The Regulation of Floral Colour Change in Pleroma raddianum (DC.) Gardner
Abstract
1. Introduction
2. Results
2.1. Chemical Profile
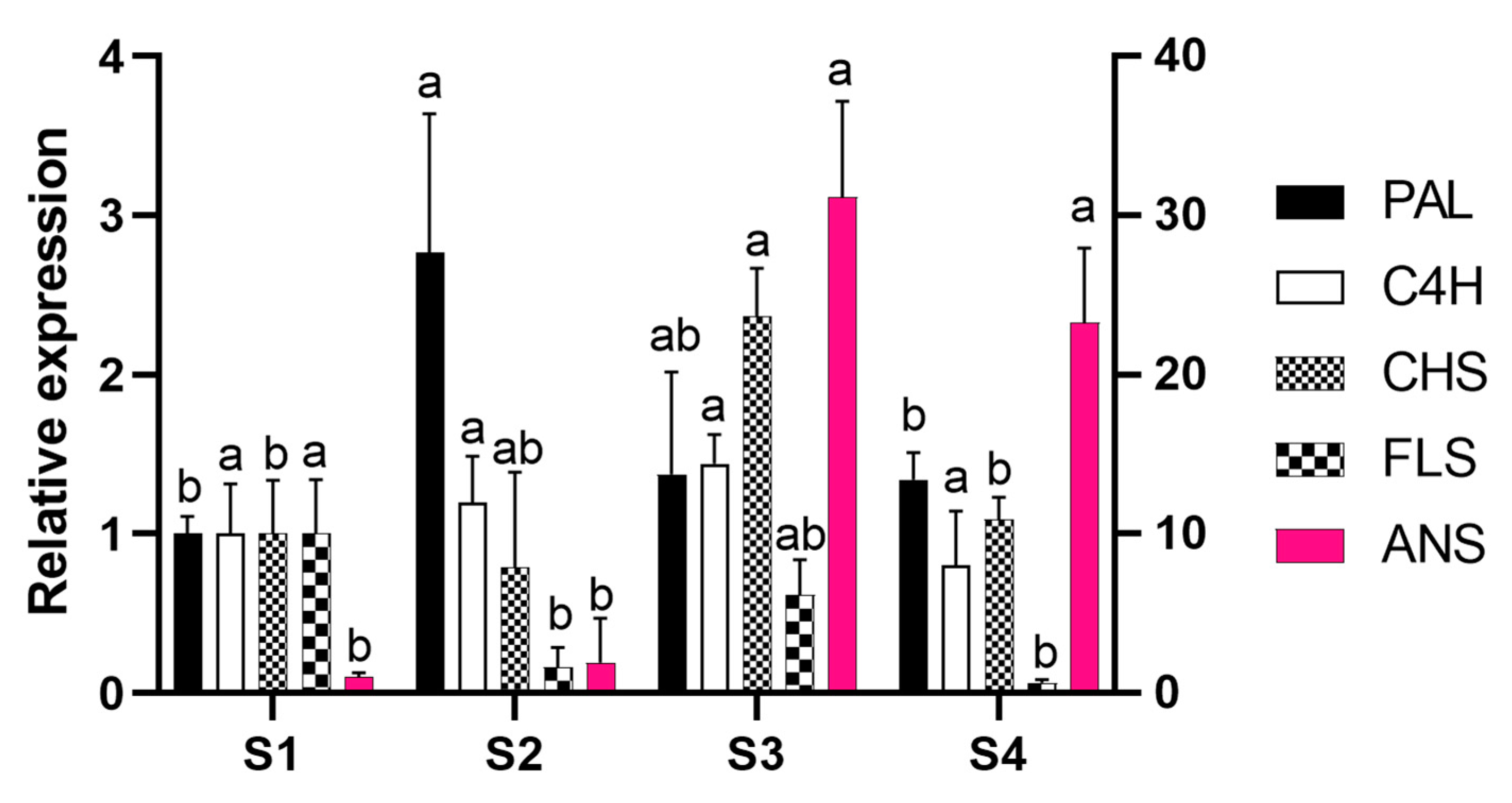
2.2. Gene Cloning and Expression Analysis
2.3. Metal Content
2.4. Colour Characterization of Petals
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Metabolic Profile
4.2.1. Pigment Profile
4.2.2. Primary Metabolism Profile
4.3. Gene Cloning and Expression Profile
4.3.1. Sequence Analysis for Primers Design
4.3.2. RNA Extraction and cDNA Synthesis
4.3.3. Cloning
4.3.4. Sequence Analyses
4.3.5. RT-qPCR
4.4. Metal Content
4.5. Flower Colour
4.6. Data Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guimarães, P.J.F.; Michelangeli, F.A.; Sosa, K.; Gómez, J.R.S. Systematics of Tibouchina and allies (Melastomataceae: Melastomateae): A new taxonomic classification. Taxon 2019, 68, 937–1002. [Google Scholar] [CrossRef]
- Rezende, F.M.; Ferreira, M.J.P.; Clausen, M.H.; Rossi, M.; Furlan, C.M. Acylated Flavonoid Glycosides are the Main Pigments that Determine the Flower Color of the Brazilian Native Tree Tibouchina pulchra (Cham.) Cogn. Molecules 2019, 24, 718. [Google Scholar] [CrossRef] [PubMed]
- Ruxton, G.D.; Schaefer, H.M. Floral colour change as a potential signal to pollinators. Curr. Opin. Plant Biol. 2016, 32, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Macnish, A.J.; Jiang, C.Z.; Zakharov-Negre, F.; Reid, M. Physiological and molecular changes during opening and senescence of Nicotiana mutabilis flowers. Plant Sci. 2010, 179, 267–272. [Google Scholar] [CrossRef]
- Gao, L.; Yang, H.; Liu, H.; Yang, J.; Hu, Y. Extensive Transcriptome Changes Underlying the Flower Color Intensity Variation in Paeonia ostia. Front. Plant Sci. 2016, 6, 1–16. [Google Scholar] [CrossRef]
- Teppabut, Y.; Oyama, K.; Kondo, T.; Yoshida, K. Change of Petals Color and Chemical components in Oenothera Flowers during Senescence. Molecules 2018, 23, 1698. [Google Scholar] [CrossRef]
- Guo, L.; Wang, Y.; da Silva, J.A.T.; Fan, Y.; Yu, X. Transcriptome and chemical analysis reveal putative genes involved in fower color change in Paeonia ‘Coral Sunset’. Plant Physiol. Biochem. 2019, 128, 130–139. [Google Scholar] [CrossRef]
- Vaknin, H.; Bar-Akiva, A.; Ovadia, R.; Nissim-Levi, A.; Forer, I.; Weiss, D.; Oren-Shamir, M. Active anthocyanin degradation in Brunfelsia calycina (yesterday-today-tomorrow) flowers. Planta 2005, 222, 19–26. [Google Scholar] [CrossRef]
- Trouillas, P.; Sancho-garc, J.C.; Freitas, V.D.; Gierschner, J.; Otyepka, M.; Dangles, O. Stabilizing and Modulating Color by Copigmentation: Insights from Theory and Experiment. Chem. Rev. 2016, 116, 4937–4982. [Google Scholar] [CrossRef]
- Brito, L.G.; Sazima, M. Tibouchina pulchra (Melastomataceae): Reproductive biology of a tree species at two sites of an elevational gradient in the Atlantic rainforest in Brazil. Plant Syst. Evol. 2012, 7, 1271–1279. [Google Scholar] [CrossRef]
