Synthesis, Crystal Structure, and Computational Methods of Vanadium and Copper Compounds as Potential Drugs for Cancer Treatment
Abstract
:1. Introduction
2. Results
2.1. Visible Spectroscopy
2.2. FTIR Spectroscopy
2.3. Theoretical Calculations
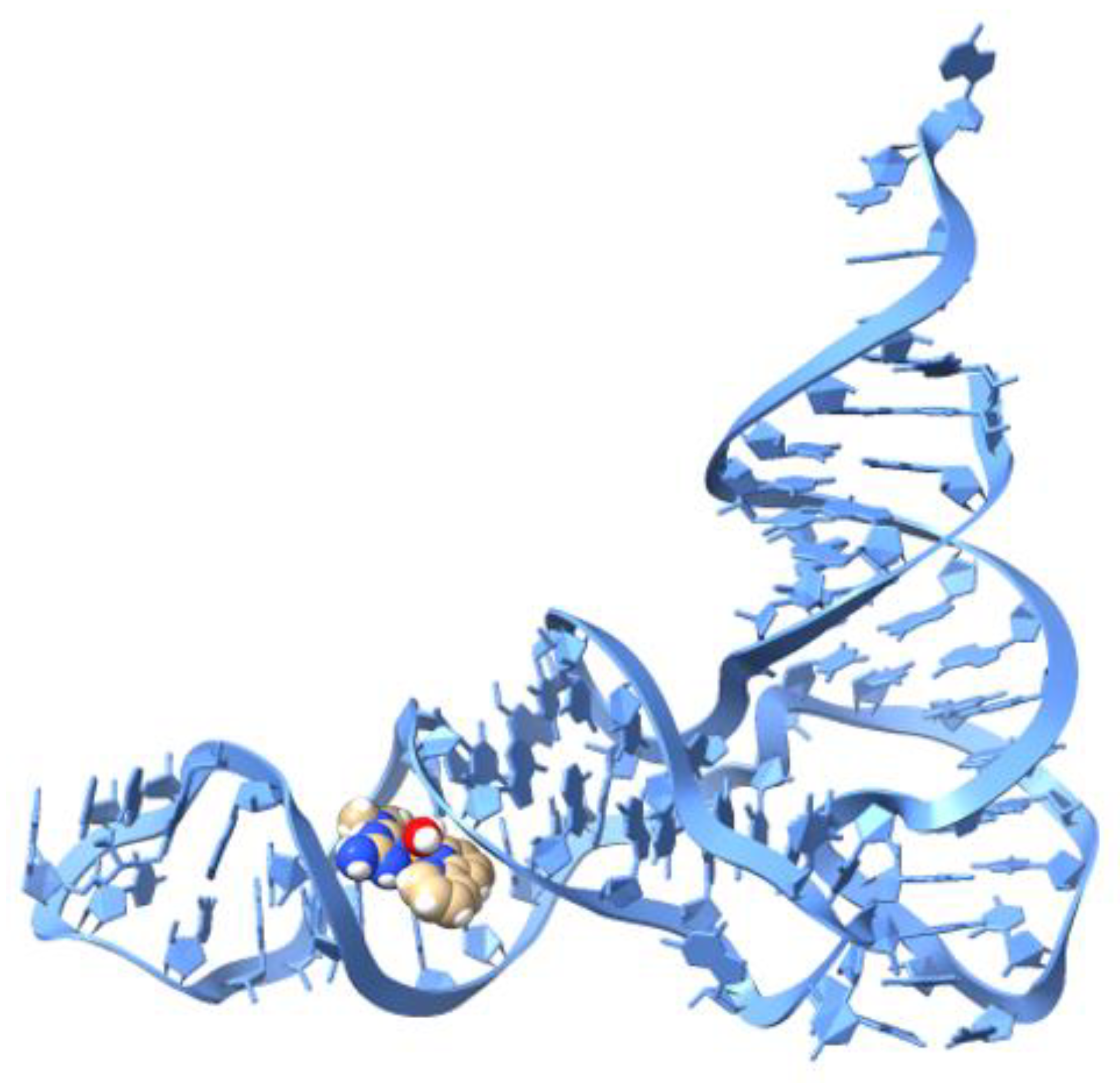
2.4. Molecular Docking (DNA)
| Ligand | Binding Energy (kcal/mol) 1BNA [52] | Binding Energy (kcal/mol) 151D [53] | Interaction |
|---|---|---|---|
| Doxorubicine | −11.09 | −11.54 | H bond, π-anion |
| 1’ [Cu(Metf)(bipy)(H2O)]2+ | −9.69 | −7.05 | H bond, salt-bridge |
| 2. [Cu(Impy)(Gly)(H2O)]+ | −8.82 | −6.73 | H bond, π-anion |
| 3. [Cu(phen)(Lys)(H2O)]2+ [37] | −11.03 | −9.98 | H Bond, π-anion, salt-bridge |
| 4. [Cu(bipy)(Orn)(H2O)]2+ [37] | −11.12 | −9.68 | H bond, salt-bridge, π-anion |
| 5. [Cu(phen)(Gly)(H2O)]+ [38] | −9.5 | −8.52 | H bond, |
| 6. [Cu(phen)(Orn))(H2O)]2+ [#] | −11.05 | −9.43 | H bond, salt-bridge, π-anion |
| 7. [Cu(bipy)(Lys)(H2O)]2+ [#] | −11.04 | −8.72 | H bond, salt-bridge |
| 8. [Cu(phen)2(H2O)]+ [54] | −8.79 | −8.53 | H bond, π-anion |
2.5. Molecular Docking (tRNA)
3. Discussion
4. Materials and Methods
4.1. Synthesis
4.2. Computational Methods
4.3. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- OPS/OMS, Programa de Cáncer. Available online: https://www.paho.org/hq/index.php?option=com_content&view=article&id=292:cancer-program&Itemid=3904&lang=es (accessed on 30 August 2020).
