Crystal Structure of Mannose Specific IIA Subunit of Phosphotransferase System from Streptococcus pneumoniae
Abstract
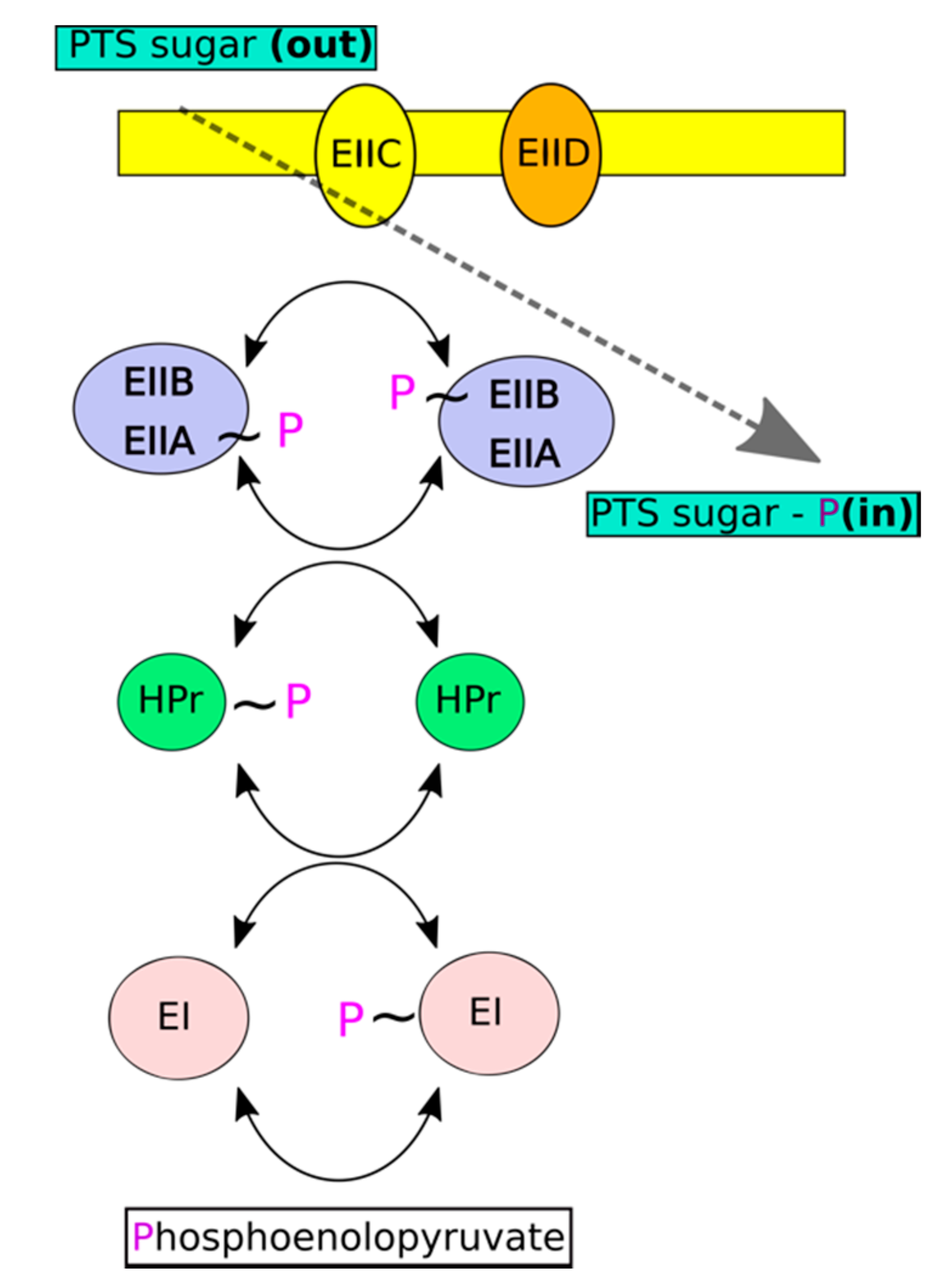
:1. Introduction
2. Results and Discussion
2.1. Expression and Crystallization of EII Components of S. pneumoniae Man-PTS
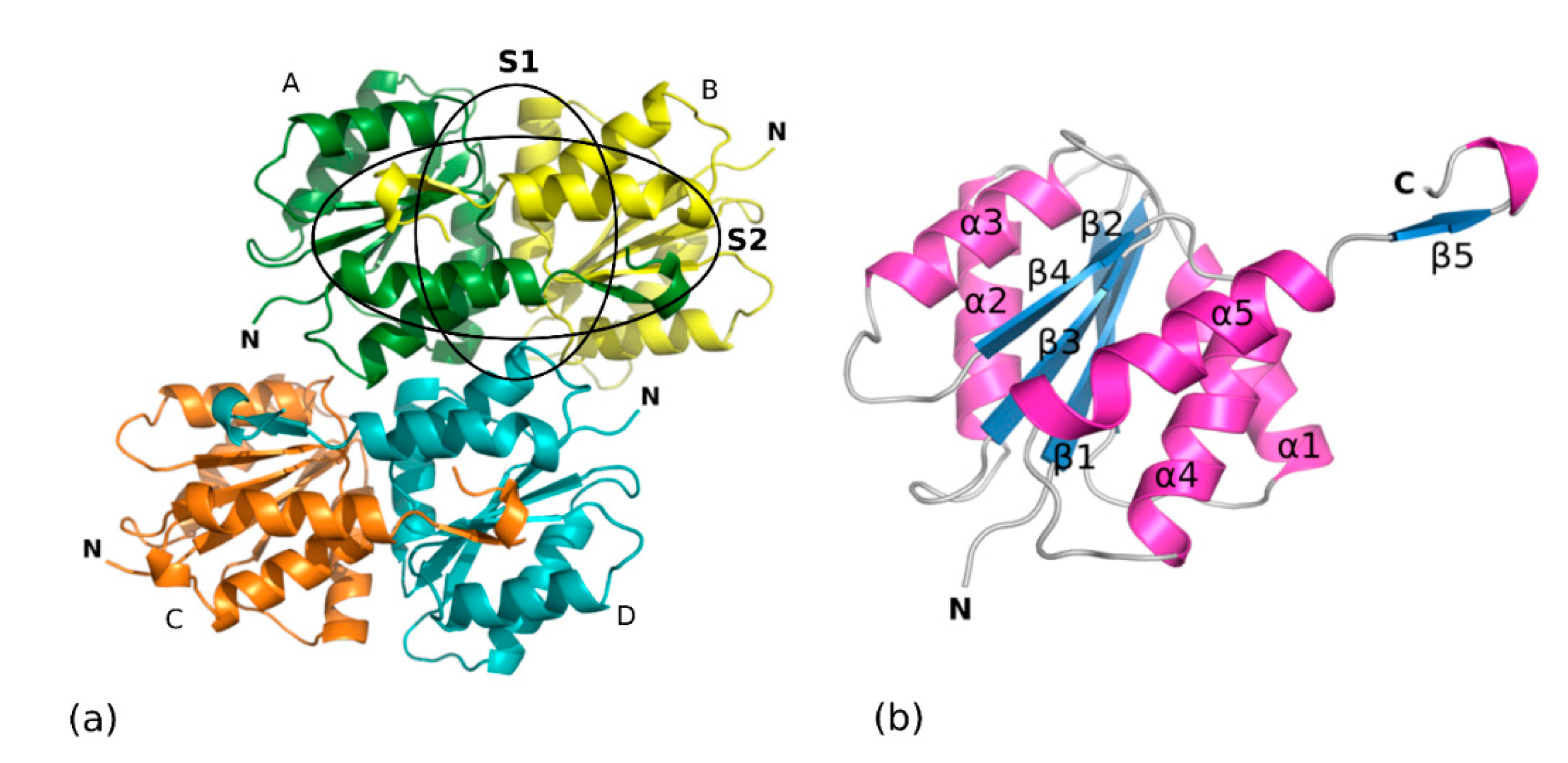
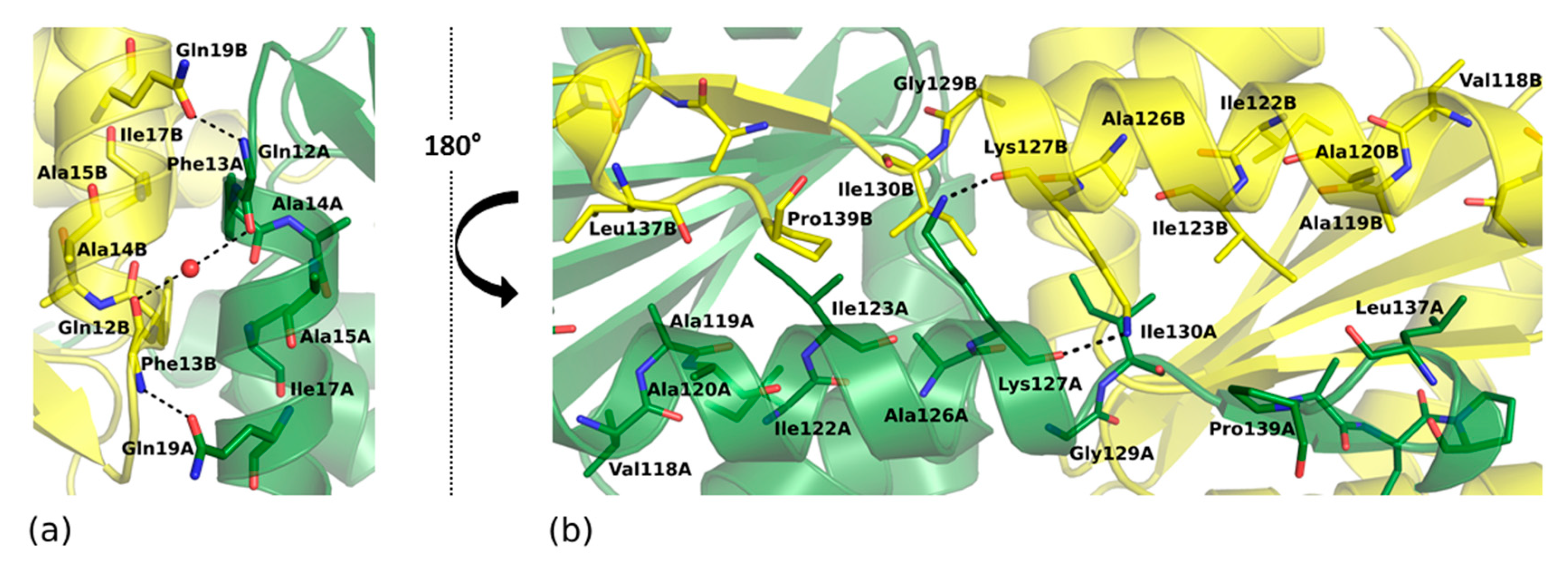
2.2. Overall Crystal Structure of SpEIIA-Man
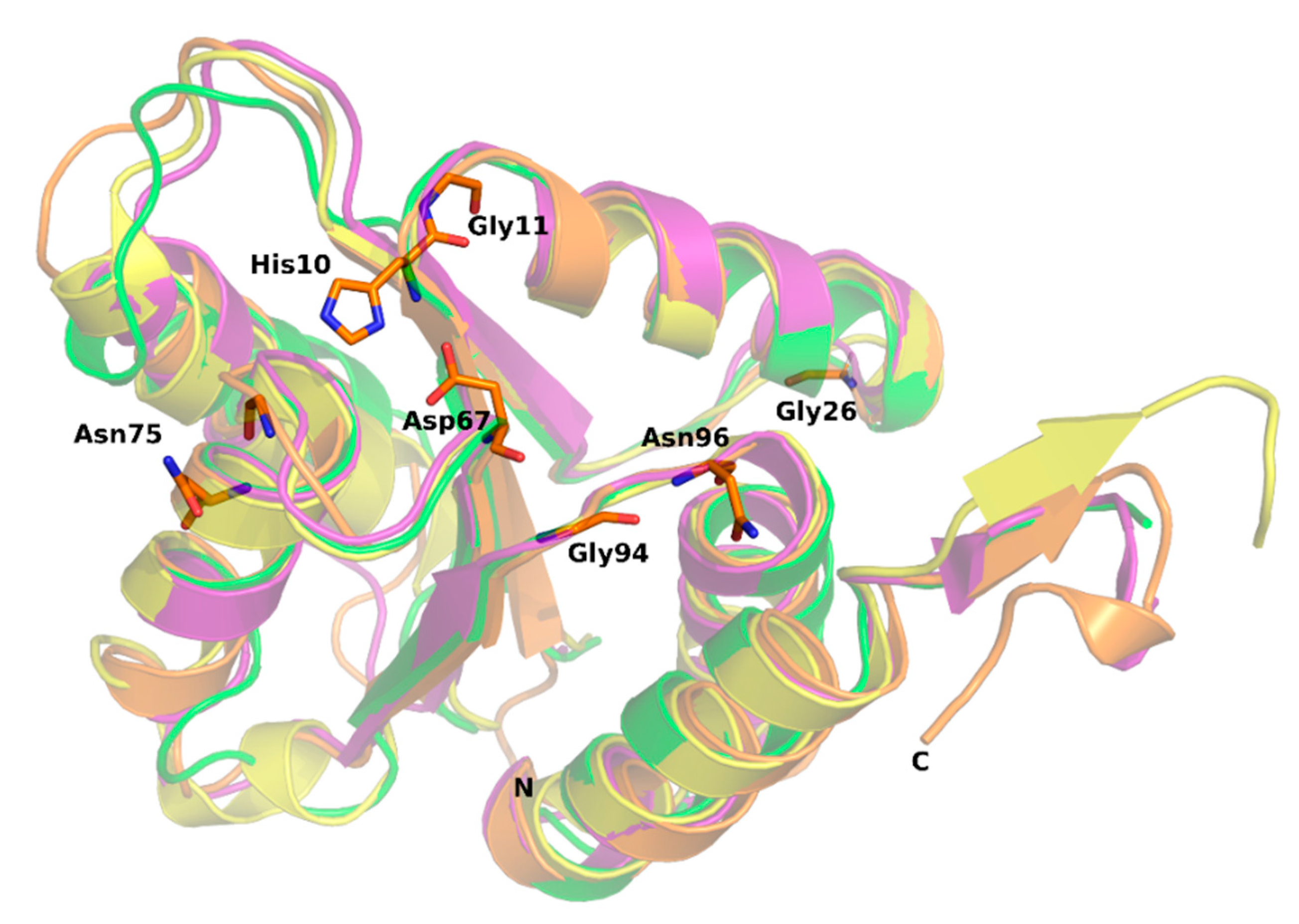
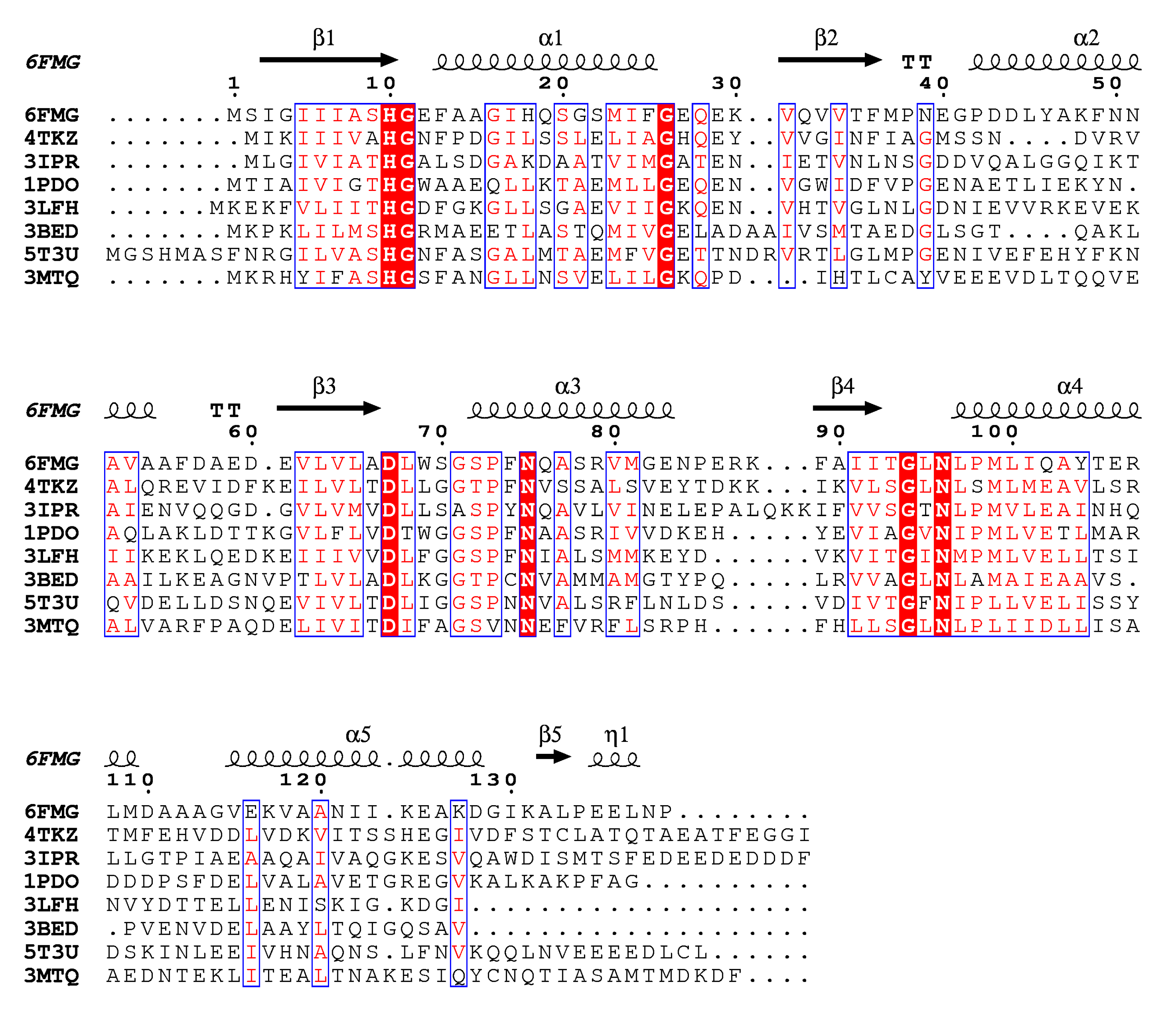
2.3. Structure Conservation among EIIA Components
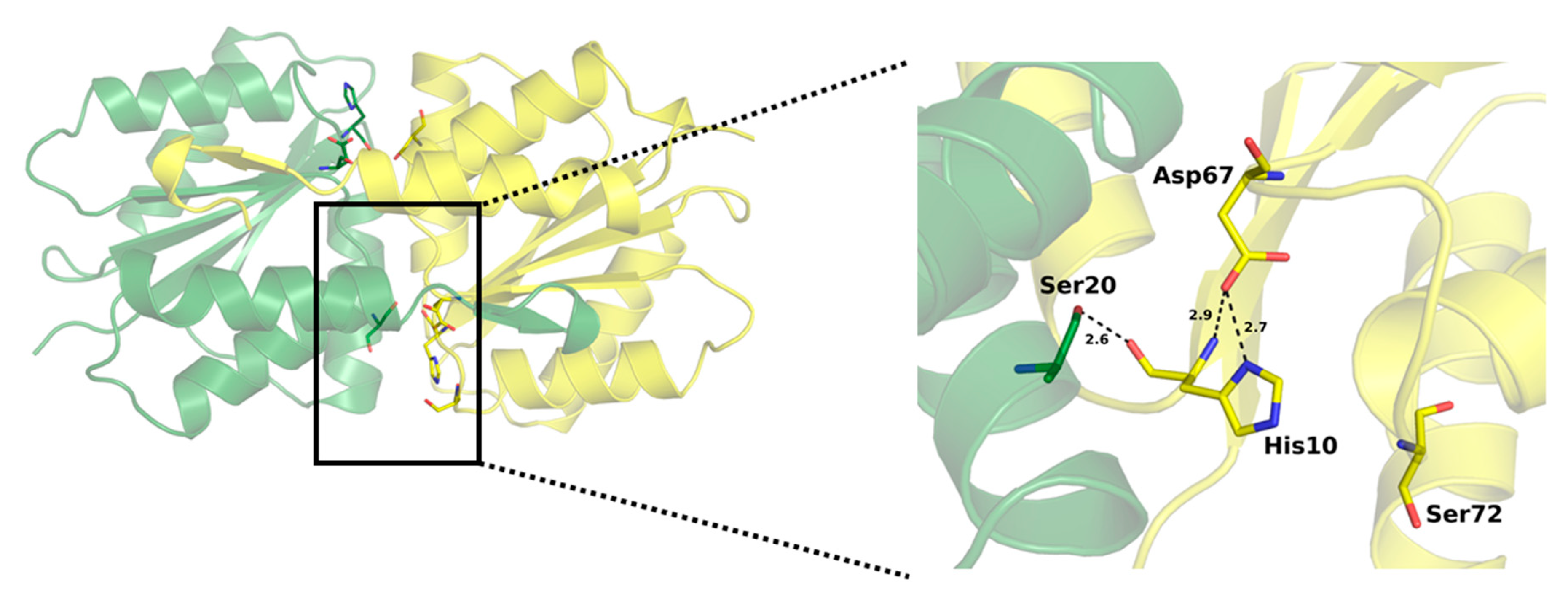
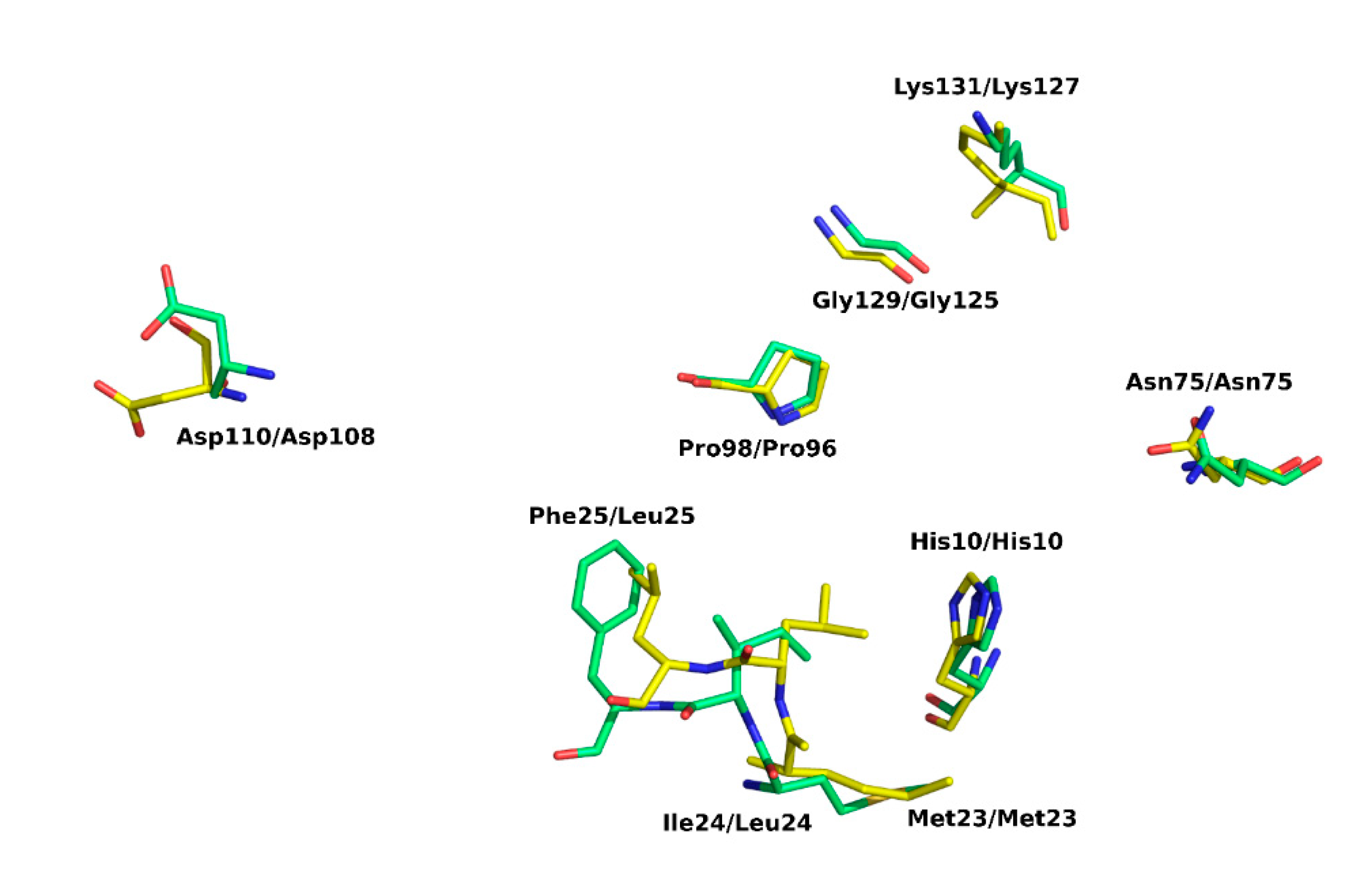
2.4. Active Site of SpEIIA-Man
3. Materials and Methods
3.1. Cloning, Expression and Purification of SpEIIAB-Man
3.2. Limited Proteolysis
3.3. Crystalization of SpEIIAB-Man
3.4. Data Collection and Crystal Structure Solution
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Walsh, R.L.; Camilli, A. Streptococcus pneumoniae is desiccation tolerant and infectious upon rehydration. MBio 2011, 2, e00092–e00111. [Google Scholar] [CrossRef] [Green Version]
- Henriques-Normark, B.; Tuomanen, E.I. The Pneumococcus: Epidemiology, microbiology, and pathogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a010215. [Google Scholar] [CrossRef]
