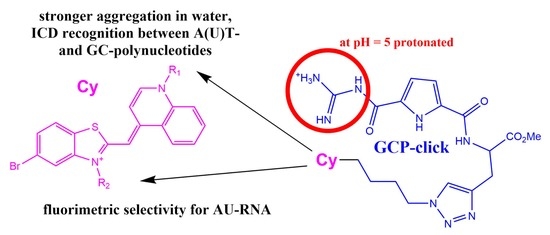
Fluorimetric and CD Recognition between Various ds-DNA/RNA Depends on a Cyanine Connectivity in Cyanine-guanidiniocarbonyl-pyrrole Conjugate
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Characterization of 1 and 2 in Aqueous Solutions
2.3. Interactions with ds-DNA, ds-RNA, and ss-RNA
2.3.1. Thermal Denaturation Experiments
2.3.2. Fluorimetric Titrations
2.3.3. Circular Dichroism (CD) Experiments
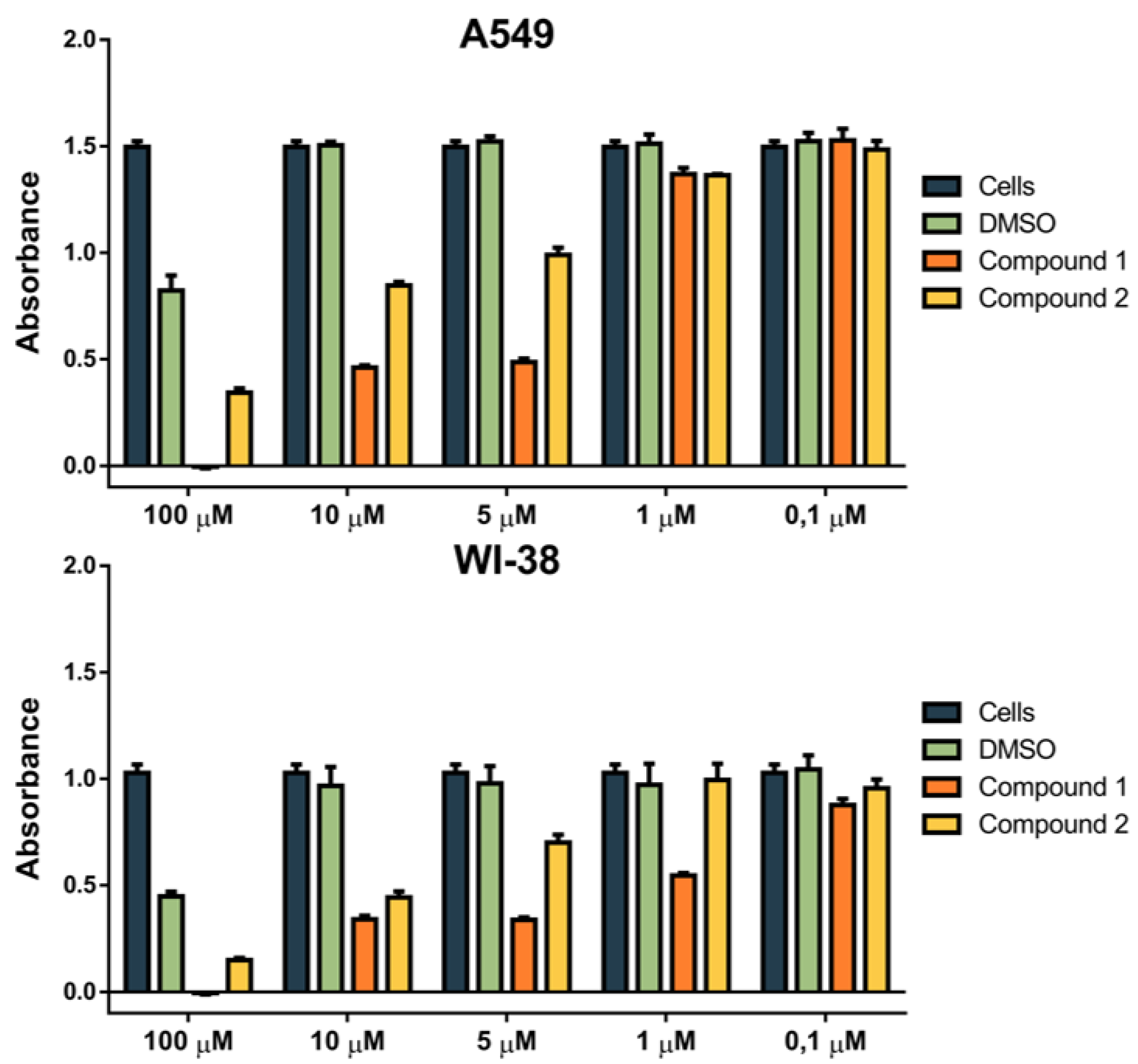
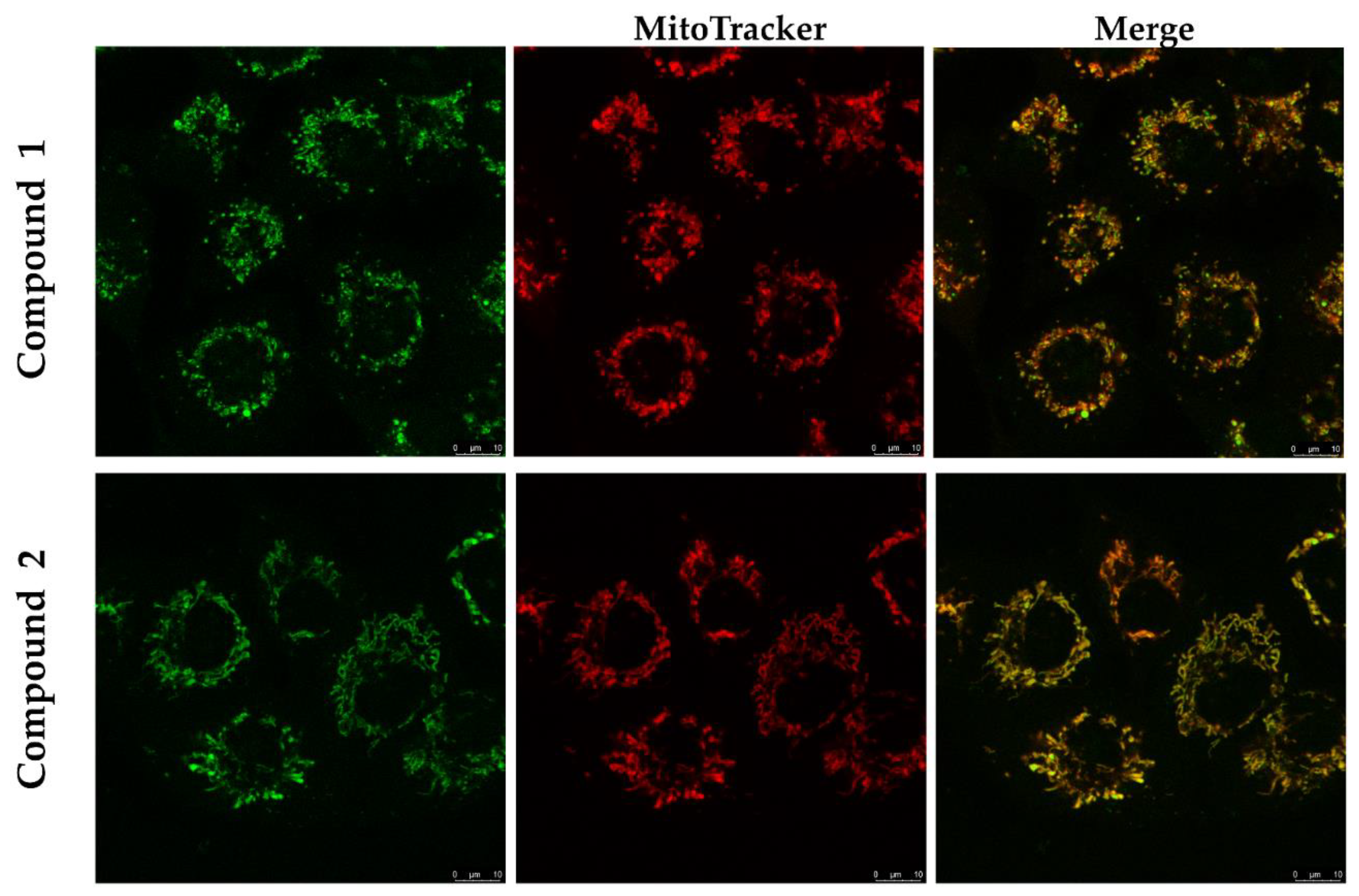
2.3.4. Biological Evaluation of 1 and 2 on Human Cell Lines
3. Conclusions
4. Materials and Methods
4.1. General Information
4.2. Synthesis
4.3. Study of DNA/RNA Interactions
4.4. Biological Evaluation of 1 and 2 on Human Cell Lines
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Drummen, G.P.C. Fluorescent Probes and Fluorescence (Microscopy) Techniques—Illuminating Biological and Biomedical Research. Molecules 2012, 17, 14067–14090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trinquet, E.; Mathis, G. Fluorescence technologies for the investigation of chemical libraries. Mol. Biosyst. 2006, 2, 381. [Google Scholar] [CrossRef] [PubMed]
- Bhaduri, S.; Ranjan, N.; Arya, D.P. An overview of recent advances in duplex DNA recognition by small molecules. Beilstein J. Org. Chem. 2018, 14, 1051–1086. [Google Scholar] [CrossRef] [PubMed]
- Demeunynck, M.; Bailly, C.; Wilson, W.D. (Eds.) DNA and RNA Binders; Wiley-VCH: Weinheim, Germany, 2002. [Google Scholar]
- Baylin, S.B.; Schuebel, K.E. The epigenomic era opens. Nature 2007, 448, 548. [Google Scholar] [CrossRef] [PubMed]
- Li, K.K.; Luo, C.; Wang, D.X.; Jiang, H.L.; Zheng, Y.G. Chemical and biochemical approaches in the study of histone methylation and demethylation. Med. Res. Rev. 2012, 32, 815. [Google Scholar] [CrossRef]
- Tatikolov, A. Polymethine dyes as spectral-fluorescent probes for biomacromolecules. J. Photochem. Photobiol. C Photochem. Rev. 2012, 13, 55–90. [Google Scholar] [CrossRef]
