Selective Targeting of αvβ5 Integrin in HepG2 Cell Line by RGDechi15D Peptide
Abstract
1. Introduction
2. Results and Discussion
2.1. Integrin Expression on HepG2
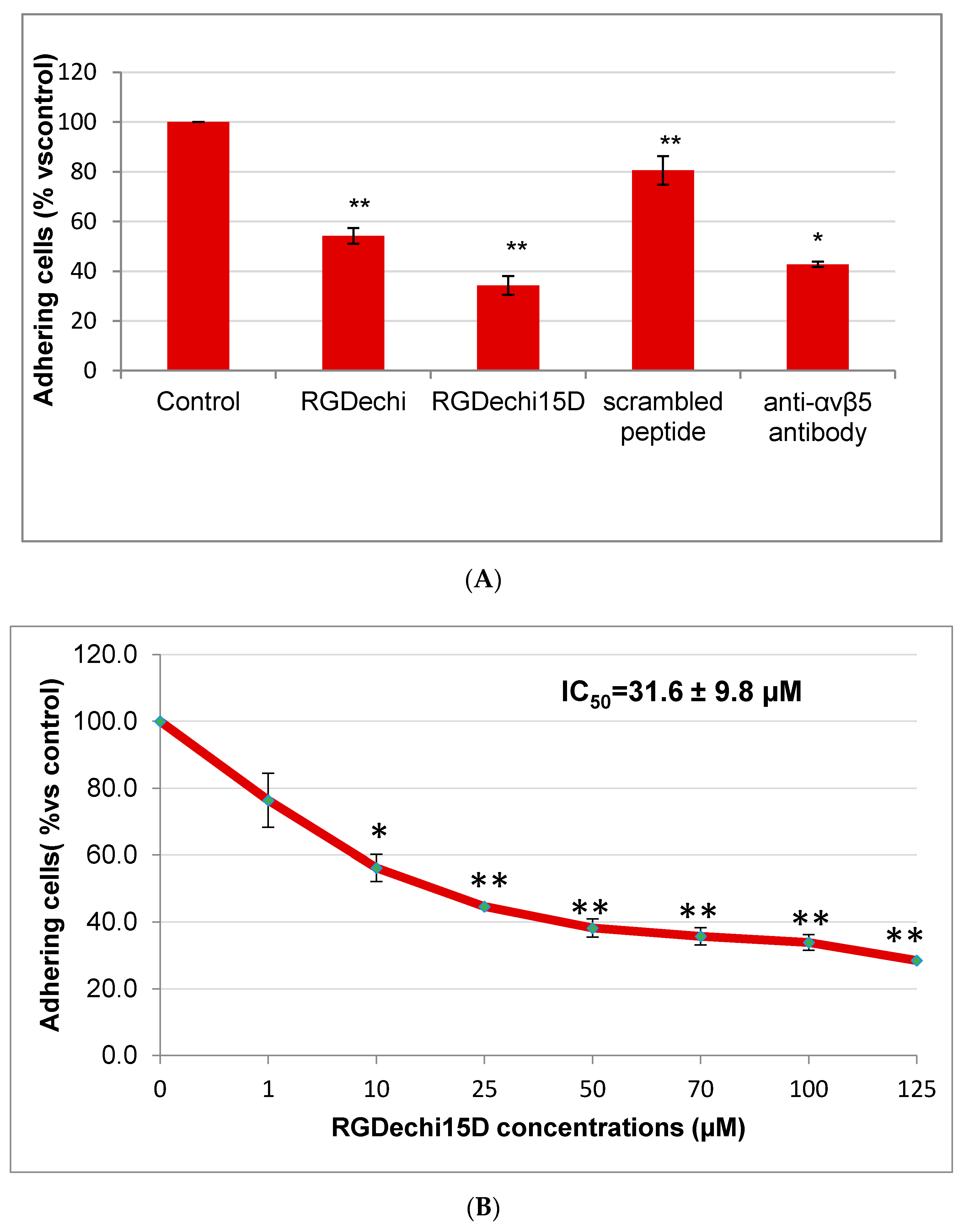
2.2. Effect of RGDechi15D on Cell Adhesion
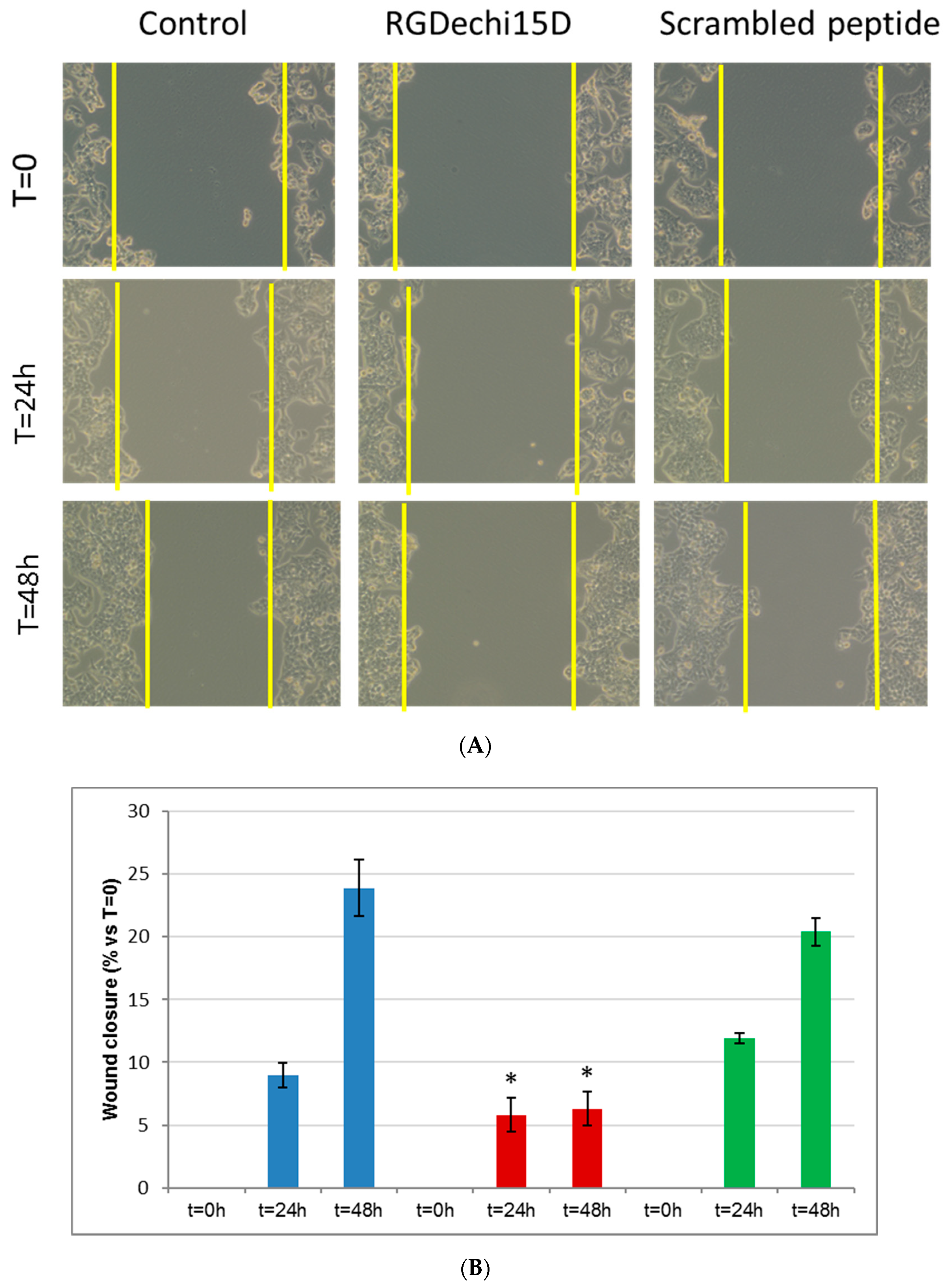
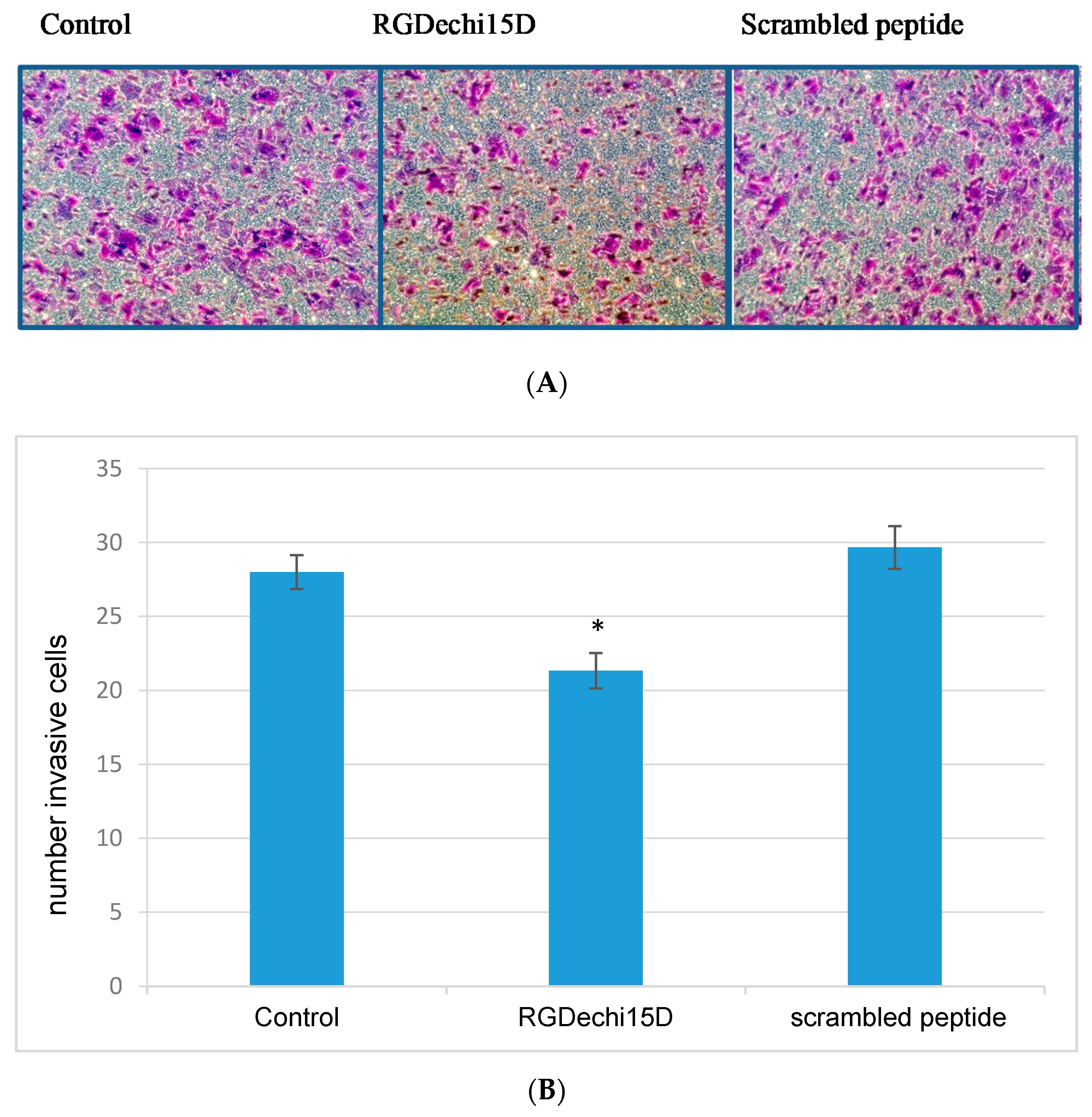
2.3. Effect of RGDechi15D on HepG2 Migration and Invasion
2.4. Evaluation of RGDechi15D Capability to Inhibit New Vessels Formation
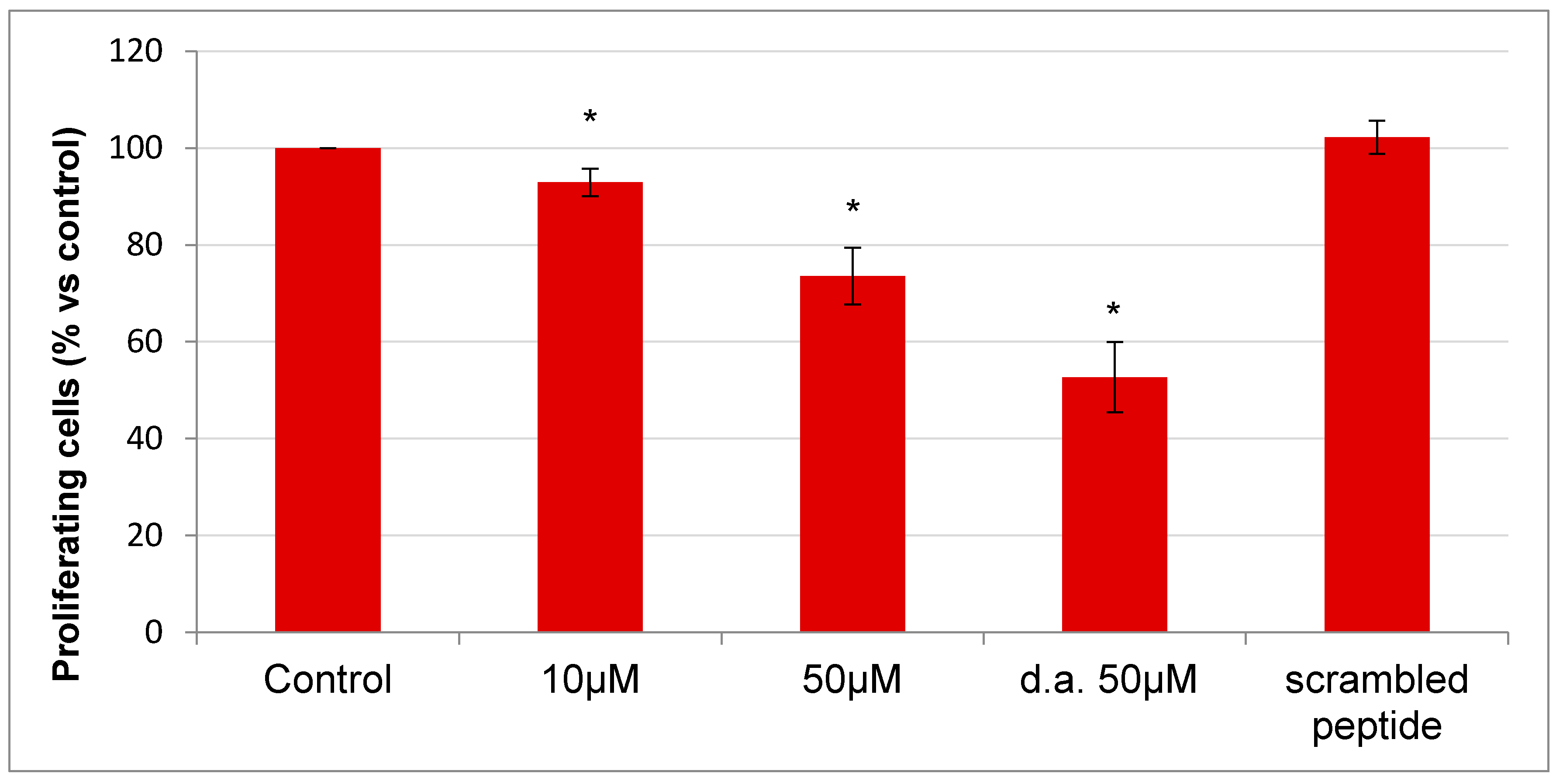
2.5. Effect of RGDechi15D on Cell Proliferation
2.6. Quantitative Analysis of the AKT Phosphorylation
2.7. Determination of Caspase-3 Activity
3. Conclusions
4. Materials and Methods
4.1. Cell Lines and Culture Conditions
4.2. FACS Analysis for αvβ5 and αvβ3 Integrins
4.3. Cell Adhesion Assay
4.4. In Vitro Scratch Assay
4.5. Invasion Assay
4.6. Angiogenesis Assay
4.7. Proliferation Assay
4.8. AKT Phosphorylation by Western Blotting
4.9. Analysis of Caspase-3 Activity
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hynes, R.O.; Lively, J.C.; McCarty, J.H.; Taverna, D.; Francis, S.E.; Hodivala-Dilke, K.; Xiao, Q. The diverse roles of integrins and their ligands in angiogenesis. Cold Spring Harb Symp. Quant. Biol. 2002, 67, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.W.; Vestal, D.J.; Irwin, S.V.; Burke, T.A.; Cheresh, D.A. Purification and functional characterization of integrin alpha v beta 5. An adhesion receptor for vitronectin. J. Biol. Chem. 1990, 265, 11008–11013. [Google Scholar] [PubMed]
- Plow, E.F.; Haas, T.A.; Zhang, L.; Loftus, J.; Smith, J.W. Ligand binding to integrins. J. Biol. Chem. 2000, 275, 21785–21788. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.A.; Walker, L.R.; Abdel-Raouf, U.M.; Desouky, S.A.; Montasser, A.K.; Akula, S.M. Beyond RGD: Virus interactions with integrins. Arch. Virol. 2015, 160, 2669–2681. [Google Scholar] [CrossRef] [PubMed]