- Ezhilarasan, D.; Arumugham, M.N. Synthesis, characterization DNA binding and biological activity of Copper(II) complexes with mixed ligands. J. Chem. Biol. Phys. Sci. 2017, 7, 896–905. [Google Scholar]
- Spreckelmeyer, S.; Orvig, C.; Casini, A. Cellular transport mechanisms of cytotoxic metallodrugs: An overview beyond cisplatin. Molecules 2014, 19, 15584–15610. [Google Scholar] [CrossRef] [Green Version]
- Seiji, K.; Casini, A. Next-generation anti-cancer metallodrugs. Curr. Trends Med. Chem. 2012, 12, 219–235. [Google Scholar]
- Mjos, K.D.; Orvig, C. Metallodrugs in medicinal inorganic chemistry. Chem. Rev. 2014, 114, 4540–4563. [Google Scholar] [CrossRef]
- Ndagi, U.; Ndumiso, M.; Mahmoud, E.S. Metal complexes in cancer therapy–an update from drug design perspective. Drug Des. Dev. Ther. 2017, 11, 599–616. [Google Scholar] [CrossRef] [Green Version]
- Volarevic, V.; Djokovic, B.; Jankovic, M.G.; Harrell, C.R.; Fellabaum, C.; Djonov, V.; Arsenijevic, N. Molecular mechanisms of cisplatin-induced nephrotoxicity: A balance on the knife edge between renoprotection and tumor toxicity. J. Biomed. Sci. 2019, 26, 1–14. [Google Scholar] [CrossRef] [Green Version]
- McWhinney, S.R.; Goldberg, R.M.; McLeod, H.L. Platinum Neurotoxicity Pharmacogenetics. Mol. Cancer Ther. 2009, 8, 10–16. [Google Scholar] [CrossRef] [Green Version]
- Wheate, N.J.; Walker, S.; Craig, G.E.; Oun, R. The Status of Platinum Anticancer Drugs in the Clinic and in Clinical Trials. Dalton Trans. 2010, 39, 8113–8127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaál, A.; Orgován, G.; Mihucz, V.G.; Pape, I.; Ingerle, D.; Streli, C.; Szoboszlai, N.J. Metal Transport Capabilities of Anticancer Copper Chelators. Trace Elem. Med. Biol. 2018, 47, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Krajčiová, D.; Melník, M.; Havránek, E.; Forgácsová, A.; Mikuš, P. Copper compounds in nuclear medicine and oncology. J. Coord. Chem. 2014, 67, 1493–1519. [Google Scholar] [CrossRef]
- Dhakshanamoorthy, S.; Krishnan, M.M.; Arumugham, M.N. Synthesis, characterisation, DNA binding/cleavage, anticancer and antimicrobial activity of ternary copper(II) complexes. Asian J. Res. Chem. 2017, 10, 312–318. [Google Scholar] [CrossRef]
- Paterson, B.M.; Donnelly, P.S. Copper complexes of bis(thiosemicarbazones): From chemotherapeutics to diagnostic and therapeutic radiopharmaceuticals. Chem. Soc. Rev. 2011, 40, 3005–3018. [Google Scholar] [CrossRef]
- Ruiz-Azuara, L.; Bravo-Gómez, M.E. Copper compounds in cancer chemotherapy. Curr. Med. Chem. 2010, 17, 3606–3615. [Google Scholar] [CrossRef]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Manzano, C. Advances in copper complexes as anticancer agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef]
- Tardito, S.; Marchiò, L. Copper compounds in anti-cancer strategies. Curr. Med. Chem. 2009, 16, 1325–1348. [Google Scholar] [CrossRef]
- Shobha, C.; Thulasiram, B.; Aerva, R.R.; Nagababu, P. Recent Advances in Copper Intercalators as Anticancer Agents. J. Fluoresc. 2018, 28, 1195–1205. [Google Scholar] [CrossRef]
- Kuwabara, M.; Yoon, C.; Goyne, T.; Thederahn, T.; Sigman, D.S. Nuclease activity of 1, 10-phenanthroline-copper ion: Reaction with CGCGAATTCGCG and its complexes with netropsin and EcoRI. Biochemistry 1986, 25, 7401–7408. [Google Scholar] [CrossRef]
- Tabti, R.; Tounsi, N.; Gaiddon, C.; Bentouhami, E.; Désaubry, L. Progress in Copper Complexes as Anticancer Agents. Med. Chem. (Los Angeles) 2017, 7, 875–879. [Google Scholar] [CrossRef]
- Sigman, D.S.; Graham, D.R.; Aurora, V.D.; Stern, A.M. Oxygen-dependent cleavage of DNA by the 1,10-phenanthroline cuprous complex. Inhibition of Escherichia coli DNA polymerase I. J. Biol. Chem. 1979, 254, 12269–12272. [Google Scholar]
- Kwik, W.L.; Ang, K.P.; Chen, G. Complexes of (2,2′-bipyridyl) copper(II) and (1,10-phenanthroline) copper(II) with some amino acids. J. Inorg. Nucl. Chem. 1980, 42, 303–313. [Google Scholar] [CrossRef]
- Zelenko, O.; Gallagher, Y.X.; Sigman, D.S. Chemical Nuclease Activity of 1, 10-Phenanthroline−Copper. Isotopic Probes of Mechanism. Inorg. Chem. 1998, 37, 2198–2204. [Google Scholar] [CrossRef]
- Zhang, S.; Chun, X.; Chen, Y.; Zhou, J. Synthesis, Crystal Structure and DNA Cleavage Activity of a Ternary Copper(II) Complex of Dipyrido[3,2-d:2′,3′-f]-quinoxaline and Glycine. Chin. J. Chem. 2011, 29, 65–71. [Google Scholar] [CrossRef]
- Crans, D.C.; Henry, L.; Cardiff, G.; Posner, B.I. Developing vanadium as an antidiabetic or anti-cancer drug: A clinical and historical perspective. Met. Ions Life Sci. 2019. [Google Scholar] [CrossRef]
- Kowalski, S.; Wyrzykowski, D.; Inkielewicz-Stepniak, I. Molecular and Cellular Mechanisms of Cytotoxic Activity of Vanadium Compounds against Cancer Cells. Molecules 2020, 25, 1757. [Google Scholar] [CrossRef]
- Pessoa, J.C.; Etcheverry, S.; Gambino, D. Vanadium compounds in medicine. Coord. Chem. Rev. 2015, 301, 24–48. [Google Scholar] [CrossRef]
- Irving, E.; Stoker, A.W. Vanadium Compounds as PTP Inhibitors. Molecules 2017, 22, 2269. [Google Scholar] [CrossRef] [Green Version]
- Kioseoglou, E.; Petanidis, S.; Gabriel, C.; Salifoglou, A. The chemistry and biology of vanadium compounds in cancer therapeutics. Coord. Chem. Rev. 2015, 301, 87–105. [Google Scholar] [CrossRef]
- Correia, I.; Roy, S.; Matos, C.P.; Borovic, S.; Butenko, N.; Cavaco, I.; Marques, F.; Lorenzo, J.; Rodríguez, A.; Moreno, V.; et al. Vanadium(IV) and copper(II) complexes of salicylaldimines and aromatic heterocycles: Cytotoxicity, DNA binding, and DNA cleavage properties. J. Inorg. Biochem. 2015, 147, 134–146. [Google Scholar] [CrossRef]
- Novotny, L.; Kombian, S.B. Vanadium: Possible Use in Cancer Chemoprevention and Therapy. J. Cancer Res. Updates 2014, 3, 97–102. [Google Scholar] [CrossRef] [Green Version]
- Ling-Pan, L.; Feng-Zhi, S.; Ya-Li, F.; Li-Li, S.; Ying, L.; Yang-Jie, L.; Kai-Ti, W. Synthesis and biological evaluation of vanadium complex as novel antitumor agents. Eur. J. Med. Chem. 2019, 176, 1–10. [Google Scholar]
- Griffin, E.; Levina, A.; Lay, P.A. Vanadium(V) tris-3,5-di-tert-butylcatecholato complex: Links between speciation and antiproliferative activity in human pancreatic cancer cells. J. Inorg. Biochem. 2019, 201, 110815. [Google Scholar] [CrossRef]
- Rozzo, C.; Sanna, D.; Garribba, E.; Serra, M.; Cantara, A.; Palmieri, G.; Pisano, M. Antitumoral effect of vanadium compounds in malignant melanoma cell lines. J. Inorg. Biochem. 2017, 174, 14–24. [Google Scholar] [CrossRef]
- Pisano, M.; Arru, C.; Serra, M.; Galleri, G.; Sanna, D.; Garribba, E.; Palmieri, G.; Rozzo, C. Antiproliferative activity of vanadium compounds: Effects on the major malignant melanoma molecular pathways. Metallomics 2019, 11, 1687–1699. [Google Scholar] [CrossRef]
- León, I.L.; Ruiz, M.C.; Franca, C.A.; Parajón-Costa, B.S.; Baran, E.J. Metvan, bis(4,7-Dimethyl-1,10-phenanthroline) sulfatooxidovanadium(IV): DFT and Spectroscopic Study—Antitumor Action on Human Bone and Colorectal Cancer Cell Lines. Biol. Trace Elem. Res. 2018. [Google Scholar] [CrossRef]
