Lindahl, U.; Lu, K.; Emerson, C.P., Jr. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J. Biol. Chem. 2006, 281, 4969–4976. [Google Scholar] [CrossRef]
- Ambasta, R.K.; Ai, X.; CP, E., Jr. Quail Sulf1 function requires asparagine-linked glycosylation. J. Biol. Chem. 2007, 282, 34492–34499. [Google Scholar] [CrossRef]
- Seffouh, A.; Milz, F.; Przybylski, C.; Laguri, C.; Oosterhof, A.; Bourcier, S.; Sadir, R.; Dutkowski, E.; Daniel, R.; Kuppevelt, T.H. HSulf sulfatases catalyze processive and oriented 6-O-desulfation of heparan sulfate that differentially regulates fibroblast growth factor activity. FASEB J. 2013, 27, 2431–2439. [Google Scholar] [CrossRef]
- Seffouh, A.; El Masri, R.; Makshakova, O.; Gout, E.; Hassoun, Z.E.O.; Andrieu, J.-P.; Lortat-Jacob, H.; Vivès, R.R. Expression and purification of recombinant extracellular sulfatase HSulf-2 allows deciphering of enzyme sub-domain coordinated role for the binding and 6-O-desulfation of heparan sulfate. Cell Mol. Life Sci. CMLS 2019, 76, 1807–1819. [Google Scholar] [CrossRef]
- Harder, A.; Möller, A.-K.; Milz, F.; Neuhaus, P.; Walhorn, V.; Dierks, T.; Anselmetti, D. Catch bond interaction between cell-surface sulfatase Sulf1 and glycosaminoglycans. Biophys. J. 2015, 108, 1709–1717. [Google Scholar] [CrossRef]
- Pempe, E.H.; Burch, T.C.; Law, C.J.; Liu, J. Substrate specificity of 6-O-endosulfatase (Sulf-2) and its implications in synthesizing anticoagulant heparan sulfate. Glycobiology 2012, 22, 1353–1562. [Google Scholar] [CrossRef] [PubMed]
- Ai, X.; Do, A.T.; Lozynska, O.; Kusche-Gullberg, M.; Lindahl, U.; Emerson, C.P., Jr. QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J. Cell Biol. 2003, 162, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Viviano, B.L.; Paine-Saunders, S.; Gasiunas, N.; Gallagher, J.; Saunders, S. Domain-specific modification of heparan sulfate by Qsulf1 modulates the binding of the bone morphogenetic protein antagonist Noggin. J. Biol. Chem. 2004, 279, 5604–5611. [Google Scholar] [CrossRef] [PubMed]
- Langsdorf, A.; Schumacher, V.; Shi, X.; Tran, T.; Zaia, J.; Jain, S.; Taglienti, M.; Kreidberg, J.A.; Fine, A.; Ai, X. Expression regulation and function of heparan sulfate 6-O-endosulfatases in the spermatogonial stem cell niche. Glycobiology 2011, 21, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Otsuki, S.; Hanson, S.R.; Miyaki, S.; Grogan, S.P.; Kinoshita, M.; Asahara, H.; Wong, C.-H.; Lotz, M.K. Extracellular sulfatases support cartilage homeostasis by regulating BMP and FGF signaling pathways. Proc. Natl. Acad. Sci. USA 2010, 107, 10202–10207. [Google Scholar] [CrossRef] [PubMed]
- Kleinschmit, A.; Koyama, T.; Dejima, K.; Hayashi, Y.; Kamimura, K.; Nakato, H. Drosophila heparan sulfate 6-O endosulfatase regulates Wingless morphogen gradient formation. Dev. Biol. 2010, 345, 204–214. [Google Scholar] [CrossRef]
- Uchimura, K.; Morimoto-Tomita, M.; Bistrup, A.; Li, J.; Lyon, M.; Gallagher, J.; Werb, Z.; Rosen, S.D. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: Effects on VEGF, FGF-1, and SDF-1. BMC BioChem. 2006, 7, 2. [Google Scholar] [CrossRef]
- Wang, S.; Ai, X.; Freeman, S.D.; Pownall, M.E.; Lu, Q.; Kessler, D.S.; CP, E., Jr. QSulf1, a heparan sulfate 6-O-endosulfatase, inhibits fibroblast growth factor signaling in mesoderm induction and angiogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 4833–4838. [Google Scholar] [CrossRef]
- Lai, J.; Chien, J.; Staub, J.; Avula, R.; Greene, E.L.; Matthews, T.A.; Smith, D.I.; Kaufmann, S.H.; Roberts, L.R.; Shridhar, V. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J. Biol. Chem. 2003, 278, 23107–23117. [Google Scholar] [CrossRef]
