Chemical Space Charting of Different Parts of Inula nervosa Wall.: Upregulation of Expression of Nrf2 and Correlated Antioxidants Enzymes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Chemicals and Primary Detection Kits
2.3. Crude Extract Preparation of Different Parts of I. nervosa Wall
2.4. HPLC-MS/MS Analysis
2.5. Molecular Networking Creation and Annotation
2.6. Multivariate Analyses
2.7. `ili Visualization
2.8. DPPH Radical Scavenging Activity Assay
2.9. Intracellular Antioxidative Activity Assay
2.10. Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.11. Statistical Analysis
3. Results and Discussion
3.1. Molecular Networking and Annotation of IEs
3.2. Multivariate Analyses of IEs
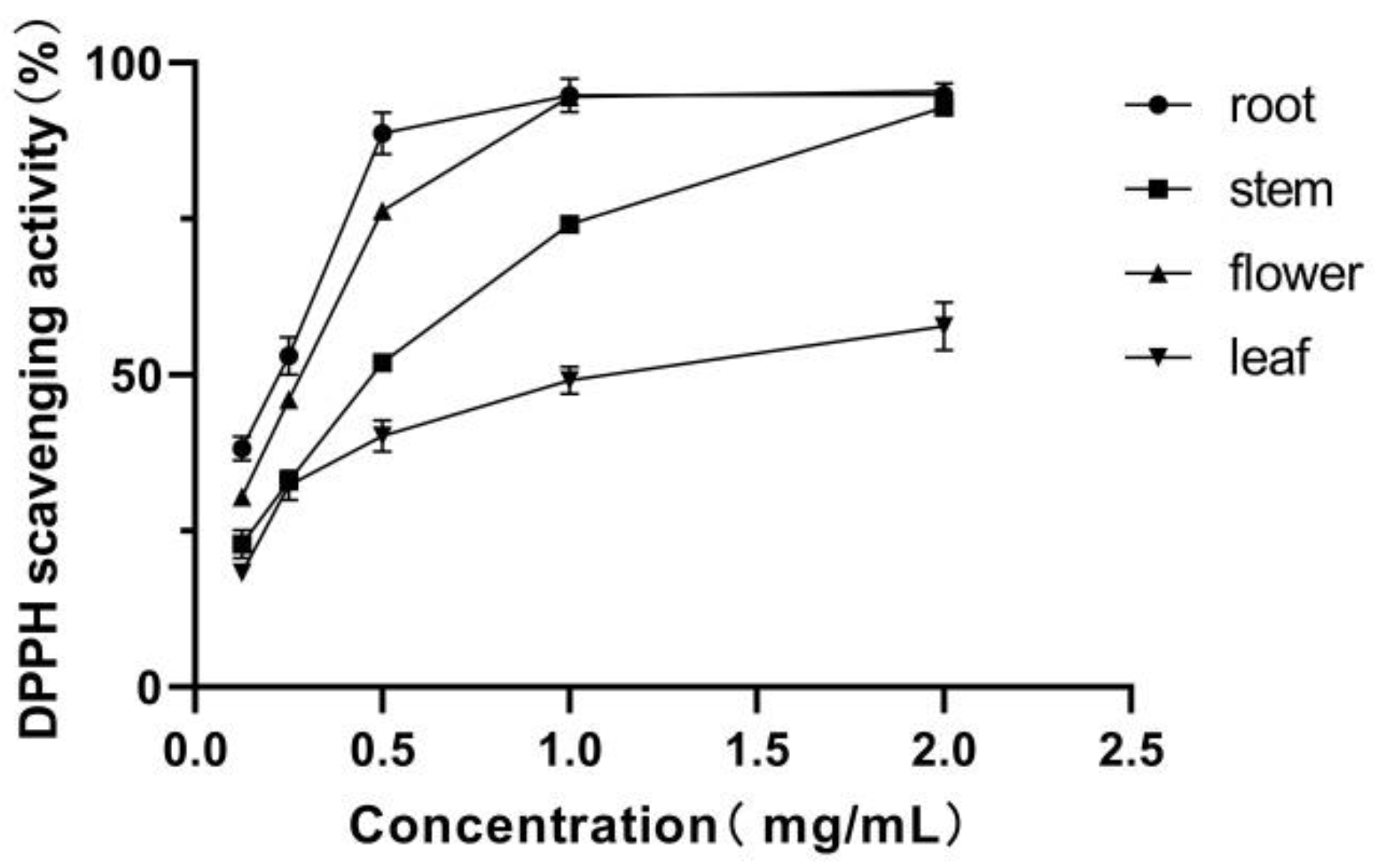
3.3. DPPH Radical Scavenging Activity
3.4. Intracellular Antioxidative Activities of IEs
3.5. Effects of IEs on Nrf2/ARE Pathway Activation
3.6. Correlations between Compound 1 and Antioxidant Parameters
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhao, L.; Xin, L.Q.; Li, Y.Q. Research advance of Inula nervosa Wall. in China. Food Drug 2007, 9, 56–57. [Google Scholar]
- Yan, L.; Cheng, X.R.; Zeng, Q.; Qin, J.J.; Zhang, W.D.; Jin, H.Z. Phytane and neoclerodane diterpenes from the aerial parts of Inula nervosa Wall. Biochem. Syst. Ecol. 2001, 39, 700–703. [Google Scholar] [CrossRef]
- Yan, L.; Huang, Y.; Fu, J.J.; Qin, J.J.; Zeng, Q.; Zhu, Y.; Yan, S.K.; Zhang, W.D.; Jin, H.Z. Three new phenylpropanoids from Inula nervosa Wall. Helv. Chim. Acta 2010, 93, 1418–1421. [Google Scholar] [CrossRef]
- Fujita, H.; Motokawa, T.; Katagiri, T.; Yokota, S.; Yamamoto, A.; Himeno, M.; Tanaka, Y. Inulavosin, a melanogenesis inhibitor, leads to mistargeting of tyrosinase to lysosomes and accelerates its degradation. J. Invest. Dermatol. 2009, 129, 1489–1499. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.R.; Zeng, Q.; Ren, J.; Qin, J.J.; Zhang, S.D.; Shen, Y.H.; Zhu, J.X.; Zhang, F.; Chang, R.J.; Zhu, Y.; et al. Sesquiterpene lactones from Inula falconeri, a plant endemic to the Himalayas, as potential anti-inflammatory agents. Eur. J. Med. Chem. 2011, 46, 5408–5415. [Google Scholar] [CrossRef]
- Cheng, X.R.; Zhang, S.D.; Wang, C.H.; Ren, J.; Qin, J.J.; Tang, X.; Shen, Y.H.; Yan, S.K.; Jin, H.Z.; Zhang, W.D. Bioactive eudesmane and germacrane derivatives from Inula wissmanniana Hand.-Mazz. Phytochemistry 2013, 96, 214–222. [Google Scholar] [CrossRef]