- Martínez-Valencia, B.; Corona-Motolinia, N.D.; Sánchez-Lara, E.; Noriega, L.; Sánchez-Gaytán, B.L.; Castro, M.E.; Meléndez-Bustamante, F.J.; González-Vergara, E. Cyclo-tetravanadate bridged copper complexes as potential double bullet pro-metallodrugs for cancer treatment. J. Inorg. Biochem. 2020, 208, 111081. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Valencia, B.; Corona-Motolinia, N.D.; Sánchez-Lara, E.; Sánchez-Gaytán, B.L.; Cerro-López, M.; Mendoza, A.; Castro, M.E.; Meléndez-Bustamante, F.J.; González-Vergara, E. Synthesis and Experimental-Computational Characterization of a Copper/Vanadium Compound with Potential Anticancer Activity. Crystals 2020, 10, 492. [Google Scholar] [CrossRef]
- Zhang, W.; Lu, X.; Wang, G.; Cheng, Y.; Zhang, B. Methyl-substituted enhancement of antitumor activity in square-planar metal complex and analysis of DE, DG, CV, UV-vis, and luminescence. New J. Chem. 2015, 39, 4869–4875. [Google Scholar] [CrossRef]
- Sciortino, G.; Maréchal, J.D.; Fábián, I.; Lihi, N.; Garribba, E. Quantitative prediction of electronic absorption spectra of copper (II)–bioligand systems: Validation and applications. J. Inorg. Biochem. 2020, 204, 110953. [Google Scholar]
- Gunasekaran, S.; Natarajan, R.K.; Renganayaki, V.; Natarajan, S. Vibrational spectra and thermodynamic analysis of metformin. Indian J. Pure Appl. Phys. 2006, 44, 495–500. [Google Scholar]
- Sharma, R.P.; Ajnesh, S.; Venugopalan, P.; Dansby-Sparks, R.; Xue, Z.L.; Rossetti, S.; Ferretti, V. Stabilization of tetrameric metavanadate ion by tris (1, 10-phenanthroline) cobalt (III): Synthesis, spectroscopic, and X-ray structural study of [Co (phen) 3] 3 (V4O12) 2Cl·27H2O. J. Coord. Chem. 2010, 63, 3016–3027. [Google Scholar] [CrossRef] [Green Version]
- Yurdakul, Ş.; Badoğlu, S. FT-IR spectra, vibrational assignments, and density functional calculations of imidazo [1, 2-a] pyridine molecule and its Zn (II) halide complexes. Struct. Chem. 2009, 20, 423–434. [Google Scholar] [CrossRef]
- Ghiyasiyan-Arani, M.; Masjedi-Arani, M.; Salavati-Niasari, M. Facile synthesis, characterization and optical properties of copper vanadate nanostructures for enhanced photocatalytic activity. J. Mater. Sci. Mater. Electron. 2016, 27, 4871–4878. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Sánchez-Lara, E.; Martínez-Valencia, B.; Corona-Motolinia, N.D.; Sanchez-Gaytan, B.L.; Castro, M.E.; Bernes, S.; Méndez-Rojas, M.A.; Meléndez-Bustamante, F.J.; González-Vergara, E. A one-dimensional supramolecular chain based on [H2V10O28]4− units decorated with 4-dimethylaminopyridinium ions: An experimental and theoretical characterization. New J. Chem. 2019, 43, 17746–17755. [Google Scholar] [CrossRef]
- Afonin, A.V.; Vashchenko, A.V.; Sigalov, M.V. Estimating the energy of intramolecular hydrogen bonds from 1H NMR and QTAIM calculations. Org. Biomol. Chem. 2016, 14, 11199. [Google Scholar] [CrossRef]
- Jenkins, S.; Morrison, I. The chemical character of the intermolecular bonds of seven phases of ice as revealed by ab initio calculation of electron densities. Chem. Phys. Lett. 2000, 317, 97–102. [Google Scholar] [CrossRef]
- Thorn, C.F.; Oshiro, C.; Marsh, S.; Hernandez-Boussard, T.; McLeod, H.; Klein, T.E.; Altman, R.B. Doxorubicin pathways: Pharmacodynamics and adverse effects. Pharm. Genom. 2011, 21, 440. [Google Scholar] [CrossRef]
- Galindo-Murillo, R.; García-Ramos, J.C.; Ruiz-Azuara, L.; Cheatham, T.E.; Cortés-Guzmán, F. Intercalation processes of copper complexes in DNA. Nucleic Acids Res. 2015, 43, 5364–5376. [Google Scholar] [CrossRef] [Green Version]
- Robertazzi, A.; Vargiu, A.V.; Magistrato, A.; Ruggerone, P.; Carloni, P.; de Hoog, P.; Reedijk, J. Copper-1, 10-phenanthroline complexes binding to DNA: Structural predictions from molecular simulations. J. Phys. Chem. B 2009, 113, 10881–10890. [Google Scholar] [CrossRef]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipscomb, L.A.; Peek, M.E.; Zhou, F.X.; Bertrand, J.A.; VanDerveer, D.; Williams, L.D. Water ring structure at DNA interfaces: Hydration and dynamics of DNA-anthracycline complexes. Biochemistry 1994, 33, 3649–3659. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.N.; Yogendra, P.S.; Yogendra, S.; Raymond, J.B. Synthesis, crystal structure, DFT computation, and bioactivity measurements of copper (II) polypyridyl complexes. Polyhedron 2016, 104, 116–126. [Google Scholar] [CrossRef]
- Santos, M.; Fidalgo, A.; Varanda, A.S.; Oliveira, C.; Santos, M.A. tRNA deregulation and its consequences in cancer. Trends Mol. Med. 2019, 25, 853–865. [Google Scholar] [CrossRef]
- Sussman, J.L.; Holbrook, S.R.; Warrant, R.W.; Church, G.M.; Kim, S.H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J. Mol. Biol. 1978, 123, 607–630. [Google Scholar] [CrossRef]
- Arjmand, F.; Yousuf, I.; Zaidi, Y.; Toupet, L. Crystal structure determination, spectroscopic characterization and biological profile of a tailored ionic molecular entity, Sn (iv) iminodiacetic acid–piperazinediium conjugate: In vitro DNA/RNA binding studies, Topo I inhibition activity, cytotoxic and systemic toxicity studies. RSC Adv. 2015, 5, 16250–16264. [Google Scholar]
- Agudelo, D.; Bourassa, P.; Beauregard, M.; Bérubé, G.; Tajmir-Riahi, H.A. tRNA binding to antitumor drug doxorubicin and its analogue. PLoS ONE 2013, 8, e69248. [Google Scholar] [CrossRef]
- Parveen, S.; Fatima, K.; Zehra, S.; Arjmand, F. RNA-targeted Cu (II)-based potential antitumor drug entity: Comprehensive structural, biological {DNA/RNA binding, cleavage, cytotoxicity} and computational studies. J. Biomol. Struct. Dyn. 2020. [Google Scholar] [CrossRef]
- Marzano, C.; Pellei, M.; Tisato, F.; Santini, C. Copper complex as anti-cancer agents. Anti-Cancer Agents Med. Chem. 2009, 9, 185–211. [Google Scholar] [CrossRef]
- Iakovidis, I.; Delimaris, I.; Piperakis, S.M. Copper and its complexes in medicine: A biochemical approach. Mol. Biol. Int. 2011, 2011, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Rajendiran, V.; Karthik, R.; Palaniandavar, M.; Stoeckli-Evans, H.; Periasamy, V.S.; Akbarsha, M.A.; Srinag, B.S.; Krishnamurthy, H. Mixed-ligand copper(II)-phenolate complexes: Effect of coligand on enhanced DNA and protein binding, DNA cleavage, and anti-cancer activity. Inorg. Chem. 2007, 46, 8208–8221. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-W.; Zheng, Y.-J.; Li, Y.-T.; Wu, Z.-Y.; Yan, C.-W. Synthesis and structure of new bicopper(II) complexes bridged by N-(2-aminopropyl)-N′-(2-oxidophenyl)oxamide: The effects of terminal ligands on structures, anti-cancer activities, and DNA-binding properties. Eur. J. Med. Chem. 2011, 46, 3851–3857. [Google Scholar] [CrossRef] [PubMed]
- Zoroddu, M.A.; Zanetti, S.; Pongi, R.; Basosi, R. An electron spin resonance study and antimicrobial activity of copper (II)-phenanthroline complexes. J. Inorg. Biochem. 1996, 63, 291–300. [Google Scholar] [CrossRef]
