Impacts of Endocrine Disruptor di-n-Butyl Phthalate Ester on Microalga Chlorella vulgaris Verified by Approaches of Proteomics and Gene Ontology
Abstract
1. Introduction
2. Materials and Methods
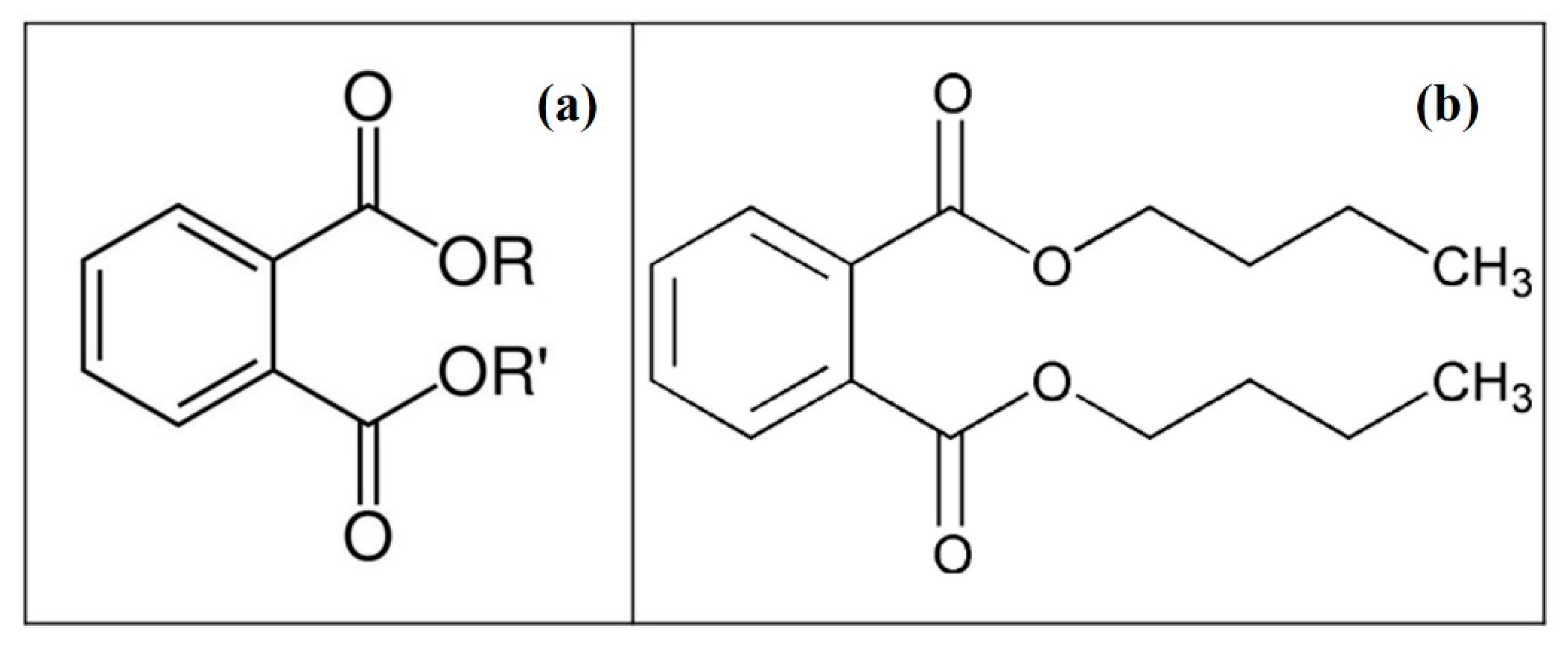
2.1. Chemicals
2.2. Cultivation of Microalgae
2.3. Experimental Design
2.4. Proteomics Analysis of C. vulgaris
2.4.1. Protein Extraction and Gel Electrophoresis
2.4.2. Nano-LC Separation and Mass Spectrometry
2.4.3. Label-Free Peptide Quantification and Identification
2.5. Proteomics and Gene Ontology Analysis
3. Results
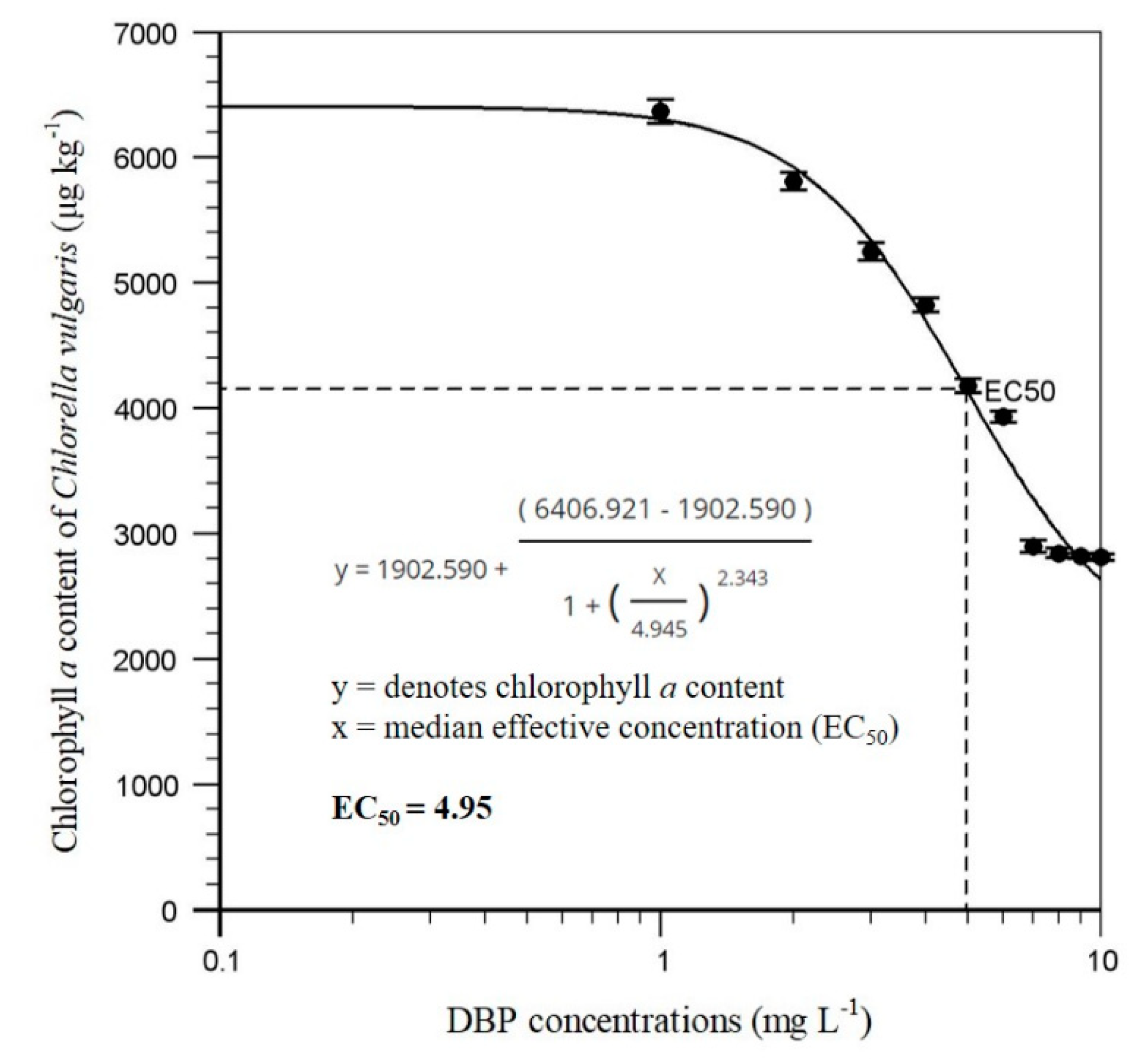
3.1. Algal Acute Biotoxicity Assays
3.2. Proteome Expression Analysis
3.3. Proteome Identification and GO Annotation and Comparison
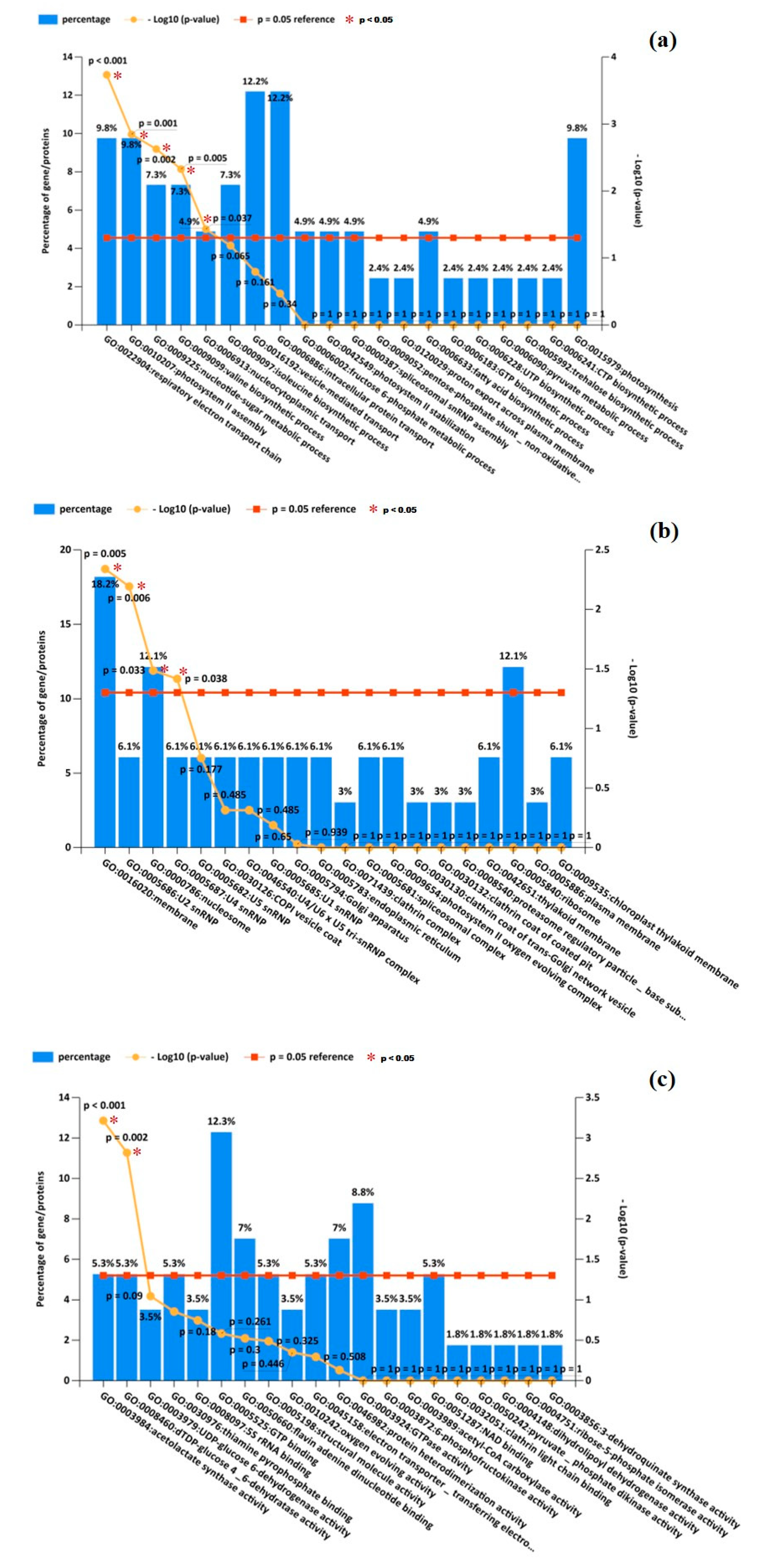
3.3.1. Biological Process (BP)
3.3.2. Cellular Component (CC)
3.3.3. Molecular Function (MF)
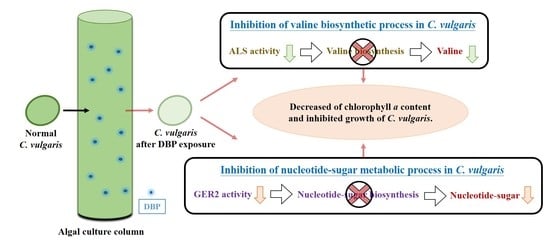
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mobin, S.; Alam, F. Some promising microalgal species for commercial applications: A review. Enegy. Proced. 2017, 110, 510–517. [Google Scholar] [CrossRef]
- Samolov, E.; Baumann, K.; Büdel, B.; Jung, P.; Leinweber, P.; Mikhailyuk, T.; Karsten, U.; Glaser, K. Biodiversity of algae and cyanobacteria in biological soil crusts collected along a climatic gradient in Chile using an integrative approach. Microorganisms 2020, 8, 1047. [Google Scholar] [CrossRef]
- Dolganyuk, V.; Belova, D.; Babich, O.; Prosekov, A.; Ivanova, S.; Katserov, D.; Patyukov, N.; Sukhikh, S. Microalgae: A Promising Source of Valuable Bioproducts. Biomolecules 2020, 10, 1153. [Google Scholar] [CrossRef]
- Oh-Hama, T.; Miyachi, S. Chlorella. In Microalgal Biotechnology; Borowitzka, M.A., Borowitzka, L.J., Eds.; Cambridge University Press: Cambridge, UK, 1988; pp. 3–26. [Google Scholar]
- Mobin, S.; Chowdhury, H.; Alam, F. Commercially important bioproducts from microalgae and their current applications-A review. Enegy. Proced. 2019, 160, 752–760. [Google Scholar] [CrossRef]
- Sadeghizadeh, A.; Moghaddasi, L.; Rahimi, R. CO2 capture from air by Chlorella vulgaris microalgae in an airlift photobioreactor. Bioresour. Technol. 2017, 243, 441–447. [Google Scholar] [CrossRef]