- Wang, G.W.; Qin, J.J.; Cheng, X.R.; Shen, Y.H.; Shan, L.; Jin, H.Z.; Zhang, W.D. Inula sesquiterpenoids: Structural diversity, cytotoxicity and anti-tumor activity. Expert Opin. Inv. Drug 2014, 23, 317–345. [Google Scholar] [CrossRef]
- Ministry of Health of PR China. Announcement of the Ministry of Health of PR China No.9 of 2010. Chin. J. Food Hyg. 2010, 22, 380. [Google Scholar]
- Dinkova-Kostova, A.T.; Talalay, P. Direct and indirect antioxidant properties of inducers of cytoprotective proteins. Mol. Nutr. Food Res. 2008, 52, S128–S138. [Google Scholar] [CrossRef]
- Ahn, Y.H.; Liu, H.; Wang, X.J.; Zhang, Y.; Stephenson, K.K.; Boronina, T.N.; Cole, R.N.; Dinkova-Kostova, A.T.; Talalay, P.; Cole, P.A. Electrophilic tuning of the chemoprotective natural product sulforaphane. Proc. Natl. Acad. Sci. USA 2010, 102, 9590–9595. [Google Scholar] [CrossRef] [Green Version]
- Gersch, M.; Kreuzer, J.; Sieber, S.A. Electrophilic natural products and their biological targets. Nat. Prod. Rep. 2012, 29, 659–682. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ernst, M.; Kang, K.B.; Caraballo-Rodriguez, A.M.; Nothias, L.F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Gowda, H.; Ivanisevic, J.; Johnson, C.H.; Kurczy, M.E.; Benton, H.P.; Rinehart, D.; Nguyen, T.; Ray, J.; Kuehl, J.; Arevalo, B.; et al. Interactive XCMS Online: Simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal. Chem. 2014, 86, 6931–6939. [Google Scholar] [CrossRef] [PubMed]
- Protsyuk, I.; Melnik, A.V.; Nothias, L.F.; Rappez, L.; Phapale, P.; Aksenov, A.A.; Bouslimani, A.; Ryazanov, S.; Dorrestein, P.C.; Alexandrov, T. 3D molecular cartography using LC-MS facilitated by Optimus and `ili software. Nat. Protoc. 2017, 13, 134–154. [Google Scholar] [CrossRef] [PubMed]
- Rosero, J.C.; Cruz, S.; Osorio, C.; Hurtado, N. Analysis of phenolic composition of byproducts (seeds and peels) of Avocado (Persea americana Mill.) cultivated in Colombia. Molecules 2019, 24, 3209. [Google Scholar] [CrossRef] [Green Version]
- Salla, S.; Sunkara, R.; Ogutu, S.; Walker, L.T.; Verghese, M. Antioxidant activity of papaya seed extracts against H2O2 induced oxidative stress in HepG2 cells. LWT-Food Sci. Technol. 2016, 66, 293–297. [Google Scholar] [CrossRef]
- Sohn, S.H.; Kim, S.K.; Kim, Y.O.; Kim, H.D.; Shin, Y.S.; Yang, S.O.; Kim, S.Y.; Lee, S.W. A comparison of antioxidant activity of Korean White and Red Ginsengs on H2O2-induced oxidative stress in HepG2 hepatoma cells. J. Ginseng Res. 2013, 37, 442–450. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=74fb92bf0553404ab0ffdc5376cfc44c (accessed on 26 April 2019).