- Narita, K.; Staub, J.; Chien, J.; Meyer, K.; Bauer, M.; Friedl, A.; Ramakrishnan, S.; Shridhar, V. HSulf-1 inhibits angiogenesis and tumorigenesis in vivo. Cancer Res. 2006, 66, 6025–6032. [Google Scholar] [CrossRef]
- Lai, J.P.; Chien, J.; Strome, S.E.; Staub, J.; Montoya, D.P.; Greene, E.L.; Smith, D.I.; Roberts, L.R.; Shridhar, V. HSulf-1 modulates HGF-mediated tumor cell invasion and signaling in head and neck squamous carcinoma. Oncogene 2004, 23, 1439–1447. [Google Scholar] [CrossRef]
- Dai, Y.; Yang, Y.; MacLeod, V.; Yue, X.; Rapraeger, A.C.; Shriver, Z.; Venkataraman, G.; Sasisekharan, R.; Sanderson, R.D. HSulf-1 and HSulf-2 are potent inhibitors of myeloma tumor growth in vivo. J. Biol. Chem. 2005, 280, 40066–40073. [Google Scholar] [CrossRef]
- Narita, K.; Chien, J.; Mullany, S.A.; Staub, J.; Qian, X.; Lingle, W.L.; Shridhar, V. Loss of HSulf-1 expression enhances autocrine signaling mediated by amphiregulin in breast cancer. J. Biol. Chem. 2007, 282, 14413–14420. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Li, X.; Nguyen, H.T.; Chin, D.R.; Sullivan, D.E.; Lasky, J.A. Transforming growth factor-beta1 induces heparan sulfate 6-O-endosulfatase 1 expression in vitro and in vivo. J. Biol. Chem. 2008, 283, 20397–20407. [Google Scholar] [CrossRef] [PubMed]
- Guimond, S.; Maccarana, M.; Olwin, B.B.; Lindahl, U.; Rapraeger, A.C. Activating and inhibitory heparin sequences for FGF-2 (basic FGF). Distinct requirements for FGF-1, FGF-2, and FGF-4. J. Biol. Chem. 1993, 268, 23906–23914. [Google Scholar]
- Pye, D.A.; Vives, R.R.; Turnbull, J.E.; Hyde, P.; Gallagher, J.T. Heparan sulfate oligosaccharides require 6-O-sulfation for promotion of basic fibroblast growth factor mitogenic activity. J. Biol. Chem. 1998, 273, 22936–22942. [Google Scholar] [CrossRef] [PubMed]
- El Masri, R.; Crétinon, Y.; Gout, E.; Vives, R. HS and inflammation: A potential playground for the Sulfs? Front. Immunol. 2020, 11, 570. [Google Scholar] [CrossRef]
- Ai, X.; Kitazawa, T.; Do, A.T.; Kusche-Gullberg, M.; Labosky, P.A.; CP, E., Jr. SULF1 and SULF2 regulate heparan sulfate-mediated GDNF signaling for esophageal innervation. Development 2007, 134, 3327–3338. [Google Scholar] [CrossRef] [PubMed]
- Danesin, C.; Agius, E.; Escalas, N.; Ai, X.; Emerson, C.; Cochard, P.; Soula, C. Ventral neural progenitors switch toward an oligodendroglial fate in response to increased Sonic hedgehog (Shh) activity: Involvement of Sulfatase 1 in modulating Shh signaling in the ventral spinal cord. J. NeuroSci. 2006, 26, 5037–5048. [Google Scholar] [CrossRef]
- Kalus, I.; Rohn, S.; Puvirajesinghe, T.M.; Guimond, S.E.; Eyckerman-Kolln, P.J.; Ten Dam, G.; Kuppevelt, T.H.; Turnbull, J.E.; Dierks, T. Sulf1 and Sulf2 Differentially Modulate Heparan Sulfate Proteoglycan Sulfation during Postnatal Cerebellum Development: Evidence for Neuroprotective and Neurite Outgrowth Promoting Functions. PLoS ONE 2015, 10, 0139853. [Google Scholar] [CrossRef]
- Oustah, A.A.; Danesin, C.; Khouri-Farah, N.; Farreny, M.-A.; Escalas, N.; Cochard, P.; Glise, B.; Soula, C. Dynamics of Sonic hedgehog signaling in the ventral spinal cord are controlled by intrinsic changes in source cells requiring Sulfatase 1. Development 2014, 141, 1392–1403. [Google Scholar] [CrossRef]
- Zhao, W.; Sala-Newby, G.B.; Dhoot, G.K. Sulf1 expression pattern and its role in cartilage and joint development. Dev. Dyn. 2006, 235, 3327–3335. [Google Scholar] [CrossRef]