- Zhou, M.; He, H.; Jin, T.; Wang, H. Power generation enhancement in novel microbial carbon capture cells with immobilized Chlorella vulgaris. J. Power Sources 2012, 214, 216–219. [Google Scholar] [CrossRef]
- Singh, A.; Nigam, P.S.; Murphy, J.D. Renewable fuels from algae: An answer to debatable land based fuels. Bioresour. Technol. 2011, 102, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.G.; Dosoky, N.S.; Khan, M.S.; Zoromba, M.S.; Mekki, L.; El-Bana, M.; Nobles, D.; Shafik, H.M. High-Throughput Screening of Chlorella Vulgaris Growth Kinetics inside a Droplet-Based Microfluidic Device under Irradiance and Nitrate Stress Conditions. Biomolecules 2019, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Romero-Villegas, G.I.; Fiamengo, M.; Acién-Fernández, F.G.; Molina-Grima, E. Utilization of centrate for the outdoor production of marine microalgae at the pilot-scale in raceway photobioreactors. J. Environ. Manag. 2018, 228, 506–516. [Google Scholar] [CrossRef] [PubMed]
- Deprá, M.C.; Mérida, L.G.; de Menezes, C.R.; Zepka, L.Q.; Jacob-Lopes, E. A new hybrid photobioreactor design for microalgae culture. Chem. Eng. Res. Des. 2019, 144, 1–10. [Google Scholar] [CrossRef]
- Guzzon, A.; Di Pippo, F.; Congestri, R. Wastewater biofilm photosynthesis in photobioreactors. Microorganisms 2019, 7, 252. [Google Scholar] [CrossRef] [PubMed]
- Gong, N.; Shao, K.; Feng, W.; Lin, Z.; Liang, C.; Sun, Y. Biotoxicity of nickel oxide nanoparticles and bio-remediation by microalgae Chlorella vulgaris. Chemosphere 2011, 83, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Rudel, R.A.; Gray, J.M.; Engel, C.L.; Rawsthorne, T.W.; Dodson, R.E.; Ackerman, J.M.; Rizzo, J.; Nudelman, J.L.; Brody, J.G. Food packaging and bisphenol A and bis (2-ethyhexyl) phthalate exposure: Findings from a dietary intervention. Environ. Health Persp. 2011, 119, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Abdel Daiem, M.M.; Rivera-Utrilla, J.; Ocampo-Pérez, R.; Méndez-Díaz, J.D.; Sánchez-Polo, M. Environmental impact of phthalic acid esters and their removal from water and sediments by different technologies-A review. J. Environ. Manag. 2012, 109, 164–178. [Google Scholar] [CrossRef]
