Significance of Competing Metabolic Pathways for 5F-APINACA Based on Quantitative Kinetics
Abstract
:1. Introduction
2. Results
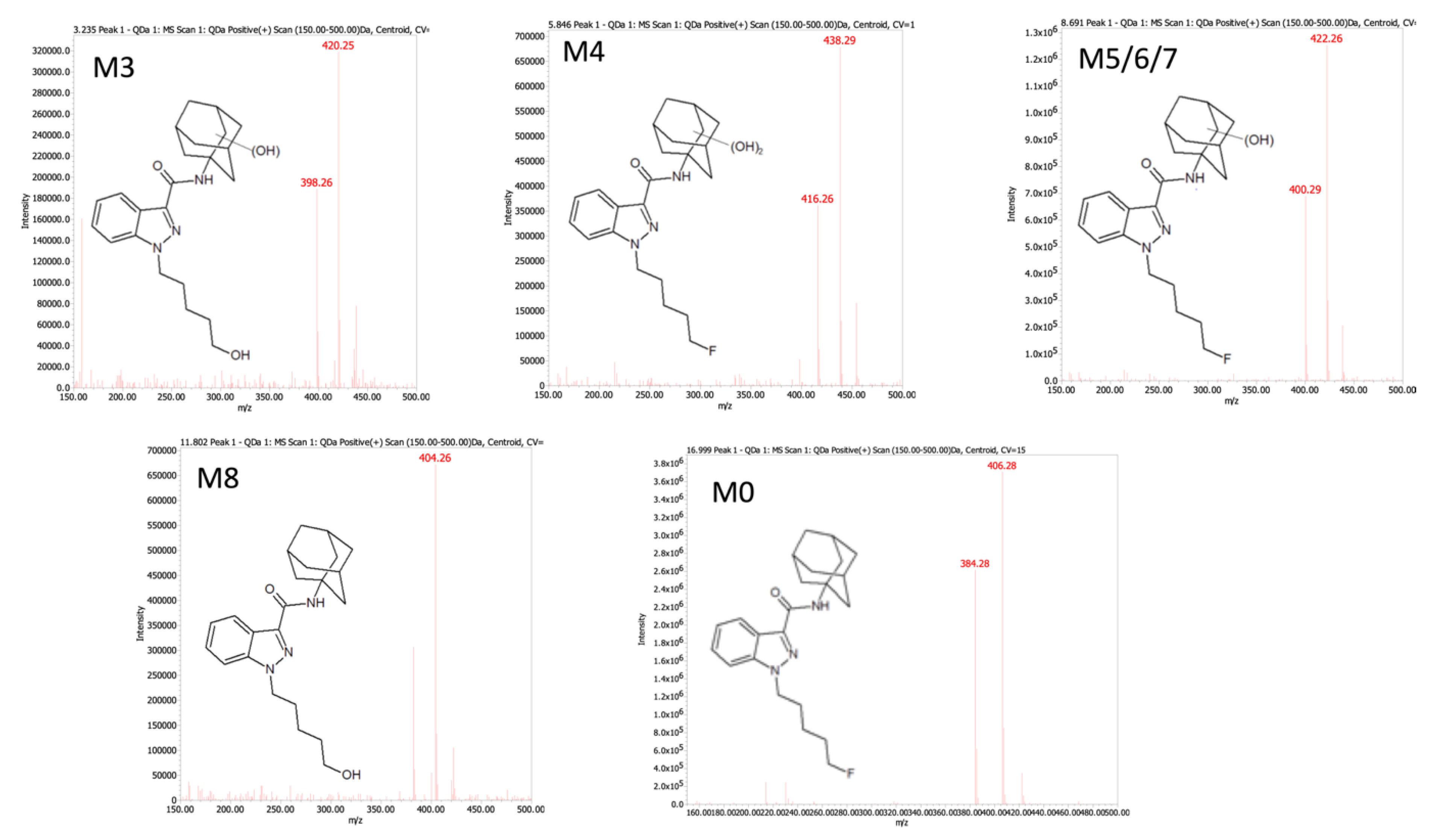
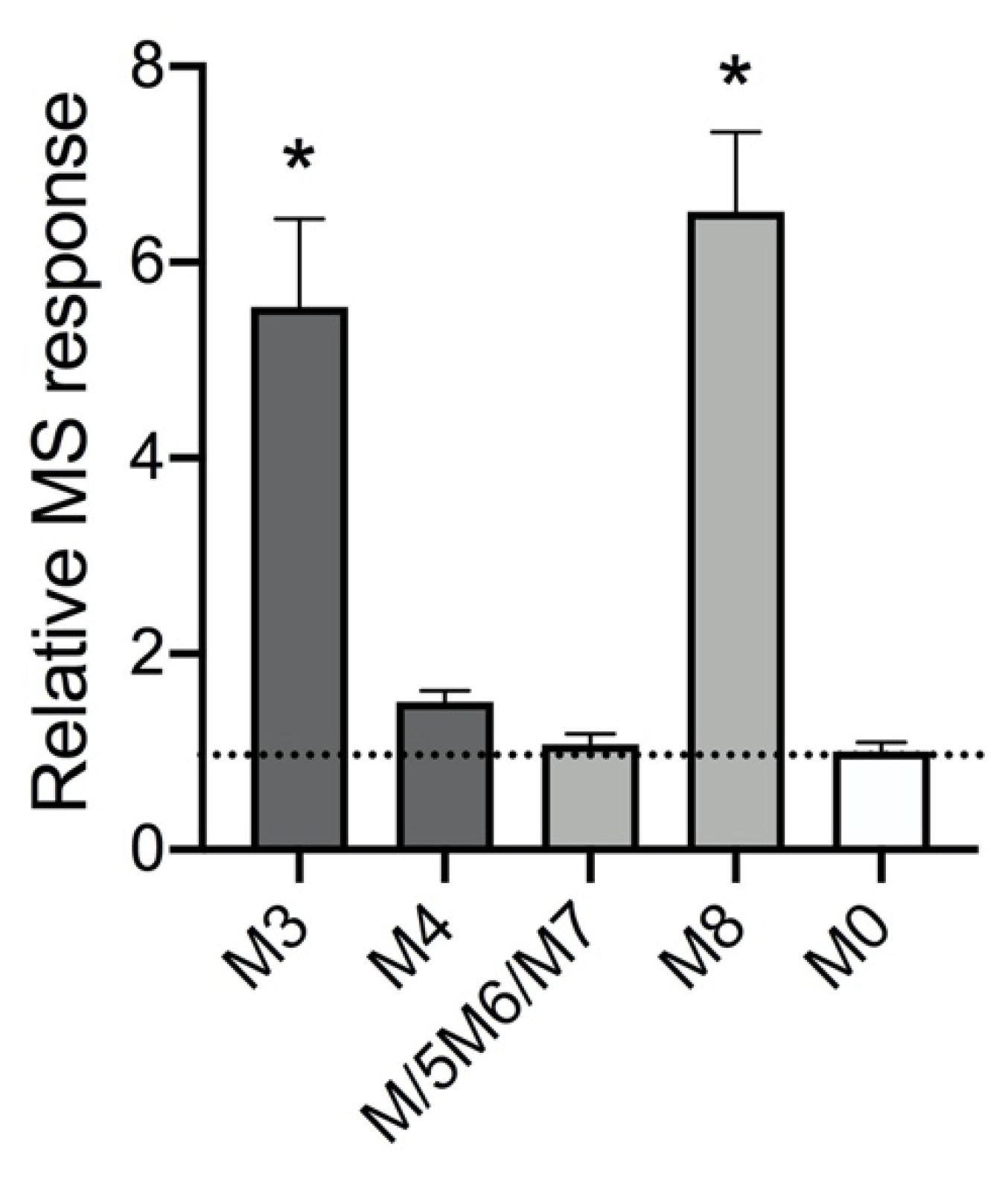
2.1. Absorbance but Not MS Proved Reliable for Inferring Quantitation of Five 5F-APINACA Metabolites
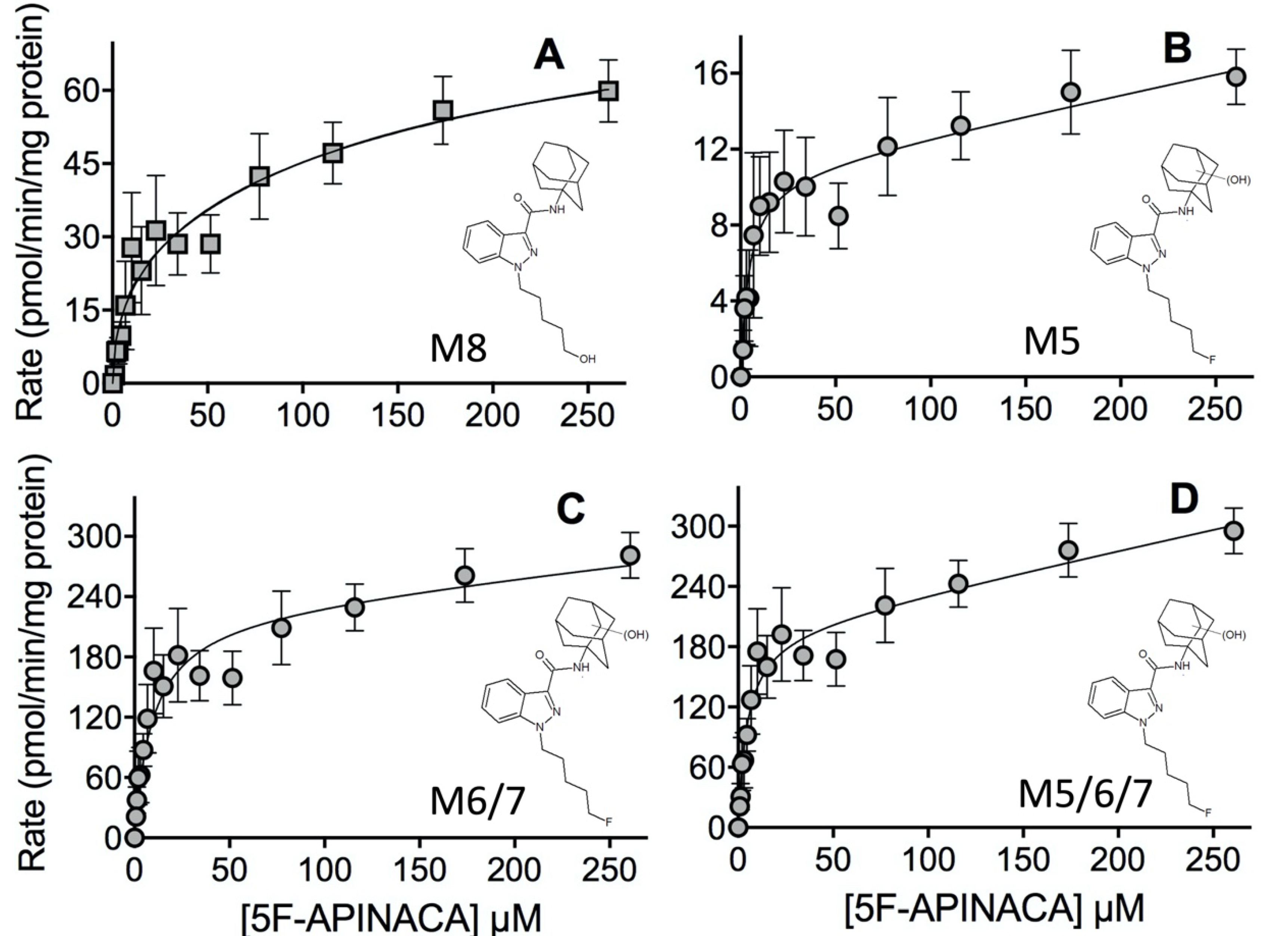
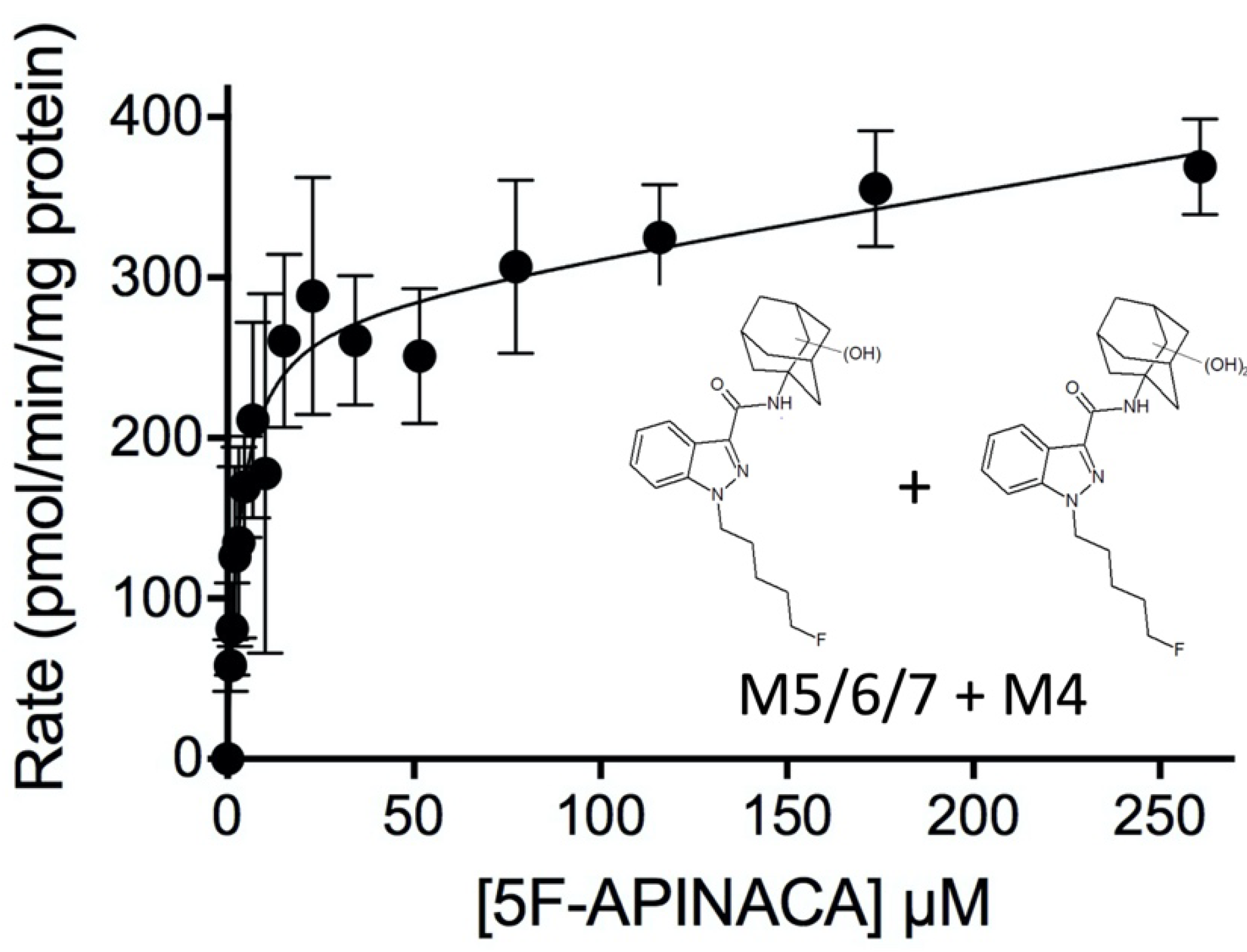
2.2. Steady-State Kinetics Revealed Relative Importance of Binding and Chemistry in Metabolism
3. Discussion
3.1. New Quantitative Approach Yielded Steady-State Kinetics for 5F-APINACA Metabolism
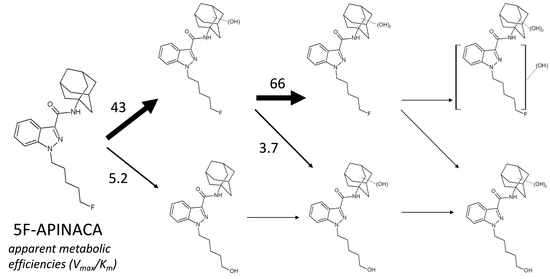
3.2. Steady-State Kinetics Revealed Major and Minor Metabolic Pathways for 5F-APINACA
3.3. 5F-APINACA Kinetics Supported Extrapolating the Potential Relevance of Metabolism on Abuser Responses
4. Materials and Methods
4.1. Materials
4.2. Steady-State Kinetics for 5F-APINACA Metabolism by Human Liver Microsomes
4.3. HPLC Resolution and Analysis of 5F-APINACA Analytes from Reactions
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A


References
- News: January 2020—UNODC-SMART: Share of NPS Stimulants and Synthetic Cannabinoids Large but Stable While Opioids Increase. Available online: https://www.unodc.org/LSS/Announcement/Details/0096a6ec-c5d0-404f-a559-d15fb84ca75c (accessed on 22 September 2020).