- Shindy, H.A. Basics, Mechanisms and Properties in the Chemistry of Cyanine Dyes: A Review Paper. Mini-Rev. Org. Chem. 2012, 9, 352–360. [Google Scholar] [CrossRef]
- Armitage, B. Cyanine Dye-DNA Interactions: Intercalation, Groove Binding and Aggregation. In DNA Binders and Related Subjects; Springer: Berlin/Heidelberg, Germany, 2005; pp. 55–76. [Google Scholar]
- Matić, J.; Šupljika, F.; Tandarić, T.; Dukši, M.; Piotrowski, P.; Vianello, R.; Brozović, A.; Piantanida, I.; Schmuck, C.; Radić Stojković, M. DNA/RNA recognition controlled by the glycine linker and the guanidine moiety of phenanthridine peptides. Int. J. Biol. Macromol. 2019, 134, 422–434. [Google Scholar] [CrossRef]
- Maity, D.; Matković, M.; Li, S.; Ehlers, M.; Wu, J.C.; Piantanida, I.; Schmuck, C. Peptide-Based Probes with an Artificial Anion-Binding Motif for Direct Fluorescence "Switch-On" Detection of Nucleic Acid in Cells. Chem. Eur. J. 2017, 23, 17356–17362. [Google Scholar] [CrossRef]
- Schmuck, C. A Journey through 12 Years of Interacting Molecules: From Artificial Amino Acid Receptors to the Recognition of Biomolecules and Switchable Nanomaterials. SYNLETT 2011, 2011, 1798–1815. [Google Scholar] [CrossRef]
- Hernandez-Folgado, L.; Baretić, D.; Piantanida, I.; Marjanović, M.; Kralj, M.; Rehm, T.; Schmuck, C. Guanidiniocarbonyl-pyrrole-aryl derivatives: Structure tuning for spectrophotometric recognition of specific DNA and RNA sequences and antiproliferative activity. Chem. A Eur. J. 2010, 16, 3036–3056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez-Folgado, L.; Schmuck, C.; Tomić, S.; Piantanida, I. A novel pyrene-guanidiniocarbonyl-pyrrole cation efficiently differentiates between ds-DNA and ds-RNA by two independent, sensitive spectroscopic methods. Bioorg. Med. Chem. Lett. 2008, 18, 2977–2981. [Google Scholar] [CrossRef] [PubMed]