- Peng, X.; Feng, L.; Li, X. Pathway of diethyl phthalate photolysis in sea-water determined by gas chromatography-mass spectrometry and compound-specific isotope analysis. Chemosphere 2013, 90, 220–226. [Google Scholar] [CrossRef]
- Gallinger, Z.R.; Nguyen, G.C. Presence of phthalates in gastrointestinal medications: Is there a hidden danger? World J. Gastroenterol. 2013, 19, 7042–7047. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, P.; Wang, L.; Sun, G.; Zhao, J.; Zhang, H.; Du, N. The influence of facility agriculture production on phthalate esters distribution in black soils of northeast China. Sci. Total Environ. 2015, 506-507, 118–125. [Google Scholar] [CrossRef]
- Staples, C.A.; Peterson, D.R.; Parkerton, T.F.; Adams, W.J. The environmental fate of phthalate esters: A literature review. Chemosphere 1997, 35, 667–749. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, H.; Liang, Y.; Wang, C.; Liang, H.; Cai, H. Distribution of phthalate esters in the groundwater of Jianghan plain, Hubei, China. Front. Earth Sci. Chin. 2009, 3, 73–79. [Google Scholar] [CrossRef]
- Kaewlaoyoong, A.; Vu, C.T.; Lin, C.; Liao, C.S.; Chen, J.R. Occurrence of phthalate esters around the major plastic industrial area in southern Taiwan. Environ. Earth Sci. 2018, 77, 457. [Google Scholar] [CrossRef]
- Chen, C.F.; Ju, Y.R.; Lim, Y.; Chang, J.H.; Chen, C.W.; Dong, C.D. Spatial and temporal distribution of di-(2-ethylhexyl) phthalate in urban river sediments. Int. J. Environ. Res. Public Health 2018, 15, 2228. [Google Scholar] [CrossRef] [PubMed]
- Škrbić, B.D.; Ji, Y.; Đurišić-Mladenović, N.; Zhao, J. Occurence of the phthalate esters in soil and street dust samples from the Novi Sad city area, Serbia, and the influence on the children’s and adults’ exposure. J. Hazard. Mater. 2016, 312, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Bergh, C.; Luongo, G.; Wise, S.; Östman, C. Organophosphate and phthalate esters in standard reference material 2585 organic contaminants in house dust. Anal. Bioanal. Chem. 2012, 402, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Das, M.T.; Ghosh, P.; Thakur, I.S. Intake estimates of phthalate esters for South Delhi population based on exposure media assessment. Environ. Pollut. 2014, 189, 118–125. [Google Scholar] [CrossRef]