- Ballana, E.; Pauls, E.; Clotet, B.; Perron-Sierra, F.; Tucker, G.C.; Este, J.A. Beta5 integrin is the major contributor to the alphaVintegrin-mediated blockade of HIV-1 replication. J. Immunol. 2011, 186, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, Q.; Tiwari, S.K.; Lichinchi, G.; Yau, E.H.; Hui, H.; Li, W.; Furnari, F.; Rana, T.M. Integrin alphavbeta5 Internalizes Zika Virus during Neural Stem Cells Infection and Provides a Promising Target for Antiviral Therapy. Cell Rep. 2020, 30, 969–983.e4. [Google Scholar] [CrossRef]
- Pirone, L.; Del Gatto, A.; Di Gaetano, S.; Saviano, M.; Capasso, D.; Zaccaro, L.; Pedone, E.M. A multi-targeting approach to fight SARS-CoV-2 attachment. Front. Mol. Biosci. 2020, 7, 186. [Google Scholar] [CrossRef]
- Friedlander, M.; Brooks, P.C.; Shaffer, R.W.; Kincaid, C.M.; Varner, J.A.; Cheresh, D.A. Definition of two angiogenic pathways by distinct alpha v integrins. Science 1995, 270, 1500–1502. [Google Scholar] [CrossRef]
- Su, G.; Hodnett, M.; Wu, N.; Atakilit, A.; Kosinski, C.; Godzich, M.; Huang, X.Z.; Kim, J.K.; Frank, J.A.; Matthay, M.A.; et al. Integrin alphavbeta5 regulates lung vascular permeability and pulmonary endothelial barrier function. Am. J. Respir. Cell. Mol. Biol. 2007, 36, 377–386. [Google Scholar] [CrossRef]
- Eliceiri, B.P.; Puente, X.S.; Hood, J.D.; Stupack, D.G.; Schlaepfer, D.D.; Huang, X.Z.; Sheppard, D.; Cheresh, D.A. Src-mediated coupling of focal adhesion kinase to integrin alpha(v)beta5 in vascular endothelial growth factor signaling. J. Cell Biol. 2002, 157, 149–160. [Google Scholar] [CrossRef]