- Radić-Stojković, M.; Piotrowski, P.; Schmuck, C.; Piantanida, I. Short, rigid linker between pyrene and guanidiniocarbonyl-pyrrole induced new set of spectroscopic responses to ds-DNA secondary structure. Org. Biomol. Chem. 2015, 13, 1629–1633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smidlehner, T.; Badovinac, M.; Piantanida, I. Pyrene-cyanine conjugates as multipurpose fluorescent probes for non-covalent recognition of ds-DNA, RNA and proteins. N. J. Chem. 2018, 42, 6655–6663. [Google Scholar] [CrossRef]
- Šmidlehner, T.; Karačić, Z.; Tomić, S.; Schmuck, C.; Piantanida, I. Fluorescent cyanine-guanidiniocarbonyl-pyrrole conjugate with pH-dependent DNA/RNA recognition and DPP III fluorescent labelling and inhibition properties. Monatsh. Chem. 2018, 149, 1307–1313. [Google Scholar] [CrossRef]
- Ćehić, M.; Suć Sajko, J.; Karačić, Z.; Piotrowski, P.; Šmidlehner, T.; Jerić, I.; Schmuck, C.; Piantanida, I.; Tomić, S. The guanidiniocarbonylpyrrole—Fluorophore conjugates as theragnostic tools for DPP III monitoring and inhibition. J. Biomol. Struct. Dyn. 2020, 38, 3790–3800. [Google Scholar] [CrossRef]
- Šmidlehner, T.; Kurutos, A.; Slade, J.; Belužić, R.; Ang, D.L.; Rodger, A.; Piantanida, I. Versatile Click Cyanine Amino Acid Conjugates Showing One-Atom-Influenced Recognition of DNA/RNA Secondary Structure and Mitochondrial Localisation in Living Cells. Eur. J. Org. Chem. 2018, 2018, 1682–1692. [Google Scholar] [CrossRef]
- Li, M.; Ehlers, M.; Schlesiger, S.; Zellermann, E.; Knauer, S.; Schmuck, C. Incorporation of a Non-Natural Arginine Analogue into a Cyclic Peptide Leads to Formation of Positively Charged Nanofibers Capable of Gene Transfection. Angew. Chem. Int. Ed. 2016, 55, 598–601. [Google Scholar] [CrossRef]
- Kuchelmeister, H.Y.; Karczewski, S.; Gutschmidt, A.; Knauer, S.; Schmuck, C. Utilizing Combinatorial Chemistry and Rational Design: Peptidic Tweezers with Nanomolar Affinity to DNA can be transformed into Efficient Vectors for Gene Delivery by Addition of a Lipophilic Tail. Angew. Chem. Int. Ed. Engl. 2013, 52, 14016–14020. [Google Scholar] [CrossRef]