- Cohen, K.; Weinstein, A.M. Synthetic and non-synthetic cannabinoid drugs and their adverse effects—A review from public health prospective. Front. Public Health 2018, 6, 162. [Google Scholar] [CrossRef]
- Banister, S.D.; Stuart, J.; Kevin, R.C.; Edington, A.; Longworth, M.; Wilkinson, S.M.; Beinat, C.; Buchanan, A.S.; Hibbs, D.E.; Glass, M.; et al. Effects of bioisosteric fluorine in synthetic cannabinoid designer drugs JWH-018, AM-2201, UR-144, XLR-11, PB-22, 5F-PB-22, APICA, and STS-135. ACS Chem. Neurosci. 2015, 6, 1445–1458. [Google Scholar] [CrossRef] [Green Version]
- Park, B.K.; Kitteringham, N.R.; O’Neill, P.M. Metabolism of fluorine-containing drugs. Annu. Rev. Pharmacol. Toxicol. 2001, 41, 443–470. [Google Scholar] [CrossRef] [PubMed]
- Gurney, S.M.R.; Scott, K.S.; Kacinko, S.L.; Presley, B.C.; Logan, B.K. Pharmacology, toxicology, and adverse effects of synthetic cannabinoid drugs. Forensic Sci. Rev. 2014, 26, 53–78. [Google Scholar] [PubMed]
- Gamage, T.F.; Farquhar, C.E.; McKinnie, R.J.; Kevin, R.C.; McGregor, I.S.; Trudell, M.L.; Wiley, J.L.; Thomas, B.F. Synthetic cannabinoid hydroxypentyl metabolites retain efficacy at human cannabinoid receptors. J. Pharmacol. Exp. Ther. 2019, 368, 414–422. [Google Scholar] [CrossRef] [Green Version]