- Available online: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=a26a132027a242fc993df75766ec784d (accessed on 6 October 2019).
- Feunang, Y.D.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef] [Green Version]
- Mohimani, H.; Gurevich, A.; Shlemov, A.; Mikheenko, A.; Korobeynikov, A.; Cao, L.; Shcherbin, E.; Nothias, L.F.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun. 2018, 9, 4035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, A.N.; She, C.W.; Zeng, J.Y.; Peng, S.X. Comparison study on in vitro and in vivo antioxidant activities of Inula nervosa Wall. extracts from different parts. Chin. Pharmacol. Bull. 2016, 32, 79–83. [Google Scholar]
- Li, K.; Shi, L.Z.; Chen, D.; Hu, Y.B.; Lu, M.F.; Li, R.C.; Li, S.X. Content determination of thymol and thymyl isobutyrate in Inula nervosa Wall from different parts, different habitats and different harvest periods by HPLC. J. Hunan Univ. Chin. Med. 2013, 33, 41–44. [Google Scholar]
- Forman, H.J.; Davies, K.J.; Ursini, F. How do nutritional antioxidants really work: Nucleophilic tone and para-hormesis versus free radical scavenging in vivo. Free Radic. Biol. Med. 2014, 66, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Joko, S.; Watanabe, M.; Fuda, H.; Takeda, S.; Furukawa, T.; Hui, S.-P.; Shrestha, R.; Chiba, H. Comparison of chemical structures and cytoprotection abilities between direct and indirect antioxidants. J. Funct. Foods 2017, 35, 245–255. [Google Scholar] [CrossRef]
- Jaiswal, A.K. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radicals Biol. Med. 2004, 36, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Lee, C. Collaborative power of Nrf2 and PPARγ activators against metabolic and drug-induced oxidative injury. Oxid. Med. Cell. Longev. 2017, 2017, 1378175. [Google Scholar] [CrossRef] [Green Version]
- Teskey, G.; Abrahem, R.; Cao, R.; Gyurjian, K.; Islamoglu, H.; Lucero, M.; Martinez, A.; Paredes, E.; Salaiz, O.; Robinson, B.; et al. Chapter five-Glutathione as a marker for human disease. Adv. Clin. Chem. 2018, 87, 141–159. [Google Scholar]
- Gipp, J.J.; Chang, C.; Mulcahy, R.T. Cloning and nucleotide sequence of a full-length cDNA for human liver gamma-glutamylcysteine synthetase. Biochem. Biophys. Res. Commun. 1992, 185, 29–35. [Google Scholar] [CrossRef]
- Litwinienko, G.; Ingold, K.U. Solvent effects on the rates and mechanisms of reaction of phenols with free radicals. Acc. Chem. Rev. 2007, 40, 222–230. [Google Scholar] [CrossRef]