- Patel, R.N.; Singh, N.; Shukla, K.K.; Niclós-Gutiérrez, J.; Castinerias, A.; Vaidyanathan, V.G.; Nair, B.U. Characterization and biological activities of two copper(II) complexes with diethylenetriamine and 2,2′-bipyridine or 1,10-phenanthroline as ligands. Spectrochim. Acta Part A 2005, 62, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Rufino-González, Y.; Ponce-Macotela, M.; García-Ramos, J.C.; Martínez-Gordillo, M.N.; Galindo-Murillo, R.; González-Maciel, A.; Reynoso-Robles, R.; Tovar-Tovar, A.; Flores-Alamo, M.; Toledano-Magaña, Y.; et al. Antigiardiasic activity of Cu(II) coordination compounds: Redox imbalance and membrane damage after a short exposure time. J. Inorg. Biochem. 2019, 195, 83–90. [Google Scholar] [CrossRef]
- Paixão, D.A.; Lopez, C.D.; Carneiro, Z.A.; Sousa, L.M.; de Oliveira, L.P.; Lopes, N.P.; Pivatto, M.; Chaves, J.D.S.; de Almeida, M.V.; Ellena, J. In vitro anti-Trypanosoma cruzi activity of ternary copper(II) complexes and in vivo evaluation of the most promising complex. Biomed. Pharm. 2019, 109, 157–166. [Google Scholar] [CrossRef] [PubMed]
- McGivern, T.J.P.; Afsharpour, S.; Marmion, C.J. Copper complexes as artificial DNA metallonucleases: From Sigman’s reagent to next-generation anti-cancer agent? Inorg. Chim. Acta 2018, 472, 12–39. [Google Scholar] [CrossRef]
- Alvarez, N.; Viña, D.; Leite, C.M.; Mendes, L.F.S.; Batista, A.A.; Ellena, J.; Costa-Filho, A.; Facchin, G. Synthesis and structural characterization of a series of ternary copper(II)-L-dipeptide-neocuproine complexes. Study of their cytotoxicity against cancer cells including MDA-MB-231, triple-negative breast cancer cells. J. Inorg. Biochem. 2019, 203, 110930. [Google Scholar] [CrossRef]
- Mroueh, M.; Daher, C.; Hariri, E.; Demirdjian, S.; Isber, S.; Choid, E.S.; Mirtamizdoust, B.; Hammud, H.H. Magnetic property, DFT calculation, and biological activity of bis [(μ2-chloro) chloro (1, 10-phenanthroline) copper (II)] complex. Chem. Biol. Interact. 2015, 231, 53–60. [Google Scholar] [CrossRef]
- Hayat, F.; Faryad, A.R.; Zia-ur, R.; Bélanger-Gariepy, F. Molecular, supramolecular, DNA-binding and biological studies of piperazine and piperidine based dithiocarbamates of biocompatible copper. Inorg. Chem. Commun. 2020, 121, 108190. [Google Scholar] [CrossRef]
- Gordon, A.T.; Abosede, O.O.; Ntsimango, S.; van Vuuren, S.; Hosten, E.C.; Ogunlaga, A.S. Synthesis, characterization, molecular docking and antimicrobial activity of T copper(II) complexes of metronidazole and 1,10 phenanthroline. Inorg. Chim. Acta 2020, 510, 119744. [Google Scholar] [CrossRef]
- İnci, D.; Aydın, R.; Vatan, Ö.; Zorlu, Y. A potent drug candidature of Cu(II) pyrazino[2,3 f][1,10]phenanthroline complexes with bioactive ligands: Synthesis, crystal structures, biomolecular interactions, radical scavenging and cytotoxicities. J. Biomol. Struct. Dyn. 2020. [Google Scholar] [CrossRef] [PubMed]
- Krasnovskaya, O.; Naumov, A.; Guk, D.; Gorelkin, P.; Erofeev, A.; Beloglazkina, E.; Majouga, A. Copper Coordination Compounds as Biologically Active Agents. Int. J. Mol. Sci. 2020, 21, 3965. [Google Scholar] [CrossRef] [PubMed]
- Alemon-Medina, R.; Brena-Valle, M.; Muñoz-Sánchez, J.L.; Gracia-Mora, M.I.; Ruiz-Azuara, L. Induction of oxidative damage by copper-based antineoplastic drugs (Casiopeínas). Cancer Chemother. Pharm. 2007, 60, 219–228. [Google Scholar] [CrossRef]
- Ruiz-Azuara, L. Process to Obtain New Mixed Copper Aminoacidate Complexes from Phenylate Phenathrolines to Be Used as Anticancerigenic Agents. European Patent EP 0434444A1, 26 June 1991. [Google Scholar]
- Bravo-Gómez, M.E.; Campero-Peredo, C.; García-Conde, D.; Mosqueira-Santillán, M.J.; Serment-Guerrero, J.; Ruiz-Azuara, L. DNA-binding mode of antitumoral copper compounds (Casiopeinas®) and analysis of its biological meaning. Polyhedron 2015, 102, 530–538. [Google Scholar] [CrossRef]
- Serment-Guerrero, J.; Cano-Sanchez, P.; Reyes-Perez, E.; Velazquez-Garcia, F.; Bravo-Gomez, M.E.; Ruiz-Azuara, L. Genotoxicity of the copper antineoplastic coordination complexes casiopeinas®. Toxicol. In Vitro 2011, 25, 1376–1384. [Google Scholar] [CrossRef]
- Ruiz-Azuara, L.; Bastian, G.; Bravo-Gómez, M.E.; Cañas, R.C.; Flores-Alamo, M.; Fuentes, I.; Mejia, C.; García-Ramos, J.C.; Serrano, A. Abstract CT408: Phase I study of one mixed chelates copper(II) compound, Casiopeína CasIIIia with antitumor activity and its mechanism of action. Cancer Res. 2014, 74. [Google Scholar] [CrossRef]
- Cobar, E.A.; Horn, P.H.; Bergman, R.G.; Head-Gordon, M. Examination of the hydrogen-bonding networks in small water clusters (n = 2–5, 13, 17) using absolutely localized molecular orbital energy decomposition analysis. Phys. Chem. Chem. Phys. 2012, 14, 15328–15339. [Google Scholar] [CrossRef]
- McLauchlan, C.C.; Murakami, H.A.; Wallace, C.A.; Crans, D.C. Coordination environment changes of the vanadium in vanadium-dependent haloperoxidase enzymes. JIB 2018, 186, 267–279. [Google Scholar] [CrossRef]
- Hawthorne, F.C.; Calvo, C. The crystal chemistry of the M+ VO3 (M+= Li, Na, K, NH4, Tl, Rb, and Cs) pyroxenes. JSSC 1977, 22, 157–170. [Google Scholar] [CrossRef]
- Pérez-Benítez, A.; Bernès, S. Redetermination of ammonium metavanadate. IUCrData 2018, 3, x181080. [Google Scholar] [CrossRef]
- Mukherjee, A.; Tothadi, S.; Desiraju, G.R. Halogen Bonds in Crystal Engineering: Like Hydrogen Bonds yet Different. Acc. Chem. Res. 2014, 47, 2514–2524. [Google Scholar] [CrossRef]
- Ferreira Da Silva, J.L.; Minas da Piedade, M.F.; Duarte, M.T. Decavanadates: A building-block for supramolecular assemblies. Inorg. Chim. Acta. 2003, 356, 222–242. [Google Scholar] [CrossRef]
- Amanchi, S.R.; Das, S.K. A Versatile Polyoxovanadate in Diverse Cation Matrices: A Supramolecular Perspective. Front. Chem. 2018, 6, 469. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; Van de Street, J.; Wood, P.A. Mercury CSD 2.0—new features for the visualization and investigation of crystal structures. J. Appl. Cryst. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement, and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Austin, A.; Peterson, G.A.; Frisch, M.; Dobek, F.; Scalmani, G.; Throssell, K. A density functional with spherical atom dispersion terms. J. Chem. Theor. Comput. 2012, 8, 4989–5007. [Google Scholar] [CrossRef]
- Raghavachari, K.; Trucks, G.W. Highly correlated systems: Excitation energies of first row transition metals Sc-Cu. J. Chem. Phys. 1989, 91, 1062–1065. [Google Scholar] [CrossRef]
- Cossi, M.; Barone, V.; Cammi, R.; Tomasi, J. Ab initio study of solvated molecules: A new implementation of the polarizable continuum model. Chem. Phys. Lett. 1996, 255, 327–335. [Google Scholar] [CrossRef]
- Simon, S.; Duran, M.; Dannenberg, J.J. How does basis set superposition error change the potential surfaces for hydrogen bonded dimers? J. Chem. Phys. 1996, 105, 11024–11031. [Google Scholar] [CrossRef] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision, B; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Turner, M.J.; MacKiinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer17. Available online: https://crystalexplorer.scb.uwa.edu.au/ (accessed on 4 October 2020).