- Canazza, I.; Ossato, A.; Trapella, C.; Fantinati, A.; De Luca, M.A.; Margiani, G.; Vincenzi, F.; Rimondo, C.; Di Rosa, F.; Gregori, A.; et al. Effect of the novel synthetic cannabinoids AKB48 and 5F-AKB48 on “tetrad”, sensorimotor, neurological and neurochemical responses in mice. In vitro and in vivo pharmacological studies. Psychopharmacology 2016, 233, 3685–3709. [Google Scholar] [CrossRef] [PubMed]
- Pinson, A.; Yarbrough, A.L.; Bush, J.M.; Cabanlong, C.V.; Shoeib, A.; Jackson, B.K.; Fukuda, S.; Gogoi, J.; Fantegrossi, W.E.; McCain, K.; et al. Metabolism, CB1 cannabinoid receptor binding and in vivo activity of synthetic cannabinoid 5F-AKB48: Implications for toxicity. Pharmacol. Biochem. Behav. 2020, 195, 172949. [Google Scholar] [CrossRef]
- Van Hout, M.C.; Hearne, E. User experiences of development of dependence on the synthetic cannabinoids, 5f-AKB48 and 5F-PB-22, and subsequent withdrawal syndromes. Int. J. Ment. Health Addict. 2017, 15, 565–579. [Google Scholar] [CrossRef]
- Holm, N.B.; Pedersen, A.J.; Dalsgaard, P.W.; Linnet, K. Metabolites of 5F-AKB-48, a synthetic cannabinoid receptor agonist, identified in human urine and liver microsomal preparations using liquid chromatography high-resolution mass spectrometry. Drug Test. Anal. 2015, 7, 199–206. [Google Scholar] [CrossRef]
- Vikingsson, S.; Josefsson, M.; Gréen, H. Identification of AKB-48 and 5F-AKB-48 metabolites in authentic human urine samples using human liver microsomes and time of flight mass spectrometry. J. Anal. Toxicol. 2015, 39, 426–435. [Google Scholar] [CrossRef]