- Albar, H.M.S.A.; Ali, N.; Shahzad, K.; Ismail, I.M.I.; Rashid, M.I.; Wang, W.; Ali, L.N.; Eqani, S.A.M.A.S. Phthalate esters in settled dust of different indoor microenvironments; source of non-dietary human exposure. Microchem. J. 2017, 132, 227–232. [Google Scholar] [CrossRef]
- Sugatt, R.H.; O’Grady, D.P.; Banerjee, S.; Howard, P.H.; Gledhill, W.E. Shake flask biodegradation of 14 commercial phthalate esters. Appl. Environ. Microbiol. 1984, 47, 601–606. [Google Scholar] [CrossRef]
- Ding, M.; Kang, Q.; Zhang, S.; Zhao, F.; Mu, D.; Zhang, H.; Yang, M.; Hu, J. Contribution of phthalates and phthalate monoesters from drinking water to daily intakes for the general population. Chemosphere 2019, 229, 125–131. [Google Scholar] [CrossRef]
- Kelley, K.E.; Hernández-Díaz, S.; Chaplin, E.L.; Hauser, R.; Mitchell, A.A. Identification of phthalates in medications and dietary supplement formulations in the United States and Canada. Environ. Health Perspect. 2011, 120, 379–384. [Google Scholar] [CrossRef]
- Ji, Y.Q.; Wang, F.M.; Zhang, L.B.; Shan, C.Y.; Bai, Z.P.; Sun, Z.R.; Liu, L.L.; Shen, B.X. A comprehensive assessment of human exposure to phthalates from environmental media and food in Tianjin, China. J. Hazard Mater. 2014, 279, 133–140. [Google Scholar] [CrossRef]
- Gao, D.W.; Wen, Z.D. Phthalate esters in the environment: A critical review of their occurrence, biodegradation, and removal during wastewater treatment processes. Sci. Total Environ. 2016, 541, 986–1001. [Google Scholar] [CrossRef]
- Chi, J.; Li, Y.; Gao, J. Interaction between three marine microalgae and two phthalate acid esters. Ecotox. Environ. Saf. 2019, 170, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, N.; Zhao, J.; Ge, F.; Xu, Y.; Chen, Y. Disturbance of photosystem II-oxygen evolution complex induced the oxidative damage in Chlorella vulgaris under the stress of cetyltrimethylammonium chloride. Chemosphere 2019, 223, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass spectrometry-based proteomics. Nature 2003, 422, 198. [Google Scholar] [CrossRef]