- Keith, T.A. TK Gristmill Software, Version 19.02.13; AIMAll: Overland Park, KS, USA, 2019. [Google Scholar]
- Gilad, Y.; Senderowitz, H. Docking studies on DNA intercalators. J. Chem. Inf. Model. 2014, 54, 96–107. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 16, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |
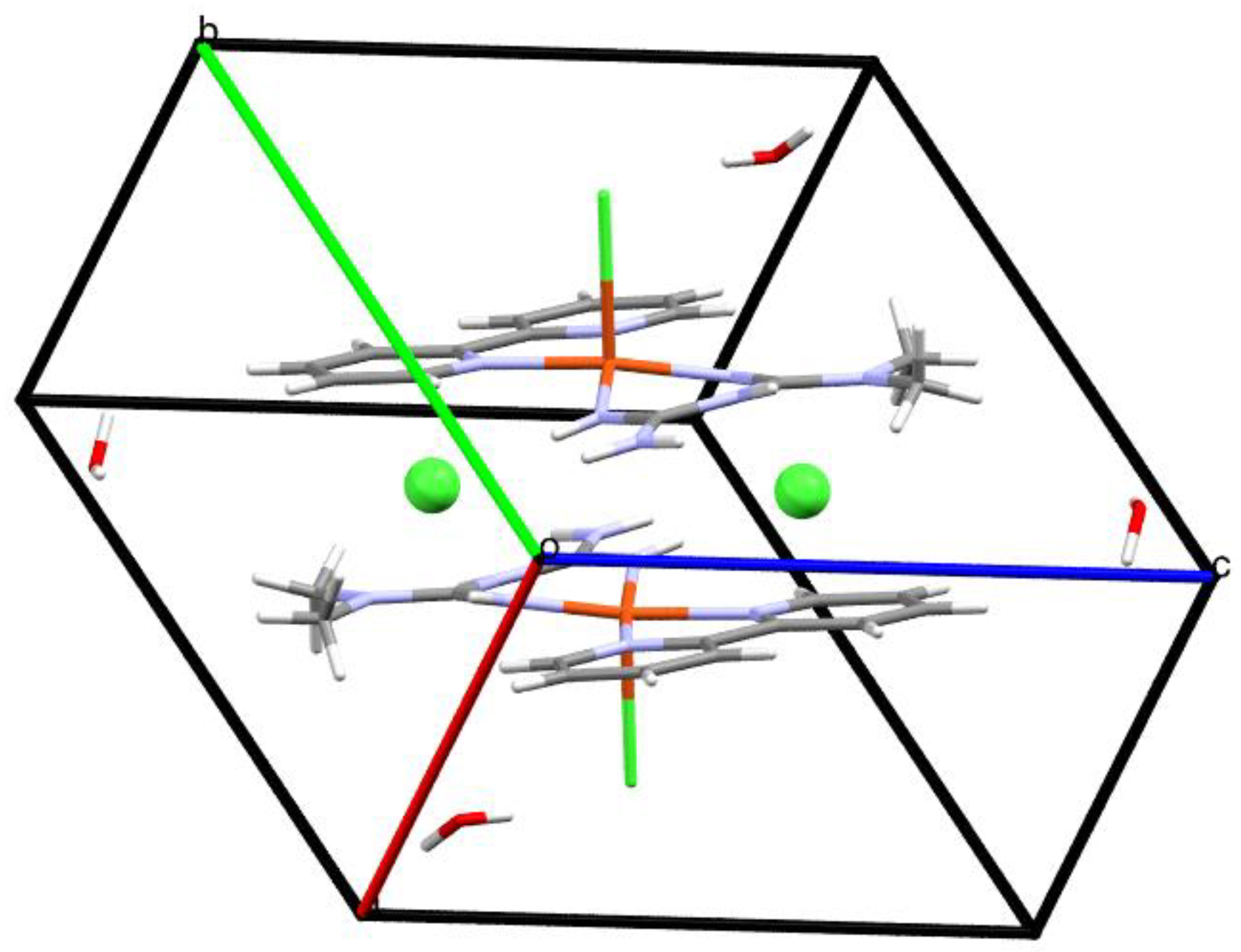
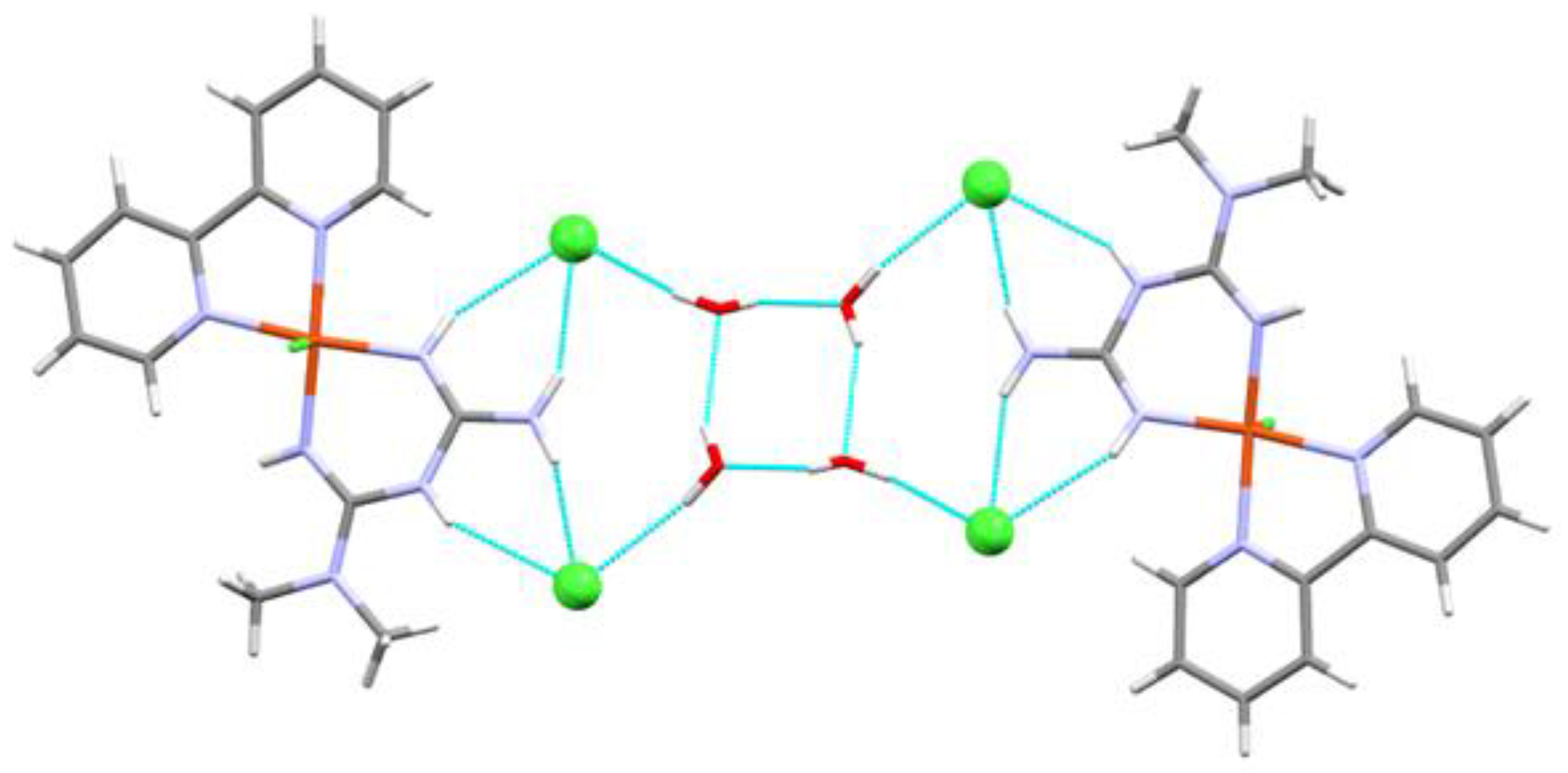
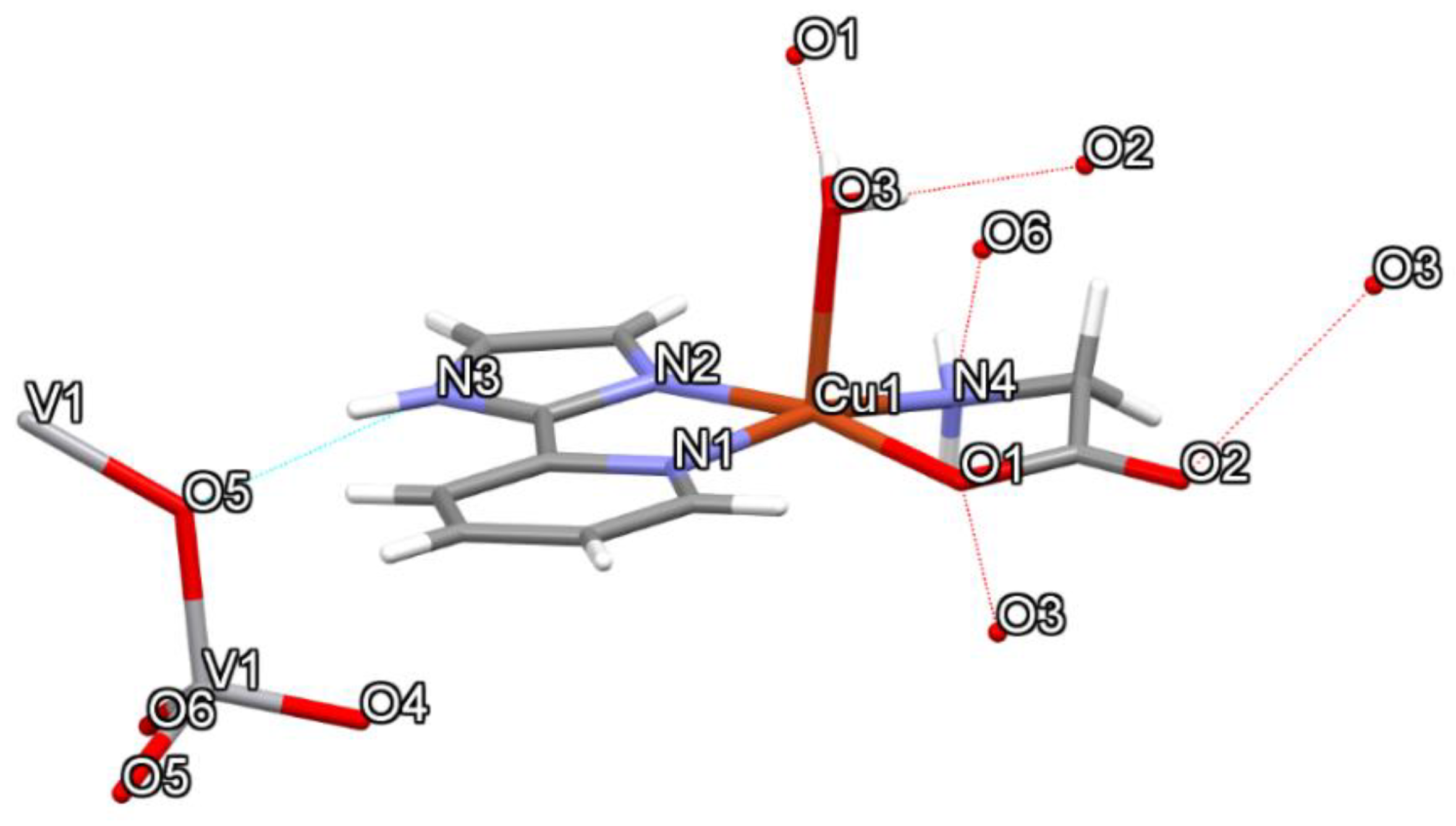

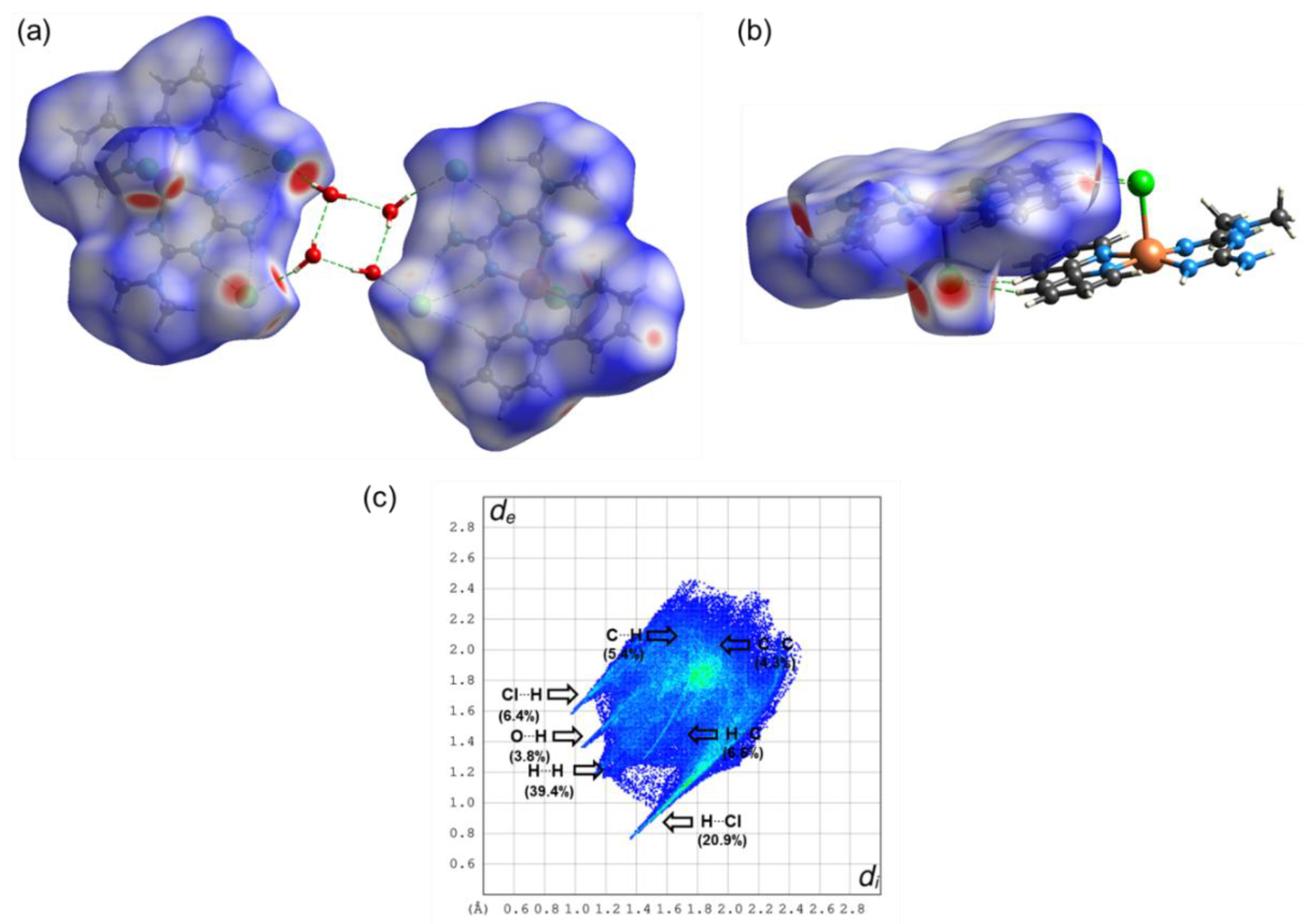
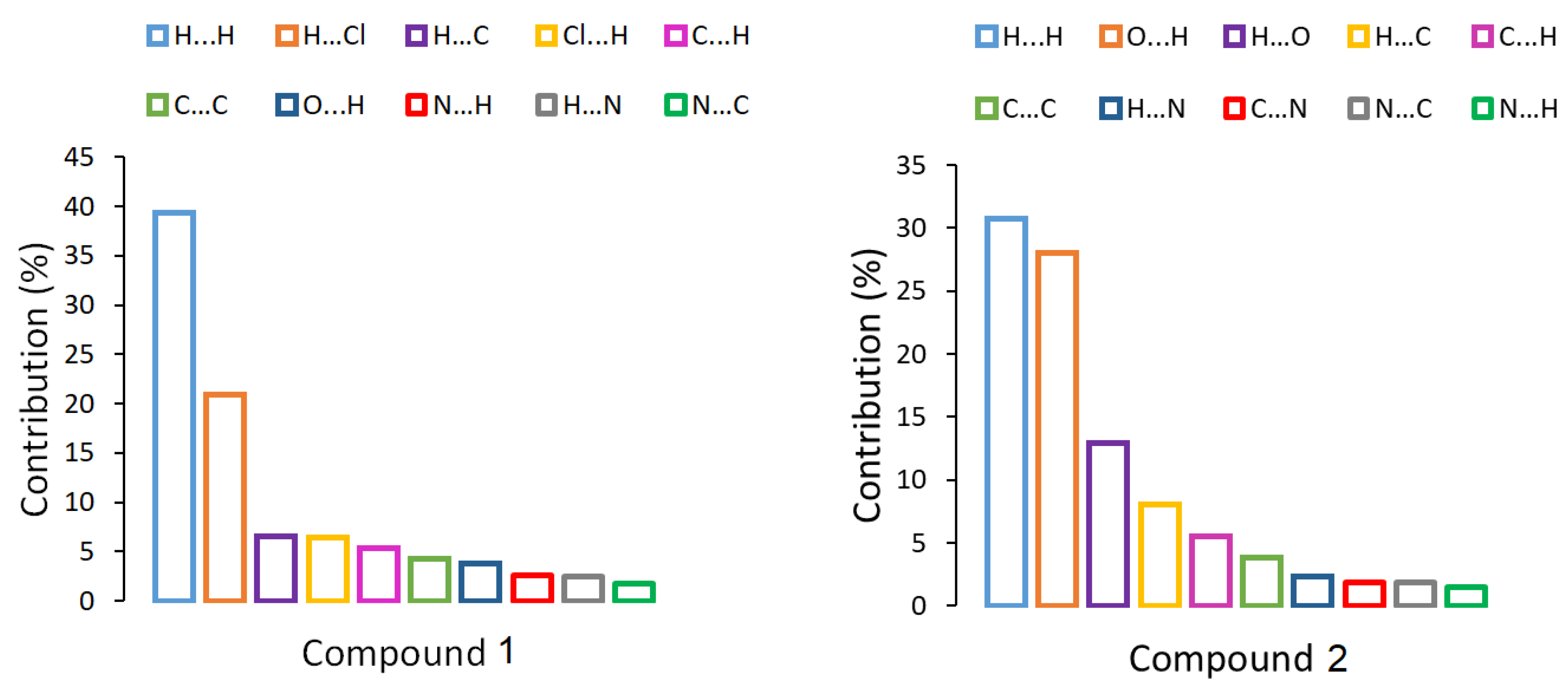


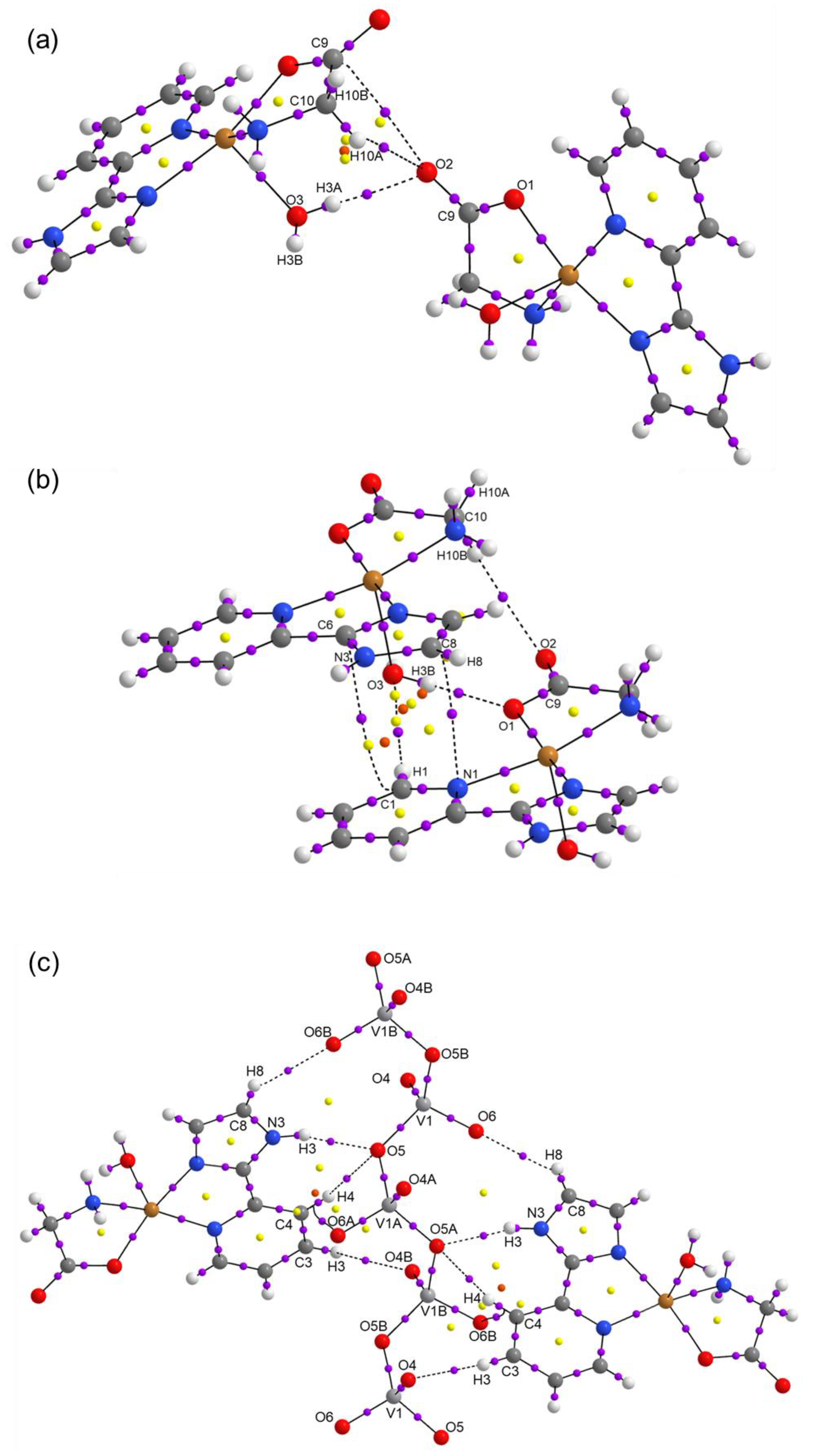

| Compound 1 | Compound 2 | |
|---|---|---|
| Empirical formula | C14H23Cl2CuN7O2 | C10H13CuN4O6V |
| Formula weight | 453.83 | 399.72 |
| Temperature/K | 293 (2) | 293 (2) |
| Crystal system | Triclinic | Monoclinic |
| Space group | P-1 | P21/c |
| a/Å | 8.4235 (3) | 13.3902 (4) |
| b/Å | 10.8688 (5) | 5.21481 (14) |
| c/Å | 11.2005 (4) | 20.6731 (5) |
| α/° | 108.249 (4) | 90 |
| β/° | 93.220 (3) | 107.316 (3) |
| γ/° | 90.608 (3) | 90 |
| Volume/Å3 | 971.91 (7) | 1378.13 (7) |
| Z | 2 | 4 |
| δcalc g/cm3 | 1.551 | 1.927 |
| μ/mm−1 | 1.421 | 2.257 |
| F(000) | 468 | 804 |
| Crystal size/mm3 | 0.67 × 0.275 × 0.12 | 0.34 × 0.211 × 0.093 |
| Radiation | Mo Kα (λ = 0.71073 Å) | Mo Kα (λ = 0.71073 Å) |
| 2Θ range for data collection/° | 5.99 to 77.408 | 5.918 to 70.408 |
| Index ranges | −13 ≤ h ≤ 13, −17 ≤ k ≤ 17, −17 ≤ l ≤ 17 | −20 ≤ h ≤ 21, −8 ≤ k ≤ 8, −33 ≤ l ≤ 33 |
| Reflections collected | 40,564 | 30,742 |
| Independent reflections | 8328 [Rint = 0.0940, Rsigma = 0.0687] | 5883 [Rint = 0.0415, Rsigma = 0.0357] |