- Galian, R.E.; Litwinienko, G.; Perez-Prieto, J.; Ingold, K.U. Kinetic solvent effects on the reaction of an aromatic ketone π,π* triplet with phenol. Rate-retarding and rate-accelerating effects of hydrogen-bond acceptor solvents. J. Am. Chem. Soc. 2007, 129, 9280–9281. [Google Scholar] [CrossRef] [PubMed]
- Litwinienko, G.; Ingold, K.U. Abnormal solvent effects on hydrogen atom abstraction. 2. Resolution of the curcumin antioxidant controversy. The role of sequential proton loss electron transfer. J. Org. Chem. 2004, 69, 5888–5896. [Google Scholar] [CrossRef] [PubMed]
- Litwinienko, G.; Ingold, K.U. Abnormal solvent effects on hydrogen atom abstraction. 3. Novel kinetics in sequential proton loss electron transfer chemistry. J. Org. Chem. 2005, 70, 8982–8990. [Google Scholar] [CrossRef] [Green Version]
- Eggler, A.L.; Liu, G.; Pezzuto, J.M.; van Breemen, R.B.; Mesecar, A.D. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. USA 2005, 102, 10070–10075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holland, R.; Hawkins, A.E.; Eggler, A.L.; Mesecar, A.D.; Fabris, D.; Fishbein, J.C. Prospective type 1 and type 2 disulfides of Keap1 protein. Chem. Res. Toxicol. 2008, 21, 2051–2060. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Eggler, A.L.; Liu, D.; Liu, G.; Mesecar, A.D.; van Breemen, R.B. Sites of alkylation of human Keap1 by natural chemoprevention agents. J. Am. Soc. Mass Spectrom. 2007, 18, 2226–2232. [Google Scholar] [CrossRef] [Green Version]





| Gene | Primer (5′→3′) | |
|---|---|---|
| Forward | Reverse | |
| β-actin | GTTGTCGACGACGAGCG | GCACAGAGCCTCGCCTT |
| Nrf2 | TCCAGTCAGAAACCAGTGGAT | GAATGTCTGCGCCAAAAGCTG |
| NQO1 | GTGGTGGAGTCGGACCTCTATG | AAGCCAGAACAGACTCGGCAG |
| HO-1 | GAGTGTAAGGACCCATCGGA | GCCAGCAACAAAGTGCAAG |
| MnSOD | CCAGAAAATGCTATGATTGATATGAC | AAGGGAGATGTTACAGCCCAGATA |
| GCLC | AGGGAGTTTATCGCAAACCA | AAGTAACTCTGGGCATTCACA |
| Compound | Cluster Index | Adduct | m/z | Mass Differ | Cosine Score | Shared Peaks | Identification |
|---|---|---|---|---|---|---|---|
| 1 | 262 | [M + H]+ | 320.997 | 0.173 | 0.84 | 6 | 10-Isobutyryloxy-8,9-epoxythymol isobutyrate |
| 2 | 357 | [M + H]+ | 354.996 | 0.004 | 0.97 | 7 | Chlorogenic acid |
| 3 | 374 | [M-H2O + H]+ | 357.101 | 0.068 | 0.78 | 6 | (2S,3R,3aS,7aR)-5-Allyl-2-(4-hydroxy-3-methoxyphenyl)-3a,7a-dimethoxy-3-methyl-3,3a,7,7a-tetrahydro-1-βenzofuran-6(2H)-one |
| 4 | 414 | [M + Na]+ | 365.172 | 0.068 | 0.86 | 7 | Galactinol |