- Liang, S.S.; Liao, W.T.; Kuo, C.J.; Chou, C.H.; Wu, C.J.; Wang, H.M. Phthalic Acid chemical probes synthesized for protein-protein interaction analysis. Int. J. Mol. Sci. 2013, 14, 12914–12930. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2016, 45, 331–338. [Google Scholar]
- Wang, Y.H.; Wu, C.M.; Wu, W.L.; Chu, C.P.; Chung, Y.J.; Liao, C.S. Survey on nitrogen removal and membrane filtration characteristics of Chlorella vulgaris Beij. on treating domestic type wastewaters. Water Sci. Technol. 2013, 68, 695–704. [Google Scholar] [CrossRef][Green Version]
- Feng, H.; Ruan, Y.; Wu, R.; Zhang, H.; Lam, P.K. Occurrence of disinfection by-products in sewage treatment plants and the marine environment in Hong Kong. Ecotox. Environ. Saf. 2019, 181, 404–411. [Google Scholar] [CrossRef]
- Arvola, L. Spectrophotometric determination of chlorophyll a and phaeopigments in ethanol extraction. Ann. Bot. Fenn. 1981, 18, 221–227. [Google Scholar]
- Wang, Y.; Sun, Q.; Li, Y.; Wang, H.; Wu, K.; Yu, C.P. Biotransformation of estrone, 17β-estradiol and 17α-ethynylestradiol by four species of microalgae. Ecotox. Environ. Saf. 2019, 180, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.S.; Nishikawa, Y.; Shih, Y.T. Characterization of di-n-butyl phthalate phytoremediation by Garden lettuce (Lactuca sativa L. var. longifolia) through Kinetics and proteome analysis. Sustainability 2019, 11, 1625. [Google Scholar] [CrossRef]
- Gu, S.; Zheng, H.; Xu, Q.; Sun, C.; Shi, M.; Wang, Z.; Li, F. Comparative toxicity of the plasticizer dibutyl phthalate to two freshwater algae. Aquat. Toxicol. 2017, 191, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.; Bisalputra, T.; Antia, N.J. Ultrastructural observation of the surface coat of Dunaliella tertiolecta from staining with cationic dyes and enzyme treatments. New Phytol. 1980, 85, 385–392. [Google Scholar] [CrossRef]
- Li, Z.; Yi, X.; Zhou, H.; Chi, T.; Li, W.; Yang, K. Combined effect of polystyrene microplastics and dibutyl phthalate on the microalgae Chlorella pyrenoidosa. Environ. Pollut. 2020, 257, 113604. [Google Scholar] [CrossRef] [PubMed]