| Data/restraints/parameters | 8328/0/247 | 5883/4/211 |
| Goodness-of-fit on F2 | 1.009 | 1.031 |
| Final R indexes [I ≥ 2σ (I)] | R1 = 0.0579, wR2 = 0.1322 | R1 = 0.0405, wR2 = 0.0837 |
| Final R indexes [all data] | R1 = 0.1070, wR2 = 0.1659 | R1 = 0.0659, wR2 = 0.0962 |
| Largest diff. peak/hole/e Å−3 | 0.58/−0.59 | 0.57/−0.85 |
| D-H···A | D-H | H···A | D···A | D-H···A |
|---|---|---|---|---|
| O25-H25B···O26 | 0.851 | 2.169 | 2.822 | 133.47 |
| O26-H26B···O25 | 0.850 | 2.002 | 2.828 | 163.68 |
| D-H···A | D-H | H···A | D···A | D-H···A |
|---|---|---|---|---|
| NH···OVO2- (N3H3···O5) | 0.861 | 2.050 | 2.877 | 160.96 |
| HOH··· OCO- (O3H3A···O2) | 0.755 | 2.019 | 2.771 | 174.53 |
| HNH···OVO2- (N4H4B···O6) | 0.788 | 2.196 | 2.914 | 151.77 |
| HOH····OCO coord (O3H3B···O1) | 0.773 | 1.995 | 2.762 | 171.17 |
| CH····OCO- (C3H3C···O2) | 0.930 | 2.569 | 3.436 | 155.30 |
| CH····OH2 (C1H1···O3) | 0.930 | 2.580 | 3.426 | 150.50 |
| CH2····OCO- (C10H10A···O2) | 0.970 | 2.587 | 3.406 | 142.16 |
| CH····OVO2- (C3H3C···O6) | 0.930 | 2.541 | 3.139 | 122.36 |
| CH····OVO2- (C4H4····O5) | 0.931 | 2.653 | 3.506 | 152.71 |
| CH····OVO2- (C8H8···O4) | 0.930 | 2.431 | 3.101 | 128.85 |
| Compound | ΔE0 (a.u.) | ΔGsol (kcal mol−1) | Eint (kcal mol−1) |
|---|---|---|---|
| 1 | 0.00 | −67.58 | −152.87 |
| 1′ | 384.03 | −137.26 | −11.33 |
| 2 | 554.52 | −62.49 | −12.50 |
| BCP | ρ(r) | ∇2ρ(r) | G (r) | V (r) | H (r) | EH…Y | Dinter |
|---|---|---|---|---|---|---|---|
| Compound 1 | |||||||
| Cl3⋯H17 | 0.0098 | 0.0031 | 0.0062 | −0.0046 | 0.0108 | 1.44 | 2.279 |
| Cl3⋯H4 | 0.0094 | 0.0325 | 0.0064 | −0.0046 | 0.0110 | 1.44 | 2.699 |