| 5 | 1409 | [M + Na]+ | 527.193 | 0.035 | 0.93 | 11 | Melezitose |
| 6 | 999 | [M + Na]+ | 467.250 | −1.954 | 0.81 | 6 | (Z)-2,6-dimethyl-7-(4-methyl-5-oxooxolan-2-yl)-3-[[3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxymethyl]hept-5-enoic acid |
| 7 | 3462 | [M + Na]+ | 691.309 | 2.099 | 0.82 | 8 | Stachyose |
| 8 | 6241 | [M + Na]+ | 851.262 | 0.007 | 0.8 | 17 | Polysaccharide Hexose x5 |
| 9 | 3590 | [M + H]+ | 701.086 | 1.521 | 0.87 | 6 | N-[(2S,3R,4E)-1-(β-d-Galactopyranosyloxy)-3-hydroxy-4-octadecen-2-yl]hexadecanamide |
| 10 | 2202 | [M + H]+ | 595.011 | 0.011 | 0.97 | 6 | Kaempferol-7-O-neohesperidoside |
| 11 | 824 | [M + Na]+ | 432.993 | −2.170 | 0.89 | 6 | 4-[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxypentan-2-yl (E)-3-(4-hydroxyphenyl) prop-2-enoate |
| 12 | 1112 | [M + K]+ | 488.030 | 0.836 | 0.87 | 8 | (2R,3S,4S,5R,6R)-2-[[(2R,3R,4R,5S)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]oxymethyl]-6-[(2E)-3,7-dimethylocta-2,6-dienoxy]oxane-3,4,5-triol |
| 13 | 826 | [M + H]+ | 435.064 | 1.954 | 0.78 | 10 | Isovitexin |
| 14 | 556 | [M + H]+ | 384.929 | 0.785 | 0.9 | 7 | Hydrastine |
| 15 | 6464 | [M + H]+ | 871.733 | 0.143 | 0.91 | 9 | Pheophytin |
| 16 | 2213 | [M + H]+ | 595.418 | 2.149 | 0.81 | 6 | Pheophorbide A |
Sample Availability: Samples of the compounds are not available from the authors. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, X.-r.; Zhao, W.; Dong, W.-l.; Le, G.-w. Chemical Space Charting of Different Parts of Inula nervosa Wall.: Upregulation of Expression of Nrf2 and Correlated Antioxidants Enzymes. Molecules 2020, 25, 4789. https://doi.org/10.3390/molecules25204789
Cheng X-r, Zhao W, Dong W-l, Le G-w. Chemical Space Charting of Different Parts of Inula nervosa Wall.: Upregulation of Expression of Nrf2 and Correlated Antioxidants Enzymes. Molecules. 2020; 25(20):4789. https://doi.org/10.3390/molecules25204789
Chicago/Turabian StyleCheng, Xiang-rong, Wei Zhao, Wen-le Dong, and Guo-wei Le. 2020. "Chemical Space Charting of Different Parts of Inula nervosa Wall.: Upregulation of Expression of Nrf2 and Correlated Antioxidants Enzymes" Molecules 25, no. 20: 4789. https://doi.org/10.3390/molecules25204789
APA StyleCheng, X.-r., Zhao, W., Dong, W.-l., & Le, G.-w. (2020). Chemical Space Charting of Different Parts of Inula nervosa Wall.: Upregulation of Expression of Nrf2 and Correlated Antioxidants Enzymes. Molecules, 25(20), 4789. https://doi.org/10.3390/molecules25204789