- Brito, V.L.; Weynans, K.; Sazima, M.; Lunau, K. Trees as huge flowers and flowers as oversized floral guides: The role of floral color change and retention of old flowers in Tibouchina pulchra. Front. Plant Sci. 2015, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Farzad, M.; Griesbach, R.; Weiss, M.R. Floral color change in Viola cornuta L. (Violaceae): A model system to study regulation of anthocyanin production. Plant Sci. 2002, 162, 225–231. [Google Scholar] [CrossRef]
- Farzad, M.; Griesbach, R.; Hammond, J.; Weiss, M.R.; Elmendorf, H.G. Differential expression of three key anthocyanin biosynthetic genes in a color-changing flower, Viola cornuta cv. Yesterday, Today and Tomorrow. Plant Sci. 2003, 165, 1333–1342. [Google Scholar] [CrossRef]
- Han, M.; Yang, C.; Zhou, J.; Zhu, J.; Meng, J.; Shen, T.; Xin, Z.; Li, H. Analysis of flavonoids and anthocyanin biosynthesis-related genes expression reveals the mechanism of petal color fading of Malus hupehensis (Rosaceae). Braz. J. Bot. 2020, 43, 81–89. [Google Scholar] [CrossRef]
- Bar-Akiva, A.; Ovadia, R.; Rogachev, I.; Bar-Or, C.; Bar, E.; Freiman, Z.; Nissim-Levi, A.; Gollop, N.; Lewinsohn, E.; Aharoni, A.; et al. Metabolic networking in Brunfelsia calycina petals after flower opening. J. Exp. Bot. 2010, 61, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Zipor, G.; Shahar, L.; Ovadia, R.; Teper-bamnolker, P.; Levin, Y.; Doron-faigenboim, A.; Oren-shamir, M. In planta anthocyanin degradation by a vacuolar class III peroxidase in Brunfelsia calycina flowers. New Phytol. 2014, 205, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Miki, N.; Momonoi, K.; Kawachi, M.; Katou, K.; Okazaki, Y.; Uozumi, N.; Meshima, M.; Kondo, T. Synchrony between flower opening and petal-color change from red to blue in morning glory Ipomoea tricolor cv. heavenly blue. Proc. Jpn. Acad. Ser. B. 2009, 85, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Verweij, W.; Spelt, C.E.; Bliek, M.; de Vries, M.; Wit, N.; Faraco, M.; Koes, R.; Quattrocchio, F.M. Functionally similar WRKY proteins regulate vacuolar acidification in Petunia and hair development in Arabidopsis. Plant Cell 2016, 28, 786–803. [Google Scholar] [CrossRef]
- Shiono, M.; Matsugaki, N.; Takeda, K. Phytochemistry: Structure of the blue cornflower pigment. Nature 2005, 436, 791. [Google Scholar] [CrossRef]
- Yoshida, K.; Toyama-Kato, Y.; Kameda, K.; Kondo, T. Sepal color variation of Hydrangea macrophylla and vacuolar pH measured with a proton-selective microelectrode. Plant Cell Physiol. 2003, 44, 262–268. [Google Scholar] [CrossRef]
- Kondo, T.; Toyama-Kato, Y.; Yoshida, K. Essential structure of co-pigment for blue sepal-color development of hydrangea. Tetrahedron Lett. 2005, 46, 6645–6649. [Google Scholar] [CrossRef]