- Karinen, R.; Tuv, S.S.; Øiestad, E.L.; Vindenes, V. Concentrations of APINACA, 5F-APINACA, UR-144 and its degradant product in blood samples from six impaired drivers compared to previous reported concentrations of other synthetic cannabinoids. Forensic Sci. Int. 2015, 246, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Hatsis, P.; Waters, N.J.; Argikar, U.A. Implications for metabolite quantification by mass spectrometry in the absence of authentic standards. Drug Metab. Dispos. 2017, 45, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Dahal, U.P.; Jones, J.P.; Davis, J.A.; Rock, D.A. Small molecule quantification by liquid chromatography-mass spectrometry for metabolites of drugs and drug candidates. Drug Metab. Dispos. 2011, 39, 2355–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laks, S.; Pelander, A.; Vuori, E.; Ali-Tolppa, E.; Sippola, E.; Ojanperä, I. Analysis of street drugs in seized material without primary reference standards. Anal. Chem. 2004, 76, 7375–7379. [Google Scholar] [CrossRef]
- Magnusson, L.-E.; Risley, D.S.; Koropchak, J.A. Aerosol-based detectors for liquid chromatography. J. Chromatogr. A 2015, 1421, 68–81. [Google Scholar] [CrossRef]
- Vishwanathan, K.; Babalola, K.; Wang, J.; Espina, R.; Yu, L.; Adedoyin, A.; Talaat, R.; Mutlib, A.; Scatina, J. Obtaining exposures of metabolites in preclinical species through plasma pooling and quantitative NMR: Addressing metabolites in safety testing (MIST) guidance without using radiolabeled compounds and chemically synthesized metabolite standards. Chem. Res. Toxicol. 2009, 22, 311–322. [Google Scholar] [CrossRef]
- Doerge, D.R.; Chang, H.C.; Churchwell, M.I.; Holder, C.L. Analysis of soy isoflavone conjugation in vitro and in human blood using liquid chromatography-mass spectrometry. Drug Metab. Dispos. 2000, 28, 298–307. [Google Scholar]
- Gandhi, A.S.; Zhu, M.; Pang, S.; Wohlfarth, A.; Scheidweiler, K.B.; Liu, H.-F.; Huestis, M.A. First characterization of AKB-48 metabolism, a novel synthetic cannabinoid, using human hepatocytes and high-resolution mass spectrometry. AAPS J. 2013, 15, 1091–1098. [Google Scholar] [CrossRef]
- Holm, N.B.; Nielsen, L.M.; Linnet, K. CYP3A4 Mediates oxidative metabolism of the synthetic cannabinoid AKB-48. AAPS J. 2015, 17, 1237–1245. [Google Scholar] [CrossRef] [Green Version]
- Kong, T.Y.; Kim, J.-H.; Kim, D.K.; Lee, H.S. Synthetic cannabinoids are substrates and inhibitors of multiple drug-metabolizing enzymes. Arch. Pharm. Res. 2018, 41, 691–710. [Google Scholar] [CrossRef]
- Chimalakonda, K.C.; Seely, K.A.; Bratton, S.M.; Brents, L.K.; Moran, C.L.; Endres, G.W.; James, L.P.; Hollenberg, P.F.; Prather, P.L.; Radominska-Pandya, A.; et al. Cytochrome P450-mediated oxidative metabolism of abused synthetic cannabinoids found in K2/Spice: Identification of novel cannabinoid receptor ligands. Drug Metab. Dispos. 2012, 40, 2174–2184. [Google Scholar] [CrossRef] [PubMed]