- Lopian, L.; Elisha, Y.; Nussbaum-Shochat, A.; Amster-Choder, O. Spatial and temporal organization of the E. coli PTS components. EMBO J. 2010, 29, 3630–3645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deutscher, J.; Francke, C.; Postma, P.W. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 939–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Montfort, R.L.M.; Pijning, T.; Halk, K.H.; Hangyi, I.; Kouwijzer, M.L.C.E.; Robillard, G.T.; Dijkstra, B.W. The structure of the Escherichia coli phosphotransferase IIA mannitol reveals a novel fold with two conformations of the active site. Structure 1998, 6, 377–388. [Google Scholar] [CrossRef] [Green Version]
- Williams, D.C.; Cai, M.; Suh, J.Y.; Peterkofsky, A.; Clore, G.M. Solution NMR structure of the 48-kDa II mannose-HPr complex of the Escherichia coli mannose phosphotransferase system. J. Biol. Chem. 2005, 280, 20775–20784. [Google Scholar] [CrossRef] [Green Version]
- Siebold, C.; Flükiger, K.; Beutler, R.; Erni, B. Carbohydrate transporters of the bacterial phosphoenolpyruvate: Sugar phosphotransferase system (PTS). FEBS Lett. 2001, 504, 104–111. [Google Scholar] [CrossRef] [Green Version]
- Paixão, L.; Oliveira, J.; Veríssimo, A.; Vinga, S.; Lourenço, E.C.; Ventura, M.R.; Kjos, M.; Veening, J.-W.; Fernandes, V.E.; Andrew, P.W.; et al. Host glycan sugar-specific pathways in Streptococcus pneumonia: Galactose as a key sugar in colonisation and infection. PLoS ONE 2015, 10, e0127483. [Google Scholar] [CrossRef] [Green Version]