- Šmidlehner, T.; Piantanida, I. Novel DNA/RNA-targeting amino acid beacon for the versatile incorporation at any position within the peptide backbone. Amino Acids 2017, 49, 1381–1388. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Lacroix, L. Analysis of Thermal Melting Curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.D.; Ratmeyer, L.; Zhao, M.; Strekowski, L.; Boykin, D. The search for structure-specific nucleic acid-interactive drugs: Effects of compound structure on RNA versus DNA interaction strength. Biochemistry 1993, 32, 4098–4104. [Google Scholar] [CrossRef] [PubMed]
- Cantor, C.R.; Schimmel, P.R. Biophysical Chemistry; WH Freeman and Company: San Francisco, CA, USA, 1980; Volume 3. [Google Scholar]
- Egli, M.; Saenger, W. Principles of Nucleic Acid Structure; Springer: Berlin/Heidelberg, Germany, 1983. [Google Scholar]
- Tumir, L.M.; Piantanida, I.; Cindrić, I.J.; Hrenar, T.; Meić, Z.; Žinić, M. New permanently charged phenanthridinium-nucleobase conjugates. Interactions with nucleotides and polynucleotides and recognition of ds-polyAH+. J. Phys. Org. Chem. 2003, 16, 891–899. [Google Scholar] [CrossRef]
- Scatchard, G. The attractions of proteins for small molecules and ions. Ann. N.Y. Acad. Sci. 1949, 51, 660–672. [Google Scholar] [CrossRef]
- McGhee, J.D.; Hippel, P.H.V. Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974, 86, 469–489. [Google Scholar] [CrossRef]
- Rodger, A.; Norden, B. Circular Dichroism and Linear Dichroism; Oxford University Press: New York, NY, USA, 1997; Chapter 2. [Google Scholar]
- Eriksson, M.; Nordén, B. Methods in Enzymology; Chaires, J.B., Waring, M.J., Eds.; Academic Press: Cambridge, MA, USA, 2001; Volume 340, pp. 68–98. [Google Scholar]
- Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Polarization spectroscopy methods in the determination of interactions of small molecules with nucleic acids–tutorial. Beil. J. Org. Chem. 2018, 14, 84–105. [Google Scholar] [CrossRef]