| Cl3⋯H12A | 0.0133 | 0.0469 | 0.0093 | −0.0069 | 0.0162 | 2.16 | 2.504 |
| Cl1⋯H12B | 0.0189 | 0.0661 | 0.0139 | −0.0114 | 0.0253 | 3.58 | 2.334 |
| Cl1⋯H6 | 0.0113 | 0.0379 | 0.0075 | −0.0056 | 0.0131 | 1.76 | 2.603 |
| Cl1⋯H9C | 0.0072 | 0.0217 | 0.0044 | −0.0034 | 0.0078 | 1.07 | 2.933 |
| Cl3⋯H26A | 0.0204 | 0.0686 | 0.0151 | −0.0130 | 0.0281 | 4.08 | 2.279 |
| O25⋯H26B | 0.0213 | 0.0885 | 0.0189 | −0.0158 | 0.0347 | 4.96 | 2.002 |
| Cl1⋯H25A | 0.0126 | 0.0428 | 0.0085 | −0.0062 | 0.0147 | 1.95 | 2.505 |
| O26⋯H25B | 0.0166 | 0.0644 | 0.0140 | −0.0119 | 0.0259 | 3.73 | 2.168 |
| Cl2⋯H24 | 0.0062 | 0.0177 | 0.0036 | −0.0028 | 0.0064 | 0.88 | 2.951 |
| Cl2⋯H14 | 0.0094 | 0.0290 | 0.0058 | −0.0044 | 0.0102 | 1.38 | 2.724 |
| Compound 2 | |||||||
| O2⋯H3A | 0.0056 | 0.0204 | 0.0043 | −0.0034 | 0.0077 | 1.07 | 2.019 |
| O2⋯H10A | 0.0194 | 0.0906 | 0.0187 | −0.0147 | 0.0334 | 4.61 | 2.767 |
| O6B⋯H8 | 0.0096 | 0.0362 | 0.0075 | −0.0060 | 0.0135 | 1.88 | 2.431 |
| O2⋯H10B | 0.0071 | 0.0232 | 0.0050 | −0.0042 | 0.0092 | 1.32 | 2.587 |
| O1⋯H3B | 0.0211 | 0.0966 | 0.0203 | −0.0165 | 0.0368 | 5.18 | 1.995 |
| O5⋯H3 | 0.0193 | 0.0794 | 0.0167 | −0.0136 | 0.0303 | 4.27 | 2.049 |
| O5⋯H4 | 0.0064 | 0.0211 | 0.0045 | −0.0038 | 0.0083 | 1.19 | 2.653 |
| O4B⋯H3 | 0.0083 | 0.0304 | 0.0064 | −0.0052 | 0.0116 | 1.63 | 2.541 |
| Ligand | Binding Energy (kcal/mol) 6TNA | Interaction |
|---|---|---|
| Doxorubicin | −9.82 | H bond, van der Waals, π-anion |
| [Cu(hydroxynaphthaldehyde)(H2O)] | −7.98 | H bond, van der Waals, π-anion, π-π |
| 1′ [Cu(Metf)(bipy)(H2O)]2+ | −12.76 | H bond, van der Waals, π-anion |