- Chimalakonda, K.C.; James, L.P.; Radominska-Pandya, A.; Moran, J.H. Sulfaphenazole and α-naphthoflavone attenuate the metabolism of the synthetic cannabinoids JWH-018 and AM2201 found in K2/spice. Drug Metab. Lett. 2013, 7, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Holm, N.B.; Noble, C.; Linnet, K. JWH-018 ω-OH, a shared hydroxy metabolite of the two synthetic cannabinoids JWH-018 and AM-2201, undergoes oxidation by alcohol dehydrogenase and aldehyde dehydrogenase enzymes in vitro forming the carboxylic acid metabolite. Toxicol. Lett. 2016, 259, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Schellinger, A.P.; Carr, P.W. Solubility of buffers in aqueous–organic eluents for reversed-phase liquid chromatography. LCGC N. Am. 2004, 22, 3. [Google Scholar]
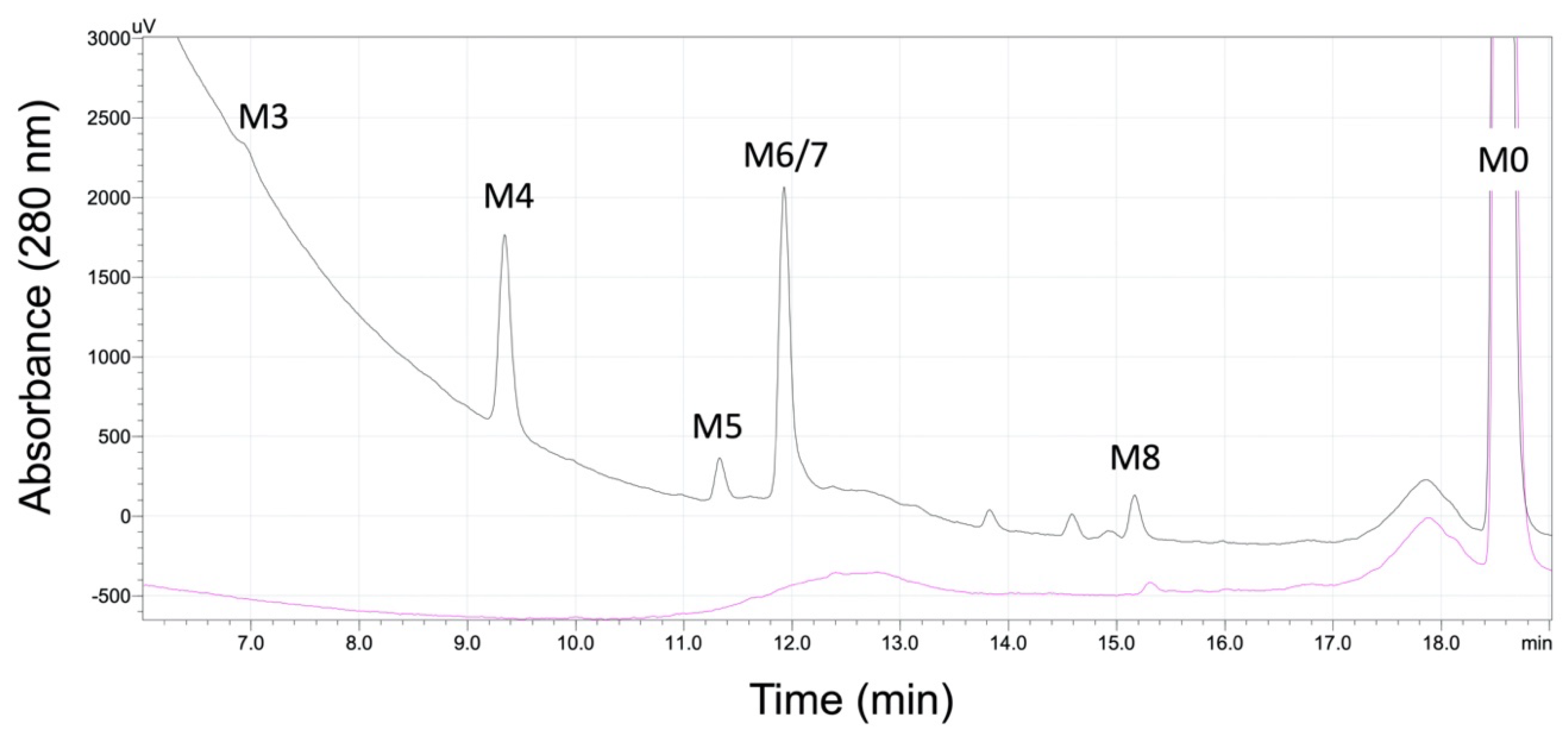
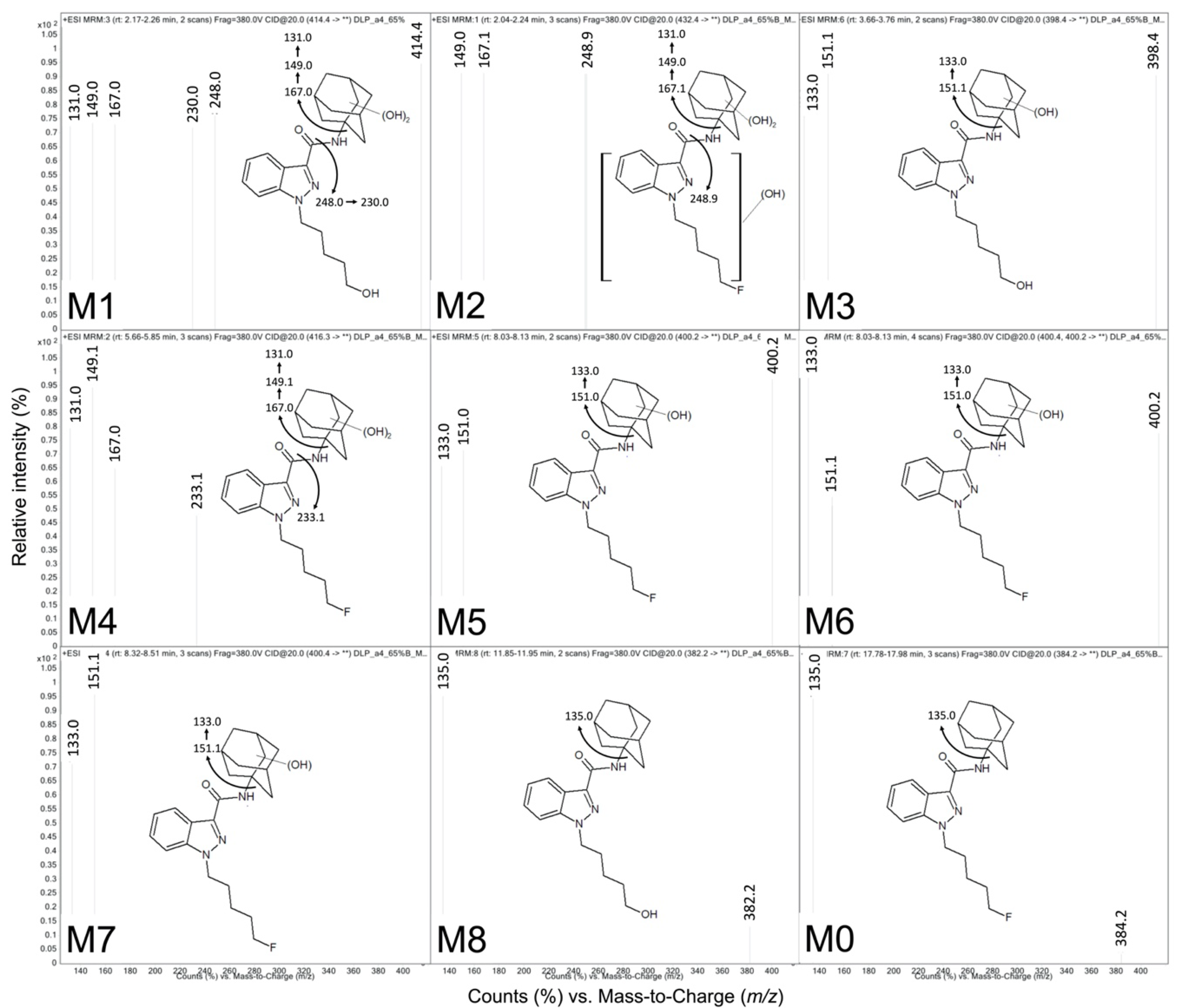

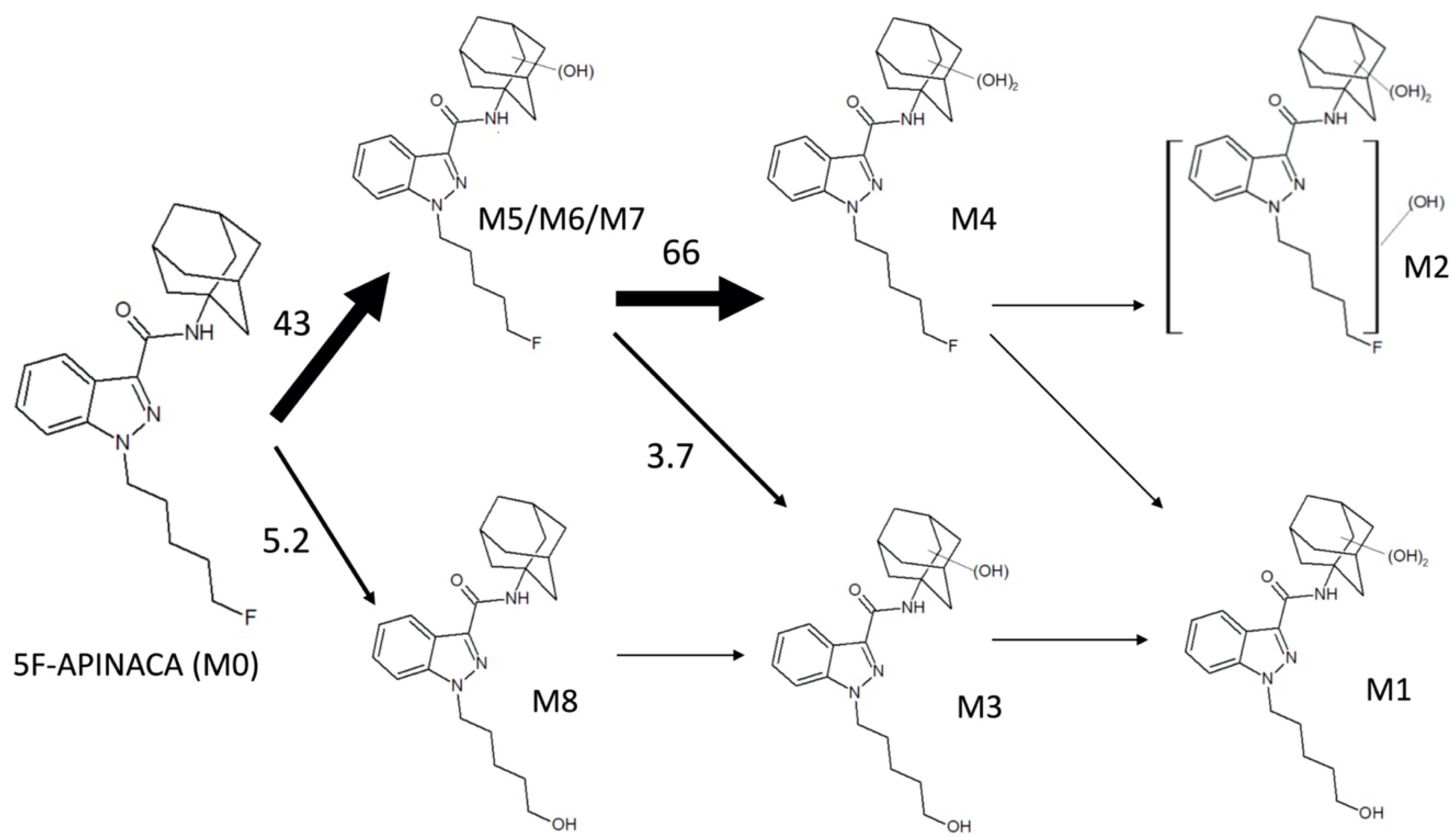
| ID | tR a | QDa b Precursor [M + H]+ (m/z) | QQQ c Precursor [M + H]+ (m/z) | Product Ions (m/z) | Proposed Modifications | Metabolite Order |
|---|---|---|---|---|---|---|
| M1 | 2.17 | ND | 414.4 | 131.0, 149.0, 167.0, 230.0, 248.0 | Dihydroxy adamantyl Oxidative defluorination | Tertiary |
| M2 | 2.24 | ND | 432.4 | 248.9, 167.1, 149.0 | Dihydroxy adamantyl Other hydroxylation | Tertiary |