- Holst, C.R.; Bou-Reslan, H.; Gore, B.B.; Wong, K.; Grant, D.; Chalasani, S.; Carano, R.A.; Frantz, G.D.; Tessier-Lavigne, M.; Bolon, B. Secreted sulfatases Sulf1 and Sulf2 have overlapping yet essential roles in mouse neonatal survival. PLoS ONE 2007, 2, 575. [Google Scholar] [CrossRef] [PubMed]
- Freeman, S.D.; Keino-Masu, K.; Masu, M.; Ladher, R.K. Expression of the heparan sulfate 6-O-endosulfatases, Sulf1 and Sulf2, in the avian and mammalian inner ear suggests a role for sulfation during inner ear development. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2015, 244, 168–180. [Google Scholar] [CrossRef]
- Hayano, S.; Kurosaka, H.; Yanagita, T.; Kalus, I.; Milz, F.; Ishihara, Y.; Islam, M.N.; Kawanabe, N.; Saito, M.; Kamioka, H. Roles of heparan sulfate sulfation in dentinogenesis. J. Biol. Chem. 2012, 287, 12217–12229. [Google Scholar] [CrossRef] [PubMed]
- Kalus, I.; Salmen, B.; Viebahn, C.; Figura, K.; Schmitz, D.; D’Hooge, R.; Dierks, T. Differential involvement of the extracellular 6-O-endosulfatases Sulf1 and Sulf2 in brain development and neuronal and behavioural plasticity. J. Cell Mol. Med. 2009, 13, 4505–4521. [Google Scholar] [CrossRef]
- Lum, D.H.; Tan, J.; Rosen, S.D.; Werb, Z. Gene trap disruption of the mouse heparan sulfate 6-O-endosulfatase gene, Sulf2. Mol. Cell Biol. 2007, 27, 678–688. [Google Scholar] [CrossRef]
- Rosen, S.D.; Lemjabbar-Alaoui, H. Sulf-2: An extracellular modulator of cell signaling and a cancer target candidate. Exp. Opin. Ther. Targets 2010, 14, 935–949. [Google Scholar] [CrossRef]
- Liu, J.; Moon, A.F.; Sheng, J.; Pedersen, L.C. Understanding the substrate specificity of the heparan sulfate sulfotransferases by an integrated biosynthetic and crystallographic approach. Curr. Opin. Struct. Biol. 2012, 22, 550–557. [Google Scholar] [CrossRef]
- Xu, Y.; Moon, A.F.; Xu, S.; Krahn, J.M.; Liu, J.; Pedersen, L.C. Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases. ACS Chem. Biol. 2017, 12, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Song, T.; Lindahl, U.; Li, J.-P. Enzyme overexpression - an exercise toward understanding regulation of heparan sulfate biosynthesis. Sci. Rep. 2016, 6, 31242. [Google Scholar] [CrossRef]
- Rong, J.; Habuchi, H.; Kimata, K.; Lindahl, U.; Kusche-Gullberg, M. Expression of heparan sulphate L-iduronyl 2-O-sulphotransferase in human kidney 293 cells results in increased D-glucuronyl 2-O-sulphation. Biochem. J. 2000, 346(Pt. 2), 463–468. [Google Scholar] [CrossRef]
- Do, A.-T.; Smeds, E.; Spillmann, D.; Kusche-Gullberg, M. Overexpression of heparan sulfate 6-O-sulfotransferases in human embryonic kidney 293 cells results in increased N-acetylglucosaminyl 6-O-sulfation. J. Biol. Chem. 2006, 281, 5348–5356. [Google Scholar] [CrossRef] [PubMed]
- Dick, G.; Akslen-Hoel, L.K.; Grøndahl, F.; Kjos, I.; Maccarana, M.; Prydz, K. PAPST1 regulates sulfation of heparan sulfate proteoglycans in epithelial MDCK II cells. Glycobiology 2015, 25, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Crawford, B.E.; Olson, S.K.; Esko, J.D.; Pinhal, M.A. Cloning, Golgi localization, and enzyme activity of the full-length heparin/heparan sulfate-glucuronic acid C5-epimerase. J. Biol. Chem. 2001, 276, 21538–21543. [Google Scholar] [CrossRef]
- Multhaupt, H.A.B.; Couchman, J.R. Heparan sulfate biosynthesis: Methods for investigation of the heparanosome. J. Histochem. Cytochem. 2012, 60, 908–915. [Google Scholar] [CrossRef]