- Oyama, K.I.; Yamada, T.; Ito, D.; Kondo, T.; Yoshida, K. Metal Complex Pigment Involved in the Blue Sepal Color Development of Hydrangea. J. Agric. Food Chem. 2015, 63, 7630–7635. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Tao, J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 2015, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Neta, I.; Shoseyov, O.; Weiss, D. Sugars enhance the expression of gibberellin-induced genes in developing petunia flowers. Physiol. Plant. 2000, 109, 196–202. [Google Scholar] [CrossRef]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.-P.; Matros, A.; Peterek, S.; Schijlen, E.G.W.M.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotech. 2008, 26, 1301–1308. [Google Scholar] [CrossRef]
- Zheng, G.; Fan, C.; Di, S.; Wang, X.; Gao, L.; Dzyubenko, N.; Chapurin, V.; Pang, Y. Ectopic expression of tea MYB genes alter spatial flavonoid accumulation in alfafa (Medicago sativa). PLoS ONE 2019, 14, e0218336. [Google Scholar] [CrossRef]
- Mallona, I.; Lischewski, S.; Weiss, J.; Hause, B.; Egea-Cortines, M. Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol. 2010, 10, 1–11. [Google Scholar] [CrossRef]
- Saito, R.; Fukuta, N.; Ohmiya, A.; Itoh, Y.; Ozeki, Y.; Kuchitsu, K.; Nakayama, M. Regulation of anthocyanin biosynthesis involved in the formation of marginal picotee petals in Petunia. Plant Sci. 2006, 170, 828–834. [Google Scholar] [CrossRef]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef]
- Tornielli, G.; Koes, R.; Quattrocchio, F. The genetics of flower color. In Petunia: Evolutionary, Developmental and Physiological Genetics; Gerats, T., Strommer, J., Eds.; Springer: New York, NY, USA, 2009; pp. 269–299. ISBN 978-0-387-84796-2. [Google Scholar]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Chalker-Scott, L. Environmental Significance of Anthocyanins in Plant Stress Responses. Photochem. Photobiol. 1999, 70, 1–9. [Google Scholar] [CrossRef]
- Maier, A.; Schrader, A.; Kokkelink, L.; Falke, C.; Welter, B.; Iniesto, E.; Rubio, V.; Uhrig, J.F.; Hülskamp; Hoecker, U. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013, 74, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Maier, A.; Hoecker, U. COP1/SPA ubiquitin ligase complexes repress anthocyanin accumulation under low light and high light conditions. Plant Signal. Behav. 2015, 10, e970440. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gao, J.; Yin, K.; Wang, R.; Wang, C.; Petersen, M.; Mundy, J.; Qiu, J.L. MYB75 Phosphorylation by MPK4 Is Required for Light-Induced Anthocyanin Accumulation in Arabidopsis. Plant Cell 2016, 18, 2866–2883. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Hwang, G.; Lee, S.; Zhu, J.Y.; Paik, I.; Nguyen, T.T.; Oh, E. High ambient temperature represses anthocyanin biosynthesis through degradation of HY5. Front. Plant Sci. 2017, 8, 1787. [Google Scholar] [CrossRef]