- Ray, T.B. Site of action of chlorsulfuron: Inhibition of valine and isoleucine biosynthesis in plants. Plant Physiol. 1984, 75, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Ware, G.W.; Whitacre, D.M. An Introduction to Insecticides. The Pesticide Book; Meister Pro Information Resources: Willoughby, OH, USA, 2004; pp. 61–72. [Google Scholar]
- Heap, I. Herbicide resistant weeds. In Integrated Pest Management; Springer: Dordrecht, The Netherlands, 2014; pp. 281–301. [Google Scholar]
- Ginsburg, V.; Kirkman, H.N. Isolation of guanosine diphosphate fucose from Aerobacter aerogenes. J. Am. Chem. Soc. 1958, 80, 3481. [Google Scholar] [CrossRef]
- Rhomberg, S.; Fuchsluger, C.; Rendić, D.; Paschinger, K.; Jantsch, V.; Kosma, P.; Wilson, I.B. Reconstitution in vitro of the GDP-fucose biosynthetic pathways of Caenorhabditis elegans and Drosophila melanogaster. FEBS J. 2006, 273, 2244–2256. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: The freshwater unicellular green alga C. vulgaris Beij. (strain #3001) are available from the authors. |
| Sample | Concentration (μg μL−1) | Amount (μg) a |
|---|---|---|
| Con.1 | 9.97 | 997 |
| Con.2 | 13.09 | 5236 |
| Con.3 | 15.74 | 6296 |
| Exp.1 | 17.4 | 1740 |
| Exp.2 | 21.93 | 8772 |
| Exp.3 | 19.30 | 7720 |
| Expressed Proteins | Differential Expressed Proteins a | |||
|---|---|---|---|---|
| Exp. vs. Con. | Upregulation | Downregulation | Upregulation | Downregulation |
| 659 | 598 | 31 | 37 | |
| Protein-ID | Protein Name | Exp. Group Average a | Con. Group Average a | Exp. vs. Con. | Biological Process | Cellular Component | Molecular Function |
|---|---|---|---|---|---|---|---|
| A0A2P6TB75 | Small nuclear ribonucleoprotein E | 2,360,960.93 | 690,303.1496 | 3.42 | U2 snRNP (GO:0005686) U4 snRNP (GO:0005687) | ||
| E1ZJG0 | Small nuclear ribonucleoprotein E | 2,360,960.93 | 690,303.1496 | 3.42 | U2 snRNP (GO:0005686) U4 snRNP (GO:0005687) | ||
| E1Z2X3 | Histone H2A | 1,447,145,818 | 528,512,569.9 | 2.74 | nucleosome (GO:0000786) | ||
| E1ZGZ5 | Histone H2A | 1,447,145,818 | 528,512,569.9 | 2.74 | nucleosome (GO:0000786) | ||