| M3 | 3.76 | 398.26 | 398.4 | 151.1, 133.0 | Monohydroxy adamantyl Oxidative defluorination | Secondary |
| M4 | 5.66 | 416.26 | 416.3 | 233.1, 167.0, 149.1, 131.0 | Dihydroxy adamantyl | Secondary |
| M5 | 8.32 | 400.29 d | 400.4 | 151.1, 133.0 | Monohydroxy adamantyl | Primary |
| M6 | 8.42 | 400.29 d | 400.2 | 151.0, 133.0 | Monohydroxy adamantyl | Primary |
| M7 | 9.10 | 400.29 d | 400.2 | 151.0, 133.0 | Monohydroxy adamantyl | Primary |
| M8 | 11.9 | 404.28 | 382.2 | 135.0 | Oxidative defluorination | Primary |
| M0 | 17.9 | 384.28 | 384.2 | 135.0 | None | Parent |
| Peak ID | Mechanism | Kinetic Constants | ||||
|---|---|---|---|---|---|---|
| Vmax | Km (μM) | Ki (μM) | Vmax2 | Km2 (μM) | ||
| M3 | Substrate inhibition | 16 | 4.3 | 92 | - | - |
| (12–19) | (1.8–6.8) | (39–145) | ||||
| M4 | Substrate inhibition | 106 | 1.6 | 480 | - | - |
| (98–114) | (1.1–2.1) | (270–700) | ||||
| M5 | Michaelis–Menten + linear | 11 | 4.4 | - | 0.021 | - |
| (9.0–12) | (2.7–6.9) | (0.012–0.030) | ||||
| M6/M7 b | Michaelis–Menten + linear | 230 | 9.7 | - | 0.18 | - |
| (227–234) | (9.1–10) | (0.16–0.20) | ||||
| M5 + M6/M7 c | Michaelis–Menten + linear | 200 | 4.6 | - | 0.41 | - |
| (170–230) | (2.8–7.4) | (0.20–0.60) | ||||
| M4 + M5 + M6/M7 d | Michaelis–Menten + linear | 280 | 3.1 | - | 0.38 | - |
| (250–320) | (2.0–4.6) | (0.16–0.60) | ||||