- Shin, D.H.; Choi, M.G.; Kang, C.S.; Park, C.S.; Choi, S.B.; Park, Y.I. Overexpressing the wheat dihydroflavonol 4-reductase gene TaDFR increases anthocyanin accumulation in an Arabidopsis dfr mutant. J. Genet. Genom. 2016, 38, 333–340. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Y.; Wang, J.; Li, P.; Zhao, C.; Chen, Y.; Bi, Y. Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light in Arabidopsis seedlings. Plant Sci. 2015, 238, 64–72. [Google Scholar] [CrossRef]
- Pham, V.N.; Kathare, P.K.; Hug, E. Phytochromes and Phytochrome Interacting Factors. Plant Physiol. 2018, 176, 1025–1038. [Google Scholar] [CrossRef]
- Czemmel, S.; Heppel, S.C.; Bogs, J. R2R3 MYB transcription factors: Key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma 2012, 249, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.N.; Li, W.C.; Wang, H.C.; Shi, S.Y.; Shu, B.; Liu, L.Q. Transcriptome profiling of light-regulated anthocyanin biosynthesis in the pericarp of Litchi. Front. Plant Sci. 2016, 7, 963. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Xi, X.; Li, S.; Chen, W.; Zhang, B.; Liu, D.; Zhang, H. Allelic Variation and Transcriptional Isoforms of Wheat TaMYC1 Gene Regulating Anthocyanin Synthesis in Pericarp. Front. Plant Sci. 2017, 8, 1645. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Wang, M.; Tu, L.; Nie, Y.; Lin, Y.; Zhang, X. The Flavonoid Pathway Regulates the Petal Colors of Cotton Flower. PLoS ONE 2013, 8, e72364. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Mori, M.; Kondo, T. Blue flower color development by anthocyanins:from chemical structure to cell physiology. Nat. Prod. Rep. 2009, 26, 857–964. [Google Scholar] [CrossRef] [PubMed]
- Sigurdson, G.T.; Robbins, R.J.; Collins, T.M.; Giusti, M.M. Spectral and colorimetric characteristics of metal chelates of acylated cyanidin derivatives. Food Chem. 2017, 221, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Kitahara, S.; Ito, D.; Kondo, T. Ferric ions involved in the flower color development of the Himalayan blue poppy, Meconopsis grandis. Phytochemistry 2006, 67, 992–998. [Google Scholar] [CrossRef]
- Park, C.H.; Yeo, H.J.; Kim, N.S.; Park, Y.E.; Park, S.; Kim, J.K.; Park, S.U. Metabolomic profiling of the white, violet and red flowers of Rhododendron schlippenbachii Maxim. Molecules 2018, 23, 827. [Google Scholar] [CrossRef]
- Zhu, H.; Yang, J.; Xiao, C.; Mao, T.; Zhang, J.; Zhang, H. Differences in flavonoid pathway metabolites and transcripts affect yellow petal coloration in the aquatic plant Nelumbo nucifera. BMC Plant Biol. 2019, 19, 277. [Google Scholar] [CrossRef]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef]
- Hsieh, L.C.; Lin, S.I.; Shih, A.C.C.; Chen, J.W.; Lin, W.Y.; Tseng, C.Y.; Li, W.H.; Chiou, T.J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.-Y.; Felippes, F.F.; Liu, C.-J.; Weigel, D.; Wang, J.-W. Negative Regulation of Anthocyanin Biosynthesis in Arabidopsis by a miR156-Targeted SPL Transcription Factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef]
