Revisiting the Formation of a Native Disulfide Bond: Consequences for Protein Regeneration and Beyond
Abstract
1. Introduction
2. General Considerations
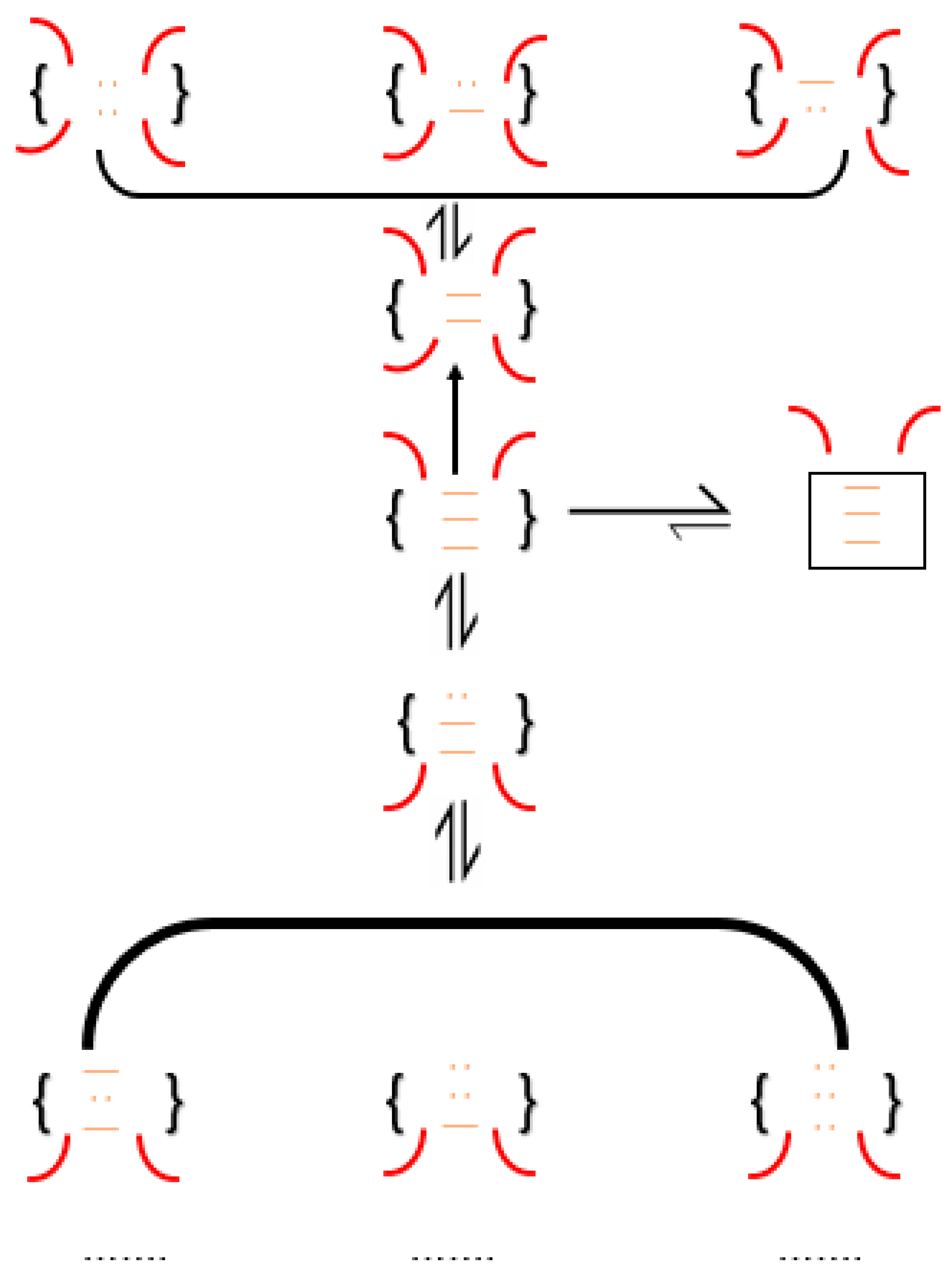
3. The Oxidative Folding Pathways of Disulfide-Bond-Containing Proteins
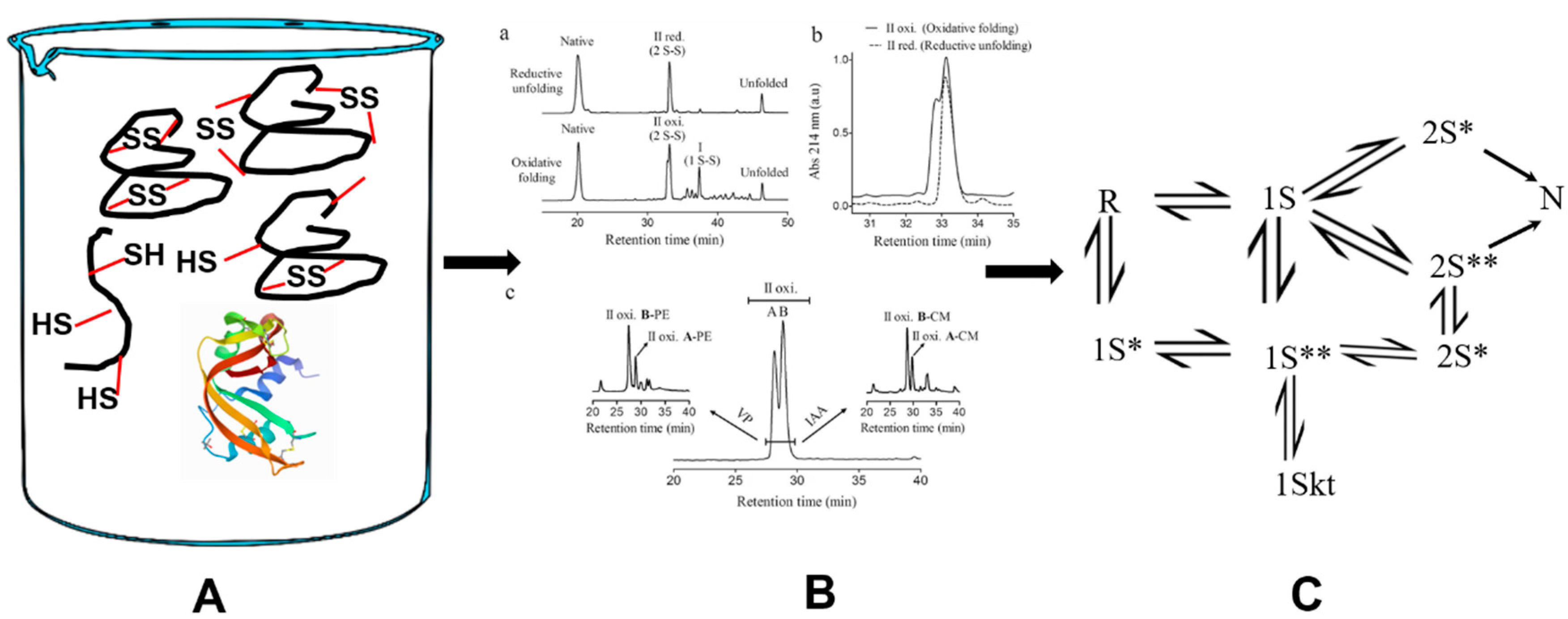
3.1. Bovine Pancreatic Ribonuclease A (RNase A)
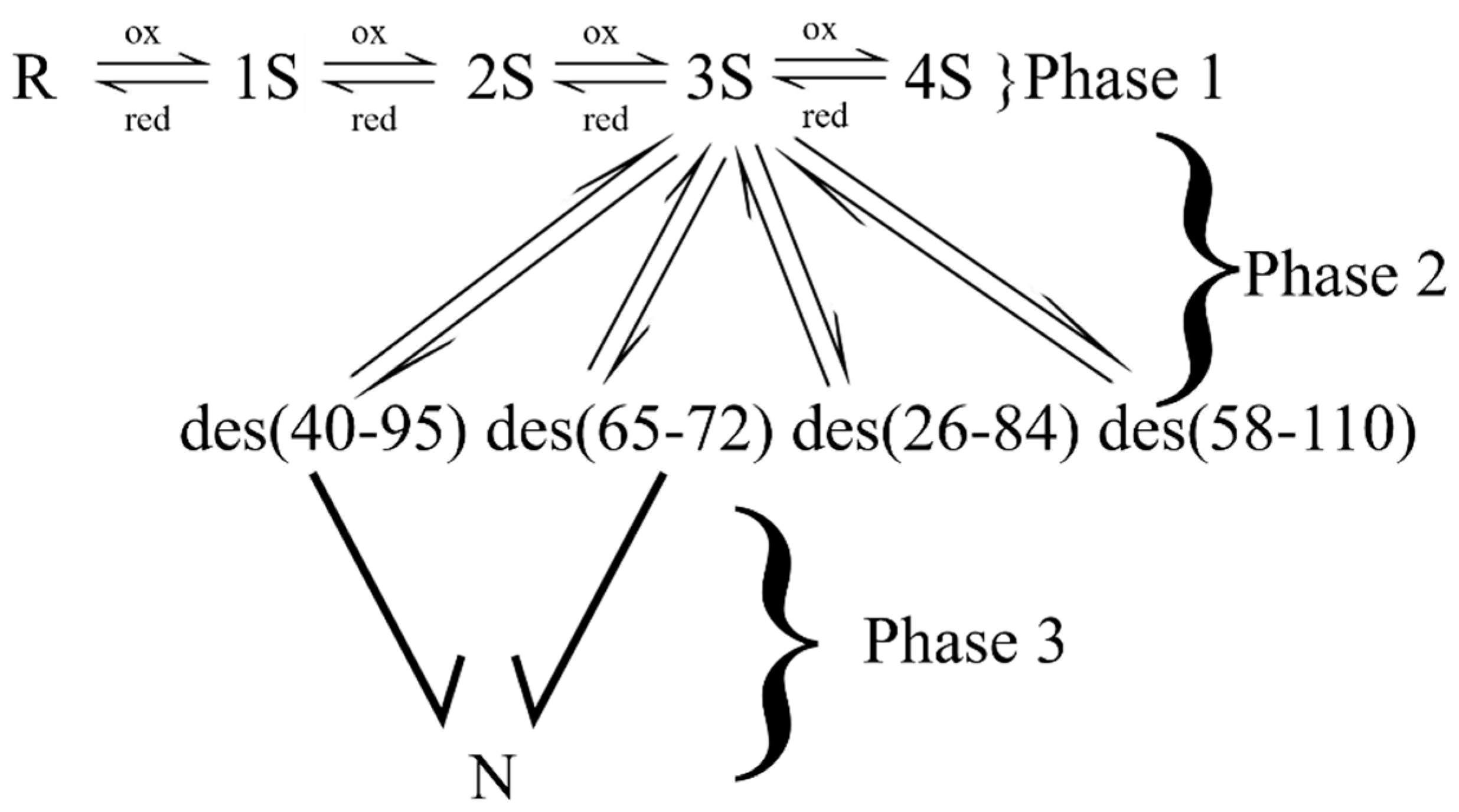
3.2. Bovine Pancreatic Trypsin Inhibitor (BPTI)
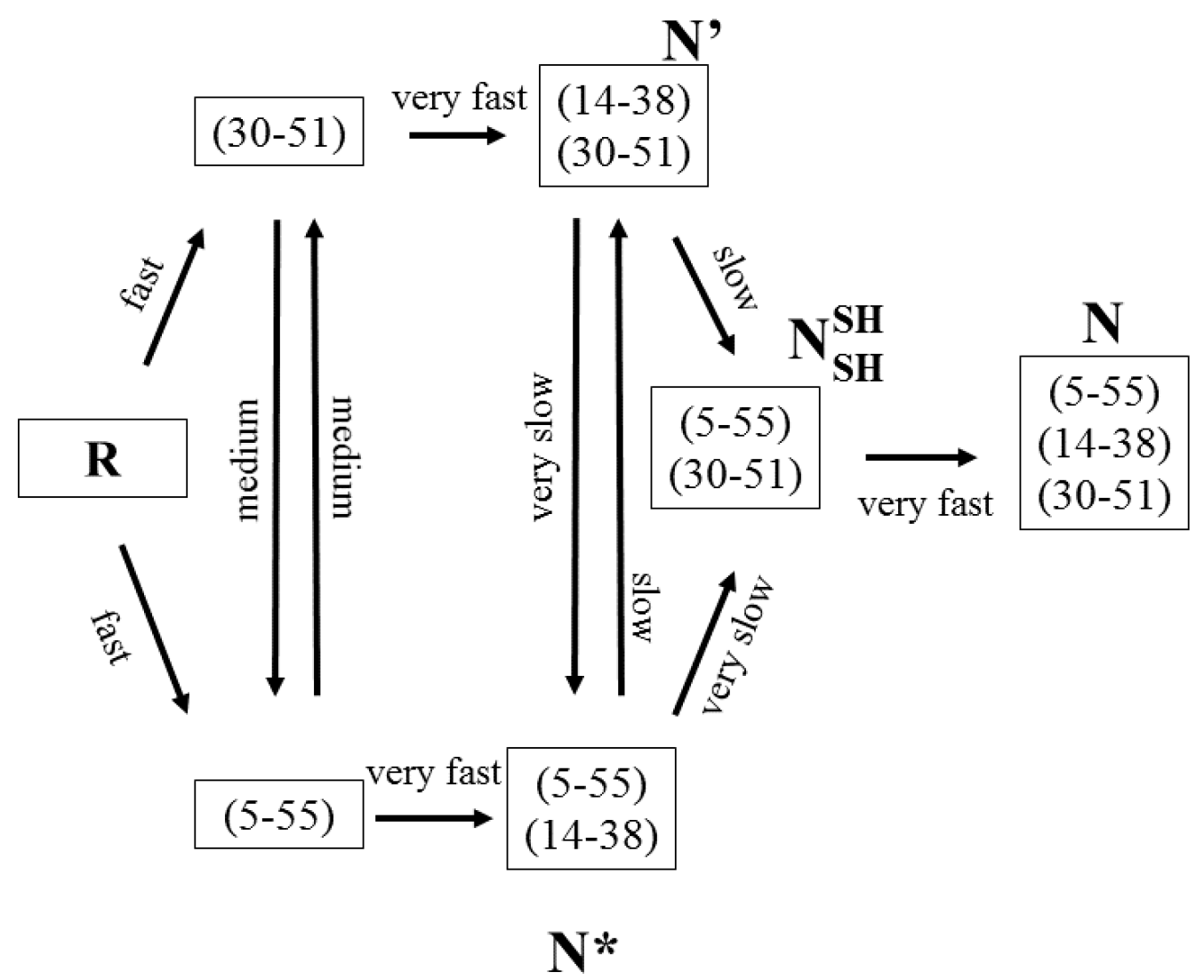
3.3. Hen-Egg White Lysozyme
4. The Role of Structure in Oxidative Folding
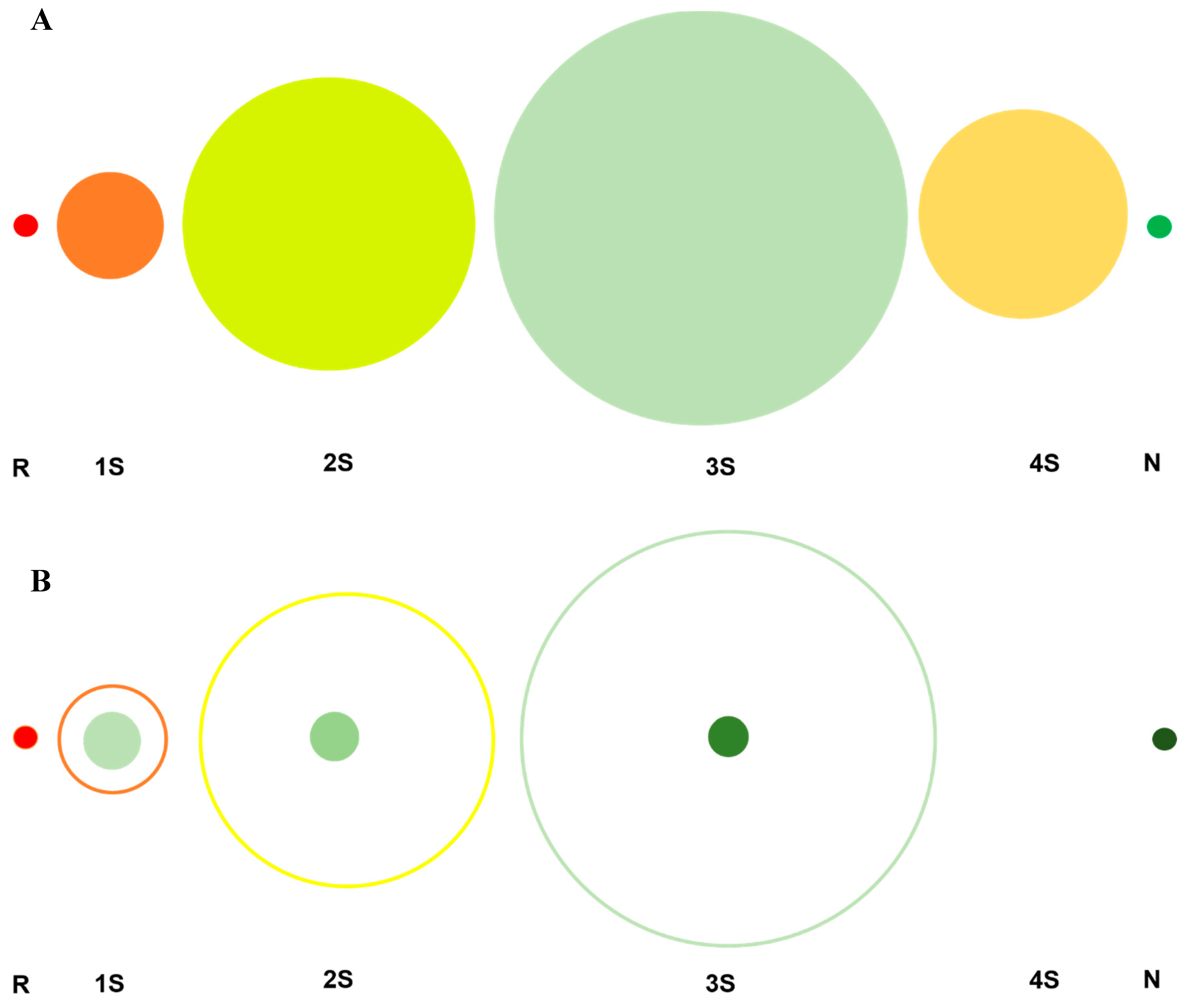
5. The Impact of Futile vs. Fruitful Reactions on the Regeneration Trajectory