| M8 | Biphasic | 22 | 4.2 | - | 60.1 | 149 |
| (21–23) | (3.9–4.6) | (59.7–60.6) | (143–157) | |||
Sample Availability: Not available. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinson, A.O.; Pouncey, D.L.; Schleiff, M.A.; Fantegrossi, W.E.; Prather, P.L.; Radominska-Pandya, A.; Boysen, G.; Miller, G.P. Significance of Competing Metabolic Pathways for 5F-APINACA Based on Quantitative Kinetics. Molecules 2020, 25, 4820. https://doi.org/10.3390/molecules25204820
Pinson AO, Pouncey DL, Schleiff MA, Fantegrossi WE, Prather PL, Radominska-Pandya A, Boysen G, Miller GP. Significance of Competing Metabolic Pathways for 5F-APINACA Based on Quantitative Kinetics. Molecules. 2020; 25(20):4820. https://doi.org/10.3390/molecules25204820
Chicago/Turabian StylePinson, Anna O., Dakota L. Pouncey, Mary A. Schleiff, William E. Fantegrossi, Paul L. Prather, Anna Radominska-Pandya, Gunnar Boysen, and Grover P. Miller. 2020. "Significance of Competing Metabolic Pathways for 5F-APINACA Based on Quantitative Kinetics" Molecules 25, no. 20: 4820. https://doi.org/10.3390/molecules25204820
APA StylePinson, A. O., Pouncey, D. L., Schleiff, M. A., Fantegrossi, W. E., Prather, P. L., Radominska-Pandya, A., Boysen, G., & Miller, G. P. (2020). Significance of Competing Metabolic Pathways for 5F-APINACA Based on Quantitative Kinetics. Molecules, 25(20), 4820. https://doi.org/10.3390/molecules25204820