- Koseki, M.; Goto, K.; Masuta, C.; Kanazawa, A. The star-type color pattern in Petunia hybrida “Red Star” flowers is induced by sequence-specific degradation of chalcone synthase RNA. Plant Cell Physiol. 2005, 46, 1879–1883. [Google Scholar] [CrossRef] [PubMed]
- Pairoba, C.F.; Walbot, V. Post-transcriptional regulation of expression of the Bronze2 gene of Zea mays L. Plant Mol. Biol. 2003, 53, 75. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gou, M.; Liu, C.-J. Arabidopsis Kelch Repeat F-Box Proteins Regulate Phenylpropanoid Biosynthesis via Controlling the Turnover of Phenylalanine Ammonia-Lyase. Plant Cell 2013, 25, 4994–5010. [Google Scholar] [CrossRef]
- Suguiyama, V.F.; Silva, E.A.; Meirelles, S.T.; Centeno, D.C.; Braga, M.R. Leaf metabolite profile of the Brazilian resurrection plant Barbacenia purpurea Hook. (Velloziaceae) shows two time-dependent responses during desiccation and recovering. Front. Plant Sci. 2014, 5, 1–14. [Google Scholar] [CrossRef]
- Viegas, M.C.; Bassoli, D.G. Utilização do índice de retenção linear para caracterização de compostos voláteis em café solúvel utilizando GC-MS e coluna HP-Innowax. Quím. Nova 2007, 30, 2031–2034. [Google Scholar] [CrossRef]
- Wanner, L.A.; Li, G.; Ware, D.; Somssich, I.E.; Davis, K.R. The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 1995, 27, 327–338. [Google Scholar] [CrossRef]
- Bell-Lelong, D.A.; Cusumano, J.C.; Meyer, K.; Chapple, C. Cinnamate-4-hydroxylase expression in Arabidopsis (regulation in response to development and the environment). Plant Physiol. 1997, 113, 729–738. [Google Scholar] [CrossRef]
- Pelletier, M.K.; Murrell, J.R.; Shirley, B.W. Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis (Further evidence for differential regulation of “early” and “late” genes). Plant physiol. 1997, 113, 1437–1445. [Google Scholar] [CrossRef]
- Feinbaum, R.L.; Ausubel, F.M. Transcriptional Regulation of the Arabidopsis thaliana Chalcone Synthase Gene. Mol. Cell Biol. 1988, 8, 1985–1992. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D. Gapped BLAST and PSLBLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Plant Comparative Genomics Portal of the Department of Energy’s Joint Genome Institute. Phytozome v9.0. Available online: http://www.phytozome.net/ (accessed on 2 October 2014).
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Oligo Analyzer Tool. Available online: https://www.idtdna.com/pages/tools/oligoanalyzer (accessed on 4 October 2014).
- Chang, S.; Puryear, J.; Cairney, J. A simple and Efficient Method for Isolating RNA from Pine Trees. Plant. Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Bertani, G. Studies on lysogenesis: The Mode of Phage Liberation by Lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293. [Google Scholar] [CrossRef]
- Plant Comparative Genomics portal of the Department of Energy’s Joint Genome Institute. Phytozome v12.1. Available online: http://www.phytozome.net/ (accessed on 24 November 2017).