6. The Formation of Native Disulfide Bonds and Structure in Other Disulfide-Bond-Containing Proteins
7. The Impact of Native Disulfide-Bonds and Early Structure Acquisition in Oxidative Protein Folding
8. Shortcomings: The Kinetically Trapped Species
9. Oxidative Folding in Biology
10. Learnings from Oxidative Protein Folding
Funding
Acknowledgments
Conflicts of Interest
References
- Thornton, J.M. Disulphide bridges in globular proteins. J. Mol. Biol. 1981, 151, 261–287. [Google Scholar] [CrossRef]
- Gilbert, H.F. Molecular and cellular aspects of thiol-disulfide exchange. Adv. Enzymol. Relat. Areas Mol. Biol. 1990, 63, 69–172. [Google Scholar] [CrossRef] [PubMed]
- Creighton, T.E. Protein folding coupled to disulphide bond formation. Biol. Chem. 1997, 378, 731–744. [Google Scholar]
- Wedemeyer, W.J.; Welker, E.; Narayan, M.; Scheraga, H.A. Disulfide bonds and protein folding. Biochemistry 2000, 39, 4207–4216. [Google Scholar] [CrossRef]
- Welker, E.; Wedemeyer, W.J.; Narayan, M.; Scheraga, H.A. Coupling of conformational folding and disulfide-bond reactions in oxidative folding of proteins. Biochemistry 2001, 40, 9059–9064. [Google Scholar] [CrossRef] [PubMed]
- Narayan, M.; Welker, E.; Wedemeyer, W.J.; Scheraga, H.A. Oxidative folding of proteins. Acc. Chem. Res. 2000, 33, 805–812. [Google Scholar] [CrossRef]
- Wedemeyer, W.J.; Scheraga, H.A. Protein folding: Overview of pathways. In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd.: Chichester, UK, 2001. [Google Scholar]