- Ban, Ž.; Žinić, B.; Vianello, R.; Schmuck, C.; Piantanida, I. Nucleobase–Guanidiniocarbonyl-Pyrrole Conjugates as Novel Fluorimetric Sensors for Single Stranded RNA. Molecules 2017, 22, 2213. [Google Scholar] [CrossRef] [Green Version]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol.-Cell Phys. 2011, 300, C723–C742. [Google Scholar] [CrossRef] [Green Version]
- Adler, J.; Pagakis, S.N.; Parmryd, I. Replicate-based noise corrected correlation for accurate measurements of colocalization. J. Microsc. 2008, 230, 121–133. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Liu, Z.N.; Lian, P.; Qian, J.; Li, X.W.; Wang, L.; Fu, W.; Chen, L.; Wei, X.B.; Li, C. Selective imaging and cancer cell death via pH switchable near-infrared fluorescence and photothermal effects. Chem. Sci. 2016, 7, 5995–6005. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Yang, C.X.; Chen, L.G.; Yan, X.P. Dual-stimuli responsive and reversibly activatable theranostic nanoprobe for precision tumor-targeting and fluorescence-guided photothermal therapy. Nat. Commun. 2017, 8, ARTN 14998. [Google Scholar] [CrossRef] [Green Version]
- Tan, X.; Luo, S.L.; Wang, D.C.; Su, Y.P.; Cheng, T.M.; Shi, C.M. A NIR heptamethine dye with intrinsic cancer targeting, imaging and photosensitizing properties. Biomaterials 2012, 33, 2230–2239. [Google Scholar] [CrossRef]
- Chaires, J.B.; Dattagupta, N.; Crothers, D.M. Studies on interaction of anthracycline antibiotics and deoxyribonucleic acid: Equilibrium binding studies on interaction of daunomycin with deoxyribonucleic. Biochemistry 1982, 21, 3933–3940. [Google Scholar] [CrossRef] [PubMed]
- Tumir, L.-M.; Piantanida, I.; Novak, P.; Žinić, M. Interactions of novel phenanthridinium-nucleobase conjugates with complementary and non-complementary nucleotides in aqueous media. J. Phys. Org. Chem. 2002, 15, 599–607. [Google Scholar] [CrossRef]
- Tumir, L.M.; Piantanida, I.; Juranović, I.; Meić, Z.; Tomić, S.; Žinić, M. Recognition of homo-polynucleotides containing adenine by phenanthridinium bis-uracil conjugate in aqueous media. Chem. Commun. 2005, 2561–2563. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |





| b r | ct-DNA | p(dAdT)2 | pApU | |
|---|---|---|---|---|
| 1, pH 7.0 | 0.1 | 2.0 | 0.6 | 1.0 |
| 0.2 | 2.0 | 0.3 | 1.0 | |
| 1, pH 5.0 | 0.1 | 2.0 | 1.3 | c 7.0; 2.0 |
| 0.2 | 2.0 | 2.6 | c 6.0; 1.0 | |
| 2, pH 7.0 | 0.1 | 1.1 | 1.0 | 2.0 |
| 0.2 | 1.7 | 1.1 | 3.0 | |
| 2, pH 5.0 | 0.1 | 2.0 | 1.0 | c 5.0; 2.0 |
| 0.2 | 6.0 | 1.1 | c 8.0; 1.0 |
| ctDNA | p(dAdT)2 | p(dGdC)2 | pApU | |
|---|---|---|---|---|
| 1, pH 7.0 | 6.1/120 | 6.2/40 | 6.4/160 | 6.2/450 |
| 1, pH 5.0 | 6.2/600 | 6.3/610 | 6.4/220 | 6.5/600 |
| 2, pH 7.0 | 6.2/300 | 6.1/270 | 6.2/480 | 5.6/500 |
| 2, pH 5.0 | 6.5/870 | 6.5/420 | 6.5/760 | 6.0/930 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šmidlehner, T.; Košćak, M.; Božinović, K.; Majhen, D.; Schmuck, C.; Piantanida, I. Fluorimetric and CD Recognition between Various ds-DNA/RNA Depends on a Cyanine Connectivity in Cyanine-guanidiniocarbonyl-pyrrole Conjugate. Molecules 2020, 25, 4470. https://doi.org/10.3390/molecules25194470
Šmidlehner T, Košćak M, Božinović K, Majhen D, Schmuck C, Piantanida I. Fluorimetric and CD Recognition between Various ds-DNA/RNA Depends on a Cyanine Connectivity in Cyanine-guanidiniocarbonyl-pyrrole Conjugate. Molecules. 2020; 25(19):4470. https://doi.org/10.3390/molecules25194470
Chicago/Turabian StyleŠmidlehner, Tamara, Marta Košćak, Ksenija Božinović, Dragomira Majhen, Carsten Schmuck, and Ivo Piantanida. 2020. "Fluorimetric and CD Recognition between Various ds-DNA/RNA Depends on a Cyanine Connectivity in Cyanine-guanidiniocarbonyl-pyrrole Conjugate" Molecules 25, no. 19: 4470. https://doi.org/10.3390/molecules25194470
APA StyleŠmidlehner, T., Košćak, M., Božinović, K., Majhen, D., Schmuck, C., & Piantanida, I. (2020). Fluorimetric and CD Recognition between Various ds-DNA/RNA Depends on a Cyanine Connectivity in Cyanine-guanidiniocarbonyl-pyrrole Conjugate. Molecules, 25(19), 4470. https://doi.org/10.3390/molecules25194470