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; Van Den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef]
- Quadrana, L.; Almeida, J.; Otaiza, S.N.; Duffy, T.; Corrêa da Silva, J.V.; de Godoy, F.; Asís, R.; Bermúdez, L.; Fernie, A.R.; Carrari, F.; et al. Transcriptional regulation of tocopherol biosynthesis in tomato. Plant Mol. Biol. 2013, 81, 309–325. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Di Rienzo, J.A.; Casanoves, F.; Balzarini, M.G.; Gonzalez, L.; Tablada, M.; Robledo, Y.C. InfoStat Versión 2011. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. 2011, Volume 8, pp. 195–199. Available online: http://www. infostat.com.ar (accessed on 13 November 2014).
- Gonnet, J.F. Color effects of co-pigmentation of anthocyanins revisited-1. A colorimetric definition using the CIELab scale. Food Chem. 1998, 63, 409–415. [Google Scholar] [CrossRef]
Sample Availability: Samples of S1 to S4 (~20 mg each n = 4) and freeze-dried pink petals (~1 g of mixed S3, and S4) are available from the authors. |


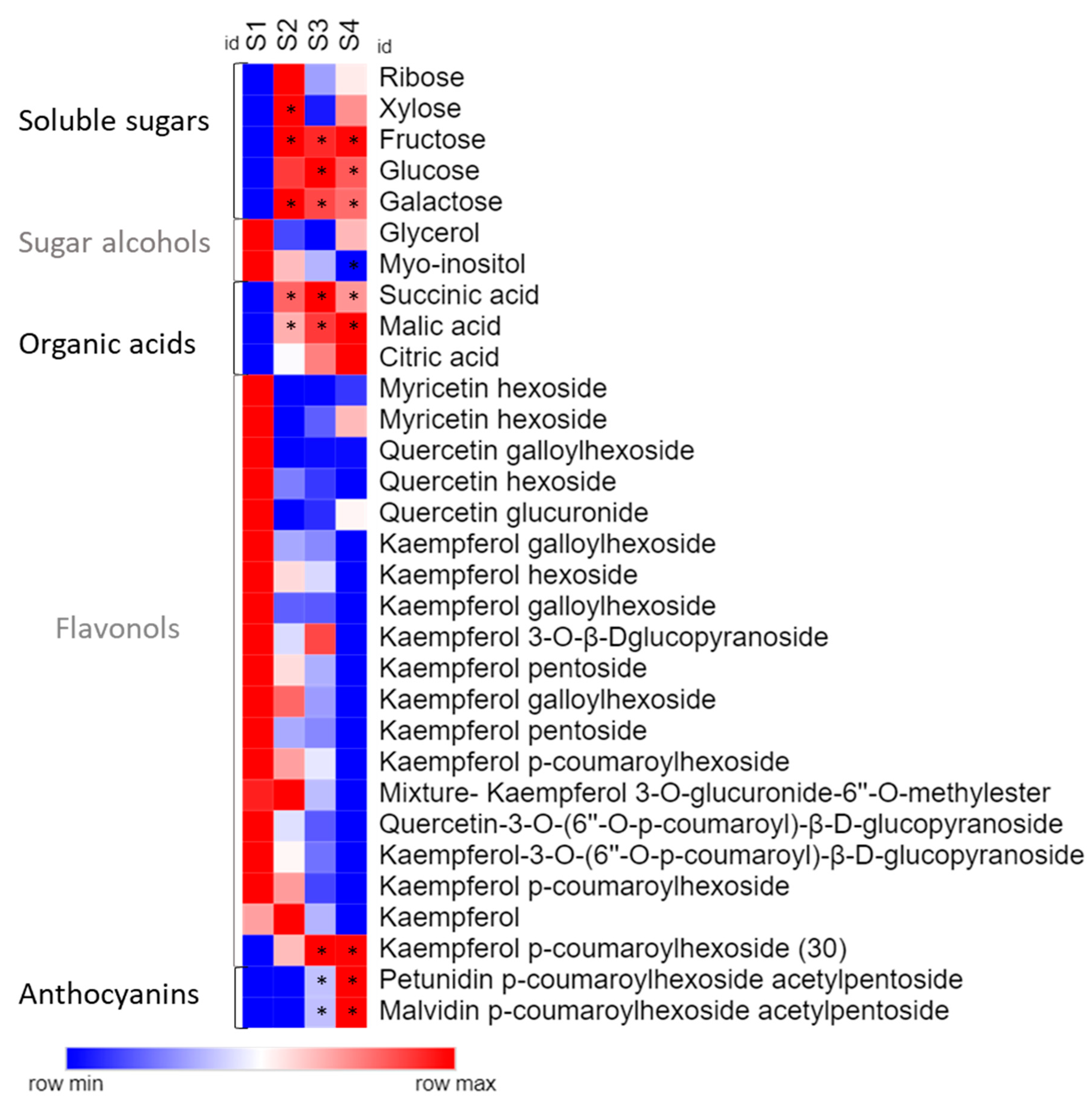

| Micronutrients | S1 (mg g−1 DW) | S2 (mg g−1 DW) | S3 (mg g−1 DW) | S4 (mg g−1 DW) |
| Cu2+ | 0.006 ± 0.001 a | 0.007 ± 0.002 a | 0.006 ± 0.001 a | 0.008 ± 0.002 a |
| Fe3+ | 0.062 ± 0.026 b | 0.116 ± 0.040 b | 0.114 ± 0.024 b | 0.220 ± 0.043 a |
| Mn2+ | 0.012 ± 0.007 a | 0.010 ± 0.006 a | 0.007 ± 0.002 a | 0.010 ± 0.004 a |
| Zn | 0.019 ± 0.003 a | 0.025 ± 0.011 a | 0.021 ± 0.003 a | 0.025 ± 0.004 a |
| Macronutrients | S1 (mg g−1 DW) | S2 (mg g−1 DW) | S3 (mg g−1 DW) | S4 (mg g−1 DW) |