- Welker, E.; Narayan, M.; Wedemeyer, W.J.; Scheraga, H.A. Structural determinants of oxidative folding in proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 2312–2316. [Google Scholar] [CrossRef]
- Tu, B.P.; Weissman, J.S. Oxidative protein folding in eukaryotes: Mechanisms and consequences: Mechanisms and consequences. J. Cell Biol. 2004, 164, 341–346. [Google Scholar] [CrossRef]
- Moroder, L.; Buchner, J. (Eds.) Oxidative Folding of Peptides and Proteins; Royal Society of Chemistry: Cambridge, UK, 2009; pp. 236–252. ISBN 978-0-85404-148-0. [Google Scholar]
- Steiner, A.M.; Woycechowsky, K.J.; Olivera, B.M.; Bulaj, G. Reagentless oxidative folding of disulfide-rich peptides catalyzed by an intramolecular diselenide. Angew. Chem. (Int. Ed. Engl.) 2012, 51, 5580–5584. [Google Scholar] [CrossRef]
- Qin, M.; Wang, W.; Thirumalai, D. Protein folding guides disulfide bond formation. Proc. Natl. Acad. Sci. USA 2015, 112, 11241–11246. [Google Scholar] [CrossRef]
- Fass, D. Disulfide bonding in protein biophysics. Annu. Rev. Biophys. 2012, 41, 63–79. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.-Y. Diverse pathways of oxidative folding of disulfide proteins: Underlying causes and folding models. Biochemistry 2011, 50, 3414–3431. [Google Scholar] [CrossRef] [PubMed]
- Hudson, D.A.; Gannon, S.A.; Thorpe, C. Oxidative protein folding: From thiol-disulfide exchange reactions to the redox poise of the endoplasmic reticulum. Free Radic. Biol. Med. 2015, 80, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, O.B.; Hedman, R.; Marino, J.; Wickles, S.; Bischoff, L.; Johansson, M.; Müller-Lucks, A.; Trovato, F.; Puglisi, J.D.; O’Brien, E.P.; et al. Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep. 2015, 12, 1533–1540. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, Y.; Shimamoto, S.J. Folding of peptides and proteins: role of disulfide bonds, recent developments. Biomol. Concepts 2013, 4, 597–604. [Google Scholar] [CrossRef]
- Robinson, P.J.; Pringle, M.A.; Woolhead, C.A.; Bulleid, N.J. Folding of a single domain protein entering the endoplasmic reticulum precedes