| E1Z345 | Histone H2A | 1,763,428,632 | 695,597,118.3 | 2.54 | nucleosome (GO:0000786) | ||
| A0A2P6U370 | Putative histone H2A variant 3 | 1,763,428,632 | 695,597,118.3 | 2.54 | nucleosome (GO:0000786) | ||
| A0A2P6TN97 | Chloroplast ATP synthase subunit delta | 94,378,441.93 | 44,558,769.92 | 2.12 | membrane (GO:0016020) | ||
| E1ZUG4 | Uncharacterized protein | 94,378,441.93 | 44,558,769.92 | 2.12 | membrane (GO:0016020) | ||
| A0A2P6TDB7 | Dihydrolipoyl dehydrogenase | 188,274,201.3 | 95,700,812.66 | 1.97 | photosystem II assembly (GO:0010207) | ||
| E1ZQR2 | Uncharacterized protein | 187,902,521.2 | 95,700,812.66 | 1.96 | photosystem II assembly (GO:0010207) | ||
| E1ZA81 | Uncharacterized protein | 23,294,602.12 | 12,323,985.51 | 1.89 | photosystem II assembly (GO:0010207) | ||
| A0A2P6TVA7 | THYLAKOID chloroplastic | 23,294,602.12 | 12,323,985.51 | 1.89 | photosystem II assembly (GO:0010207) | ||
| A0A2P6TYV5 | Prohibitin-mitochondrial-like | 26,879,102.59 | 14,485,945.79 | 1.86 | membrane (GO:0016020) | ||
| A0A2P6U113 | GTP-binding nuclear Ran | 174,458,781 | 116,477,439.7 | 1.5 | nucleocytoplasmic transport (GO:0006913) | ||
| E1ZJ35 | GTP-binding nuclear protein | 174458781 | 116,477,439.7 | 1.5 | nucleocytoplasmic transport (GO:0006913) | ||
| E1ZSU0 | Acetolactate synthase | 13,983,262 | 23,251,122 | −1.66 | valine biosynthetic process (GO:0009099) | acetolactate synthase activity (GO:0003984) | |
| A0A2P6TWL0 | Guanylate-binding 4-like | 1,129,475.663 | 2,130,899.706 | −1.89 | membrane (GO:0016020) | ||
| E1Z5A9 | Uncharacterized protein | 1,129,475.663 | 2,130,899.706 | −1.89 | membrane (GO:0016020) | ||
| A0A2P6TBH4 | Acetolactate synthase | 4,860,187.934 | 11,528,724.53 | −2.37 | valine biosynthetic process (GO:0009099) | acetolactate synthase activity (GO:0003984) | |
| A0A2P6TBH6 | Acetolactate synthase | 4,860,187.934 | 11,528,724.53 | −2.37 | valine biosynthetic process (GO:0009099) | acetolactate synthase activity (GO:0003984) | |
| F2YGL6 | Cytochrome b6 | 6,172,062.4 | 15,644,836.86 | −2.53 | respiratory electron transport chain (GO:0022904) | ||