- Higgins, M.A.; Hamilton, A.M.; Boraston, A.B. Structural characterization of the PTS IIA and IIB proteins associated with pneumococcal fucose utilization. Proteins Struct. Funct. Bioinform. 2017, 85, 963–968. [Google Scholar] [CrossRef]
- Abranches, J.; Candella, M.M.; Wen, Z.T.; Baker, H.V.; Burne, R.A. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 2006, 188, 3748–3756. [Google Scholar] [CrossRef] [Green Version]
- Stewart, R.C. Kinetic characterization of phosphotransfer between CheA and CheY in the bacterial chemotaxis signal transduction pathway. Biochemistry 1997, 36, 2030–2040. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.C.; VanBruggen, R. Phosphorylation and binding interactions of CheY studied by use of Badan-labeled protein. Biochemistry 2004, 43, 8766–8777. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.; Baliga, N.S.; Camilli, A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 2005, 187, 8340–8349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufman, G.E.; Yother, J. CcpA-dependent and-independent control of beta-galactosidase expression in Streptococcus pneumoniae occurs via regulation of an upstream phosphotransferase system-encoding operon. J. Bacteriol. 2007, 189, 5183–5192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Limoli, D.H.; Sladek, J.A.; Fuller, L.A.; Singh, A.K.; King, S.J. BgaA acts as an adhesin to mediate attachment of some pneumococcal strains to human epithelial cells. Microbiology 2011, 157, 2369–2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Görke, B.; Stülke, J. Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008, 6, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Bidossi, A.; Mulas, L.; Decorosi, F.; Colomba, L.; Ricci, S.; Pozzi, G.; Deutscher, J.; Viti, C.; Oggioni, M.R. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS ONE 2012, 7, e33320. [Google Scholar] [CrossRef] [Green Version]
- Galinier, A.; Deutscher, J. Sophisticated regulation of transcriptional factors by the bacterial phosphoenolpyruvate: Sugar phosphotransferase system. J. Mol. Biol. 2017, 429, 773–789. [Google Scholar] [CrossRef]
- Giammarinaro, P.; Paton, J.C. Role of RegM, a homologue of the catabolite repressor protein CcpA, in the virulence of Streptococcus pneumoniae. Infect. Immun. 2002, 70, 5454–5461. [Google Scholar] [CrossRef] [Green Version]
- Diep, D.B.; Skaugen, M.; Salehian, Z.; Holo, H.; Nes, I.F. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. USA 2007, 104, 2384–2389. [Google Scholar] [CrossRef] [Green Version]
- Kjos, M.; Salehian, Z.; Nes, I.F.; Diep, D.B. An extracellular loop of the mannose phosphotransferase system component IIC is responsible for specific targeting by class IIa bacteriocins. J. Bacteriol. 2010, 192, 5906–5913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundar, G.S.; Islam, E.; Gera, K.A.; Breton, Y.E.; Mclver, K.S. PTS EII mutant library in Group A Streptococcus identifies a promiscuous man-family PTS transporter influencing SLS-mediated hemolysis. Mol. Microbiol. 2017, 103, 518–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schauder, S.; Nunn, R.S.; Lanz, R.; Erni, B. Crystal structure of the IIB subunit of a fructose permease (IIBLev) from Bacillus subtilis. J. Mol. Biol. 1998, 276, 591–602. [Google Scholar] [CrossRef]
- Orriss, G.L.; Erni, B.; Schirmer, T. Crystal Structure of the IIBSor domain of the sorbose permease from Klebsiella pneumoniae solved to 1.75A resolution. J. Mol. Biol. 2003, 327, 1111–1119. [Google Scholar] [CrossRef]
- Hu, J.; Hu, K.; Williams, D.C.; Komlosh, M.E.; Cai, M.; Clore, G.M. Solution NMR structures of productive and non-productive complexes between the A and B domains of the cytoplasmic subunit of the mannose transporter of the Escherichia coli phosphotransferase system. J. Biol. Chem. 2008, 283, 11024–11037. [Google Scholar] [CrossRef] [Green Version]
- Matthews, B.W. Solvent content of protein. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef]
- Vagin, A.; Lebedev, A. MoRDa, an automatic molecular replacement pipeline. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 28. [Google Scholar] [CrossRef] [Green Version]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Nunn, R.S.; Marković-Housley, Z.; Génovésio-Taverne, J.-C.; Flükiger, K.; Rizkallah, P.J.; Jansonius, J.N.; Schirmer, T.; Erni, B. Structure of the IIA domain of the mannose transporter from Escherichia coli at 1.7 A resolution. J. Mol. Biol. 1996, 259, 502–511. [Google Scholar] [CrossRef]
- Gutknecht, R.; Flu, K.; Lanz, R.; Erni, B. Mechanism of phosphoryl transfer in the dimeric IIAB man subunit of the Escherichia coli mannose transporter. J. Biol. Chem. 1999, 274, 6091–6096. [Google Scholar] [CrossRef] [Green Version]
- Reinelt, S.; Koch, B.; Hothorn, M.; Hengstenberg, W.; Welti, S.; Scheffzek, K. Structure of the Enterococcus faecalis EIIAgnt PTS component. Biochem. Biophys. Res. Commun. 2009, 388, 626–629. [Google Scholar] [CrossRef] [PubMed]
- Holm, L. DALI and the persistence of protein shape. Protein Sci. 2020, 29, 128–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, 320–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Frey, P.A.; Whitt, S.A.; Tobin, J.B. A low-barrier hydrogen bond in the catalytic triad of serine proteases author. Science 1994, 264, 1927–1930. [Google Scholar] [CrossRef] [PubMed]
- Leskovac, V.; Trivić, S.; Peričin, D.; Popović, M.; Kandrač, J. Short hydrogen bonds in the catalytic mechanism of serine proteases. J. Serb. Chem. Soc. 2008, 73, 393–403. [Google Scholar] [CrossRef]
- Stolz, B.; Huber, M.; Markovic-Housley, Z. The Mannose Transporter of Escherichia coli. Structure and function of the IIABMan subunit. J. Bio. Chem. 1993, 268, 27094–27099. [Google Scholar]
- Kabsch, W. Xds. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 72–82. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: Samples of the compounds are not available. |

| Space Group | P 63 |
|---|---|
| Cell Parameters | |
| a, b, c (Å ) | 138.53, 138.53, 69.77 |
| α, β, γ (°) | 90, 90, 120 |
| Wavelength (Å) | 0.9184 |
| Wilson B factor (Å2) | 35.85 |
| Resolution range (Å) | 26.179–1.800 (1.864–1.800) |
| Completeness (%) | 100.0 (100.0) |
| Rmerge (%) | 14.7 (205.2) |
| Rpim (%) | 3.3 (46.0) |
| Observed reflections | 1,461,081 (213,187) |
| Unique reflections | 70,756 (10,268) |
| I/sigma (I) | 14.57 (1.8) |
| Average multiplicity | 20.6 (20.8) |
| Refinement | |
| Resolution (Å) | 1.80 |
| No. of reflections used | 70,689 |
| Rfactor (%) | 17.02 |
| Rfree (%) | 19.65 |
| Average B factor (Å2) | |
| Protein | 31.75 |
| Water | 38.87 |
| RMSD from Ideal Values | |
| Bond length (Å) | 0.011 |
| Bond angles (°) | 1.088 |
| Ramachandran Statistics (%) | |
| Most favored regions | 97.10 |
| Additionally allowed regions | 2.90 |
| Content of the Asymmetric Unit | |
| No. of protein molecules/residues/non-H atoms | 4/556/8135 |
| No. of solvent molecules | 462 |
| PDB ID | Z-Scores | RMSD Å | Sequence Identity % |
|---|---|---|---|
| 4TKZ | 21.6 | 1.5 | 34 |
| 3IPR | 20.9 | 1.6 | 30 |
| 1PDO | 20.8 | 1.5 | 36 |
| 3LFH | 19.9 | 1.6 | 30 |
| 3BED | 19.0 | 1.7 | 26 |
| 5T3U | 18.2 | 2.1 | 29 |
| 3MTQ | 17.4 | 2.2 | 30 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magoch, M.; Nogly, P.; Grudnik, P.; Ma, P.; Boczkus, B.; Neves, A.R.; Archer, M.; Dubin, G. Crystal Structure of Mannose Specific IIA Subunit of Phosphotransferase System from Streptococcus pneumoniae. Molecules 2020, 25, 4633. https://doi.org/10.3390/molecules25204633
Magoch M, Nogly P, Grudnik P, Ma P, Boczkus B, Neves AR, Archer M, Dubin G. Crystal Structure of Mannose Specific IIA Subunit of Phosphotransferase System from Streptococcus pneumoniae. Molecules. 2020; 25(20):4633. https://doi.org/10.3390/molecules25204633
Chicago/Turabian StyleMagoch, Malgorzata, Przemyslaw Nogly, Przemyslaw Grudnik, Pikyee Ma, Bozena Boczkus, Ana Rute Neves, Margarida Archer, and Grzegorz Dubin. 2020. "Crystal Structure of Mannose Specific IIA Subunit of Phosphotransferase System from Streptococcus pneumoniae" Molecules 25, no. 20: 4633. https://doi.org/10.3390/molecules25204633
APA StyleMagoch, M., Nogly, P., Grudnik, P., Ma, P., Boczkus, B., Neves, A. R., Archer, M., & Dubin, G. (2020). Crystal Structure of Mannose Specific IIA Subunit of Phosphotransferase System from Streptococcus pneumoniae. Molecules, 25(20), 4633. https://doi.org/10.3390/molecules25204633