- Li, F.; Liu, Y.; Kan, X.; Li, Y.; Liu, M.; Lu, J.G. Elevated expression of integrin alphav and beta5 subunit in laryngeal squamous-cell carcinoma associated with lymphatic metastasis and angiogenesis. Pathol. Res. Pract. 2013, 209, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Enns, A.; Korb, T.; Schluter, K.; Gassmann, P.; Spiegel, H.U.; Senninger, N.; Mitjans, F.; Haier, J. Alphavbeta5-integrins mediate early steps of metastasis formation. Eur. J. Cancer 2005, 41, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, T.; Nishikawa, Y.; Ito, R.; Kawamata, M.; Doi, Y.; Yamamoto, Y.; Yoshida, M.; Omori, Y.; Kotanagi, H.; Masuko, T.; et al. Significance of integrin alphavbeta5 and erbB3 in enhanced cell migration and liver metastasis of colon carcinomas stimulated by hepatocyte-derived heregulin. Cancer Sci. 2010, 101, 2011–2018. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Thompson, E.W.; Williams, E.D. Mesenchymal to epithelial transition in development and disease. Cells Tissues Organs 2007, 185, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Novoa, J.M.; Nieto, M.A. Inflammation and EMT: An alliance towards organ fibrosis and cancer progression. EMBO Mol. Med. 2009, 1, 303–314. [Google Scholar] [CrossRef]
- Bianchi, A.; Gervasi, M.E.; Bakin, A. Role of beta5-integrin in epithelial-mesenchymal transition in response to TGF-beta. Cell Cycle. 2010, 9, 1647–1659. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Z.; Viklund, E.K.; Stromblad, S. P21-activated kinase 4 interacts with integrin alpha v beta 5 and regulates alpha v beta 5-mediated cell migration. J. Cell Biol. 2002, 158, 1287–1297. [Google Scholar] [CrossRef]
- Majhen, D.; Stojanovic, N.; Speljko, T.; Brozovic, A.; De Zan, T.; Osmak, M.; Ambriovic-Ristov, A. Increased expression of the coxsackie and adenovirus receptor downregulates alphavbeta3 and alphavbeta5 integrin expression and reduces cell adhesion and migration. Life Sci. 2011, 89, 241–249. [Google Scholar] [CrossRef]
- Bianchi-Smiraglia, A.; Paesante, S.; Bakin, A.V. Integrin beta5 contributes to the tumorigenic potential of breast cancer cells through the Src-FAK and MEK-ERK signaling pathways. Oncogene 2013, 32, 3049–3058. [Google Scholar] [CrossRef]
- Wang, Z.; Chui, W.K.; Ho, P.C. Integrin targeted drug and gene delivery. Expert Opin. Drug Deliv. 2010, 7, 159–171. [Google Scholar] [CrossRef]
- Bruix, J.; Sherman, M. Practice Guidelines Committee, Management of hepatocellular carcinoma. Hepatology 2005, 42, 1208–1236. [Google Scholar] [CrossRef] [PubMed]
- Forner, A.; Llovet, J.M.; Bruix, J. Hepatocellular carcinoma. Lancet 2012, 379, 1245–1255. [Google Scholar] [CrossRef]
- Portolani, N.; Coniglio, A.; Ghidoni, S.; Giovanelli, M.; Benetti, A.; Tiberio, G.A.; Giulini, S.M. Early and late recurrence after liver resection for hepatocellular carcinoma: Prognostic and therapeutic implications. Ann. Surg. 2006, 243, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; He, R.; Luo, H.; Lu, C.; Ning, Z.; Wu, Y.; Han, C.; Tan, G.; Wang, Z. Integrin-beta5, a miR-185-targeted gene, promotes hepatocellular carcinoma tumorigenesis by regulating beta-catenin stabilit. J. Exp. Clin. Cancer Res. 2018, 37, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Long, Q. Elevated serum plasma fibrinogen is associated with advanced tumor stage and poor survival in hepatocellular carcinoma patients. Medicine (Baltimore) 2017, 96, e6694. [Google Scholar] [CrossRef] [PubMed]
- Kisseleva, T.; Brenner, D.A. Hepatic stellate cells and the reversal of fibrosis. J. Gastroenterol. Hepatol. 2006, 21 (Suppl. S3), S84–S87. [Google Scholar] [CrossRef]
- Masamune, A.; Kikuta, K.; Watanabe, T.; Satoh, K.; Hirota, M.; Hamada, S.; Shimosegawa, T. Fibrinogen induces cytokine and collagen production in pancreatic stellate cells. Gut 2009, 58, 550–559. [Google Scholar] [CrossRef]