| K+ | 10.207 ± 1.773 a | 11.245 ± 2.546 a | 9.731 ± 0.432 a | 11.029 ± 1.088 a |
| Ca2+ | 2.357 ± 0.402 a | 2.251 ± 0.5111 a | 2.099 ± 0.265 a | 2.051 ± 0.447 a |
| Mg2+ | 1.169 ± 0.276 a | 1.181 ± 0.289 a | 0.980 ± 0.131 a | 0.9028 ± 0.083 a |
| Na+ | 0.969 ± 0.378 a | 0.829 ± 0.244 a | 1.020 ± 0.208 a | 0.956 ± 0.483 a |
| S2 | S3 | S4 | |
|---|---|---|---|
| a * | −1.93 ± 0.60 c | 21.31 ± 1.31 b | 41.59 ± 3.97 a |
| b * | 7.544 ± 0.88 c | −10.69 ± 0.58 b | −20.47 ± 1.04 a |
| L * | 93.92 ± 0.28 a | 46.36 ± 3.99 c | 64.65 ± 2.32 b |
| hab | 104.09 ± 3.31 b | 333.35 ± 0.35 a | 333.72 ± 1.14 a |
| C * | 7.80 ± 0.98 c | 23.85 ± 1.42 b | 77.77 ± 0.68 a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rezende, F.M.; Clausen, M.H.; Rossi, M.; Furlan, C.M. The Regulation of Floral Colour Change in Pleroma raddianum (DC.) Gardner. Molecules 2020, 25, 4664. https://doi.org/10.3390/molecules25204664
Rezende FM, Clausen MH, Rossi M, Furlan CM. The Regulation of Floral Colour Change in Pleroma raddianum (DC.) Gardner. Molecules. 2020; 25(20):4664. https://doi.org/10.3390/molecules25204664
Chicago/Turabian StyleRezende, Fernanda Mendes, Mads Hartvig Clausen, Magdalena Rossi, and Cláudia Maria Furlan. 2020. "The Regulation of Floral Colour Change in Pleroma raddianum (DC.) Gardner" Molecules 25, no. 20: 4664. https://doi.org/10.3390/molecules25204664
APA StyleRezende, F. M., Clausen, M. H., Rossi, M., & Furlan, C. M. (2020). The Regulation of Floral Colour Change in Pleroma raddianum (DC.) Gardner. Molecules, 25(20), 4664. https://doi.org/10.3390/molecules25204664