| A0A2I4S6M4 | Cytochrome b6 | 6,172,062.4 | 15,644,836.86 | −2.53 | respiratory electron transport chain (GO:0022904) | ||
| C0KRE8 | Cytochrome b6 (Fragment) | 6,172,062.4 | 15,644,836.86 | −2.53 | respiratory electron transport chain (GO:0022904) | ||
| A0A2P6U4I5 | GDP-L-fucose synthase 2 | 954,277.2115 | 2,692,354.388 | −2.82 | nucleotide-sugar metabolic process (GO:0009225) | dTDP-glucose 4, 6-dehydratase activity (GO:0008460) | |
| E1Z7Y1 | Uncharacterized protein | 954,277.2115 | 2,692,354.388 | −2.82 | nucleotide-sugar metabolic process (GO:0009225) | dTDP-glucose 4, 6-dehydratase activity (GO:0008460) | |
| A0A2P6TR93 | GDP-L-fucose synthase 2 | 954,277.2115 | 2,692,354.388 | −2.82 | nucleotide-sugar metabolic process (GO:0009225) | dTDP-glucose 4, 6-dehydratase activity (GO:0008460) | |
| P56321 | Cytochrome b6 | 3,652,260.183 | 11,782,273.75 | −3.23 | Respiratory electron transport chain (GO:0022904) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, C.-S.; Hong, Y.-H.; Nishikawa, Y.; Kage-Nakadai, E.; Chiou, T.-Y.; Wu, C.-C. Impacts of Endocrine Disruptor di-n-Butyl Phthalate Ester on Microalga Chlorella vulgaris Verified by Approaches of Proteomics and Gene Ontology. Molecules 2020, 25, 4304. https://doi.org/10.3390/molecules25184304
Liao C-S, Hong Y-H, Nishikawa Y, Kage-Nakadai E, Chiou T-Y, Wu C-C. Impacts of Endocrine Disruptor di-n-Butyl Phthalate Ester on Microalga Chlorella vulgaris Verified by Approaches of Proteomics and Gene Ontology. Molecules. 2020; 25(18):4304. https://doi.org/10.3390/molecules25184304
Chicago/Turabian StyleLiao, Chien-Sen, Yong-Han Hong, Yoshikazu Nishikawa, Eriko Kage-Nakadai, Tai-Ying Chiou, and Chien-Chang Wu. 2020. "Impacts of Endocrine Disruptor di-n-Butyl Phthalate Ester on Microalga Chlorella vulgaris Verified by Approaches of Proteomics and Gene Ontology" Molecules 25, no. 18: 4304. https://doi.org/10.3390/molecules25184304
APA StyleLiao, C.-S., Hong, Y.-H., Nishikawa, Y., Kage-Nakadai, E., Chiou, T.-Y., & Wu, C.-C. (2020). Impacts of Endocrine Disruptor di-n-Butyl Phthalate Ester on Microalga Chlorella vulgaris Verified by Approaches of Proteomics and Gene Ontology. Molecules, 25(18), 4304. https://doi.org/10.3390/molecules25184304