- Nejjari, M.; Hafdi, Z.; Gouysse, G.; Fiorentino, M.; Béatrix, O.; Dumortier, J.; Pourreyron, C.; Barozzi, C.; D’Errico, A.; Grigioni, W.F.; et al. Expression, regulation, and function of alpha V integrins in hepatocellular carcinoma: An in vivo and in vitro study. Hepatology 2002, 36, 418–426. [Google Scholar] [CrossRef]
- Yan, C.; Yang, Q.; Gong, Z. Activation of Hepatic Stellate Cells During Liver Carcinogenesis Requires Fibrinogen/Integrin alphavbeta5 in Zebrafish. Neoplasia 2018, 20, 533–542. [Google Scholar] [CrossRef]
- Marinelli, L.; Gottschalk, K.E.; Meyer, A.; Novellino, E.; Kessler, H. Human integrin alphavbeta5: Homology modeling and ligand binding. J. Med. Chem. 2004, 47, 4166–4177. [Google Scholar] [CrossRef] [PubMed]
- Russo, L.; Farina, B.; Del Gatto, A.; Comegna, D.; Di Gaetano, S.; Capasso, D.; Liguoro, A.; Malgieri, G.; Saviano, M.; Fattorusso, R.; et al. Deciphering RGDechi peptide-α5β1 integrin interaction mode in isolated cell membranes. Pept. Sci. 2018, 110, e24065. [Google Scholar] [CrossRef]
- Del Gatto, A.; Zaccaro, L.; Grieco, P.; Novellino, E.; Zannetti, A.; Del Vecchio, S.; Iommelli, F.; Salvatore, M.; Pedone, C.; Saviano, M. Novel and selective alpha(v)beta3 receptor peptide antagonist: Design, synthesis, and biological behavior. J. Med. Chem. 2006, 49, 3416–3420. [Google Scholar] [CrossRef]
- Farina, B.; Del Gatto, A.; Comegna, D.; Di Gaetano, S.; Capasso, D.; Isernia, C.; Saviano, M.; Fattorusso, R.; Zaccaro, L.; Russo, L. Conformational studies of RGDechi peptide by natural-abundance NMR spectroscopy. J. Pept. Sci. 2019, 25, e3166. [Google Scholar] [CrossRef] [PubMed]
- Zannetti, A.; Del Vecchio, S.; Iommelli, F.; Del Gatto, A.; De Luca, S.; Zaccaro, L.; Papaccioli, A.; Sommella, J.; Panico, M.; Speranza, A.; et al. Imaging of alpha(v)beta(3) expression by a bifunctional chimeric RGD peptide not cross-reacting with alpha(v)beta(5). Clin. Cancer Res. 2009, 15, 5224–5233. [Google Scholar] [CrossRef]
- Farina, B.; de Paola, I.; Russo, L.; Capasso, D.; Liguoro, A.; Gatto, A.D.; Saviano, M.; Pedone, P.V.; Di Gaetano, S.; Malgieri, G.; et al. A Combined NMR and Computational Approach to Determine the RGDechi-hCit-alphav beta3 Integrin Recognition Mode in Isolated Cell Membranes. Chemistry 2016, 22, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Song, Y.; Zhuang, J.; Wang, G.; Ni, J.; Xia, W. Anticancer effects of echinacoside in hepatocellular carcinoma mouse model and HepG2 cells. J. Cell Physiol. 2019, 234, 1880–1888. [Google Scholar] [CrossRef] [PubMed]
- Davis, G.E. Affinity of integrins for damaged extracellular matrix: Alpha v beta 3 binds to denatured collagen type I through RGD sites. Biochem. Biophys. Res. Commun. 1992, 182, 1025–1031. [Google Scholar] [CrossRef]
- Kanda, S.; Kuzuya, M.; Ramos, M.A.; Koike, T.; Yoshino, K.; Ikeda, S.; Iguchi, A. Matrix metalloproteinase and alphavbeta3 integrin-dependent vascular smooth muscle cell invasion through a type I collagen lattice. Arter. Thromb. Vasc. Biol. 2000, 20, 998–1005. [Google Scholar] [CrossRef]
- Barczyk, M.; Carracedo, S.; Gullberg, D. Integrins. Cell Tissue Res. 2010, 339, 269–280. [Google Scholar] [CrossRef]
- Chen, B.; Zhou, S.; Zhan, Y.; Ke, J.; Wang, K.; Liang, Q.; Hou, Y.; Zhu, P.; Ao, W.; Wei, X.; et al. Dioscin Inhibits the Invasion and Migration of Hepatocellular Carcinoma HepG2 Cells by Reversing TGF-beta1-Induced Epithelial-Mesenchymal Transition. Molecules 2019, 24, 2222. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Yu, H.; Xia, Q.; Ma, Y.; Yin, H.; Shen, Y.; Liu, X. Cross-talk mechanism between endothelial cells and hepatocellular carcinoma cells via growth factors and integrin pathway promotes tumor angiogenesis and cell migration. Oncotarget 2017, 8, 69577–69593. [Google Scholar] [CrossRef] [PubMed]