- Nagai, N.; Habuchi, H.; Esko, J.D.; Kimata, K. Stem domains of heparan sulfate 6-O-sulfotransferase are required for Golgi localization, oligomer formation and enzyme activity. J. Cell. Sci. 2004, 117, 3331–3341. [Google Scholar] [CrossRef]
- Victor, X.V.; Nguyen, T.K.N.; Ethirajan, M.; Tran, V.M.; Nguyen, K.V.; Kuberan, B. Investigating the elusive mechanism of glycosaminoglycan biosynthesis. J. Biol. Chem. 2009, 284, 25842–25853. [Google Scholar] [CrossRef]
- Habuchi, H.; Habuchi, O.; Kimata, K. Purification and characterization of heparan sulfate 6-sulfotransferase from the culture medium of Chinese hamster ovary cells. J. Biol. Chem. 1995, 270, 4172–4179. [Google Scholar] [CrossRef]
- Esko, J.D.; Selleck, S.B. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu. Rev. BioChem. 2002, 71, 435–471. [Google Scholar] [CrossRef]
- Pegeot, M.; Sadir, R.; Eriksson, I.; Kjellen, L.; Simorre, J.-P.; Gans, P.; Lortat-Jacob, H. Profiling sulfation/epimerization pattern of full-length heparan sulfate by NMR following cell culture 13C-glucose metabolic labeling. Glycobiology 2015, 25, 151–156. [Google Scholar] [CrossRef]
- Hassinen, A.; Pujol, F.M.; Kokkonen, N.; Pieters, C.; Kihlström, M.; Korhonen, K.; Kellokumpu, S. Functional organization of Golgi N-and O-glycosylation pathways involves pH-dependent complex formation that is impaired in cancer cells. J. Biol. Chem. 2011, 286, 38329–38340. [Google Scholar] [CrossRef]
- Kellokumpu, S. Golgi pH, ion and redox homeostasis: How much do they really matter? Front. Cell Dev. Biol. 2019, 7, 93. [Google Scholar] [CrossRef] [PubMed]

| Enzymes | Function | Golgi/Cell Localization [Reference] |
|---|---|---|
| XylT-1/XylT-2 GalT-1/GalT-2 GlcAT-1 | Formation of linkage region | Cis/Medial [124] Cis/Medial [123] Cis/Medial [123] |
| EXTL EXT1/EXT2 | GlcNAc addition HS Elongation | Not yet specified Cis [125,126] |
| NDSTs | GlcNAc N-deacetylation/N-sulfation | Cis/Medial [123] |
| C5-Epi 2OST | Glc C5 Epimerization GlcA/IdoA 2-O-sulfation | Medial [123] Medial [123] |
| 6OSTs | GlcN 6-O-sulfation | Cis/Medial [127] |
| 3OSTs | GlcN 3-O-sulfation | Cis/Cell surface * [128] |
| Sulf-1/Sulf-2 | GlcNS6S 6-O-desulfation | Cell surface [129] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Annaval, T.; Wild, R.; Crétinon, Y.; Sadir, R.; Vivès, R.R.; Lortat-Jacob, H. Heparan Sulfate Proteoglycans Biosynthesis and Post Synthesis Mechanisms Combine Few Enzymes and Few Core Proteins to Generate Extensive Structural and Functional Diversity. Molecules 2020, 25, 4215. https://doi.org/10.3390/molecules25184215
Annaval T, Wild R, Crétinon Y, Sadir R, Vivès RR, Lortat-Jacob H. Heparan Sulfate Proteoglycans Biosynthesis and Post Synthesis Mechanisms Combine Few Enzymes and Few Core Proteins to Generate Extensive Structural and Functional Diversity. Molecules. 2020; 25(18):4215. https://doi.org/10.3390/molecules25184215
Chicago/Turabian StyleAnnaval, Thibault, Rebekka Wild, Yoann Crétinon, Rabia Sadir, Romain R. Vivès, and Hugues Lortat-Jacob. 2020. "Heparan Sulfate Proteoglycans Biosynthesis and Post Synthesis Mechanisms Combine Few Enzymes and Few Core Proteins to Generate Extensive Structural and Functional Diversity" Molecules 25, no. 18: 4215. https://doi.org/10.3390/molecules25184215
APA StyleAnnaval, T., Wild, R., Crétinon, Y., Sadir, R., Vivès, R. R., & Lortat-Jacob, H. (2020). Heparan Sulfate Proteoglycans Biosynthesis and Post Synthesis Mechanisms Combine Few Enzymes and Few Core Proteins to Generate Extensive Structural and Functional Diversity. Molecules, 25(18), 4215. https://doi.org/10.3390/molecules25184215