| 2 [Cu(Impy)(Gly)(H2O)]+ | −8.86 | H bond, van der Waals, π-anion |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corona-Motolinia, N.D.; Martínez-Valencia, B.; Noriega, L.; Sánchez-Gaytán, B.L.; Méndez-Rojas, M.Á.; Melendez, F.J.; Castro, M.E.; González-Vergara, E. Synthesis, Crystal Structure, and Computational Methods of Vanadium and Copper Compounds as Potential Drugs for Cancer Treatment. Molecules 2020, 25, 4679. https://doi.org/10.3390/molecules25204679
Corona-Motolinia ND, Martínez-Valencia B, Noriega L, Sánchez-Gaytán BL, Méndez-Rojas MÁ, Melendez FJ, Castro ME, González-Vergara E. Synthesis, Crystal Structure, and Computational Methods of Vanadium and Copper Compounds as Potential Drugs for Cancer Treatment. Molecules. 2020; 25(20):4679. https://doi.org/10.3390/molecules25204679
Chicago/Turabian StyleCorona-Motolinia, Nidia D., Beatriz Martínez-Valencia, Lisset Noriega, Brenda L. Sánchez-Gaytán, Miguel Ángel Méndez-Rojas, Francisco J. Melendez, María Eugenia Castro, and Enrique González-Vergara. 2020. "Synthesis, Crystal Structure, and Computational Methods of Vanadium and Copper Compounds as Potential Drugs for Cancer Treatment" Molecules 25, no. 20: 4679. https://doi.org/10.3390/molecules25204679
APA StyleCorona-Motolinia, N. D., Martínez-Valencia, B., Noriega, L., Sánchez-Gaytán, B. L., Méndez-Rojas, M. Á., Melendez, F. J., Castro, M. E., & González-Vergara, E. (2020). Synthesis, Crystal Structure, and Computational Methods of Vanadium and Copper Compounds as Potential Drugs for Cancer Treatment. Molecules, 25(20), 4679. https://doi.org/10.3390/molecules25204679