- Pankov, R.; Cukierman, E.; Clark, K.; Matsumoto, K.; Hahn, C.; Poulin, B.; Yamada, K.M. Specific beta1 integrin site selectively regulates Akt/protein kinase B signaling via local activation of protein phosphatase 2A. J. Biol. Chem. 2003, 278, 18671–18681. [Google Scholar] [CrossRef] [PubMed]
- Capasso, D.; de Paola, I.; Liguoro, A.; Del Gatto, A.; Di Gaetano, S.; Guarnieri, D.; Saviano, M.; Zaccaro, L. RGDechi-hCit: alphavbeta3 selective pro-apoptotic peptide as potential carrier for drug delivery into melanoma metastatic cells. PLoS ONE 2014, 9, e106441. [Google Scholar] [CrossRef]
- Capasso, D.; Di Gaetano, S.; Celentano, V.; Diana, D.; Festa, L.; Di Stasi, R.; De Rosa, L.; Fattorusso, R.; D’Andrea, L.D. Unveiling a VEGF-mimetic peptide sequence in the IQGAP1 protein. Mol. Biosyst. 2017, 13, 1619–1629. [Google Scholar] [CrossRef]
- Diana, D.; Russomanno, A.; De Rosa, L.; Di Stasi, R.; Capasso, D.; Di Gaetano, S.; Romanelli, A.; Russo, L.; D’Andrea, L.D.; Fattorusso, R. Functional binding surface of a beta-hairpin VEGF receptor targeting peptide determined by NMR spectroscopy in living cell. Chemistry 2015, 21, 91–95. [Google Scholar] [CrossRef]
- Comegna, D.; Zannetti, A.; Del Gatto, A.; de Paola, I.; Russo, L.; Di Gaetano, S.; Liguoro, A.; Capasso, D.; Saviano, M.; Zaccaro, L. Chemical Modification for Proteolytic Stabilization of the Selective alphavbeta3 Integrin RGDechi Peptide: In Vitro and in Vivo Activities on Malignant Melanoma Cells. J. Med. Chem. 2017, 60, 9874–9884. [Google Scholar] [CrossRef]
- Hill, B.S.; Sarnella, A.; Capasso, D.; Comegna, D.; Del Gatto, A.; Gramanzini, M.; Albanese, S.; Saviano, M.; Zaccaro, L.; Zannetti, A. Therapeutic Potential of a Novel alphavbeta(3) Antagonist to Hamper the Aggressiveness of Mesenchymal Triple Negative Breast Cancer Sub-Type. Cancers (Basel) 2019, 11, 139. [Google Scholar] [CrossRef]
- Di Gaetano, S.; Del Gatto, A.; Pirone, L.; Comegna, D.; Zaccaro, L.; Saviano, M.; Arcà, B.; Capasso, D.; Pedone, E. A selective αvβ5 integrin antagonist hidden into the anophelin family protein cE5 from the malaria vector Anopheles gambiae. Pept. Sci. 2018, 110, e24054. [Google Scholar] [CrossRef]
- Solly, K.; Wang, X.; Xu, X.; Strulovici, B.; Zheng, W. Application of real-time cell electronic sensing (RT-CES) technology to cell-based assays. Assay Drug Dev. Technol. 2004, 2, 363–372. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Capasso, D.; Del Gatto, A.; Comegna, D.; Russo, L.; Fattorusso, R.; Saviano, M.; Di Gaetano, S.; Zaccaro, L. Selective Targeting of αvβ5 Integrin in HepG2 Cell Line by RGDechi15D Peptide. Molecules 2020, 25, 4298. https://doi.org/10.3390/molecules25184298
Capasso D, Del Gatto A, Comegna D, Russo L, Fattorusso R, Saviano M, Di Gaetano S, Zaccaro L. Selective Targeting of αvβ5 Integrin in HepG2 Cell Line by RGDechi15D Peptide. Molecules. 2020; 25(18):4298. https://doi.org/10.3390/molecules25184298
Chicago/Turabian StyleCapasso, Domenica, Annarita Del Gatto, Daniela Comegna, Luigi Russo, Roberto Fattorusso, Michele Saviano, Sonia Di Gaetano, and Laura Zaccaro. 2020. "Selective Targeting of αvβ5 Integrin in HepG2 Cell Line by RGDechi15D Peptide" Molecules 25, no. 18: 4298. https://doi.org/10.3390/molecules25184298
APA StyleCapasso, D., Del Gatto, A., Comegna, D., Russo, L., Fattorusso, R., Saviano, M., Di Gaetano, S., & Zaccaro, L. (2020). Selective Targeting of αvβ5 Integrin in HepG2 Cell Line by RGDechi15D Peptide. Molecules, 25(18), 4298. https://doi.org/10.3390/molecules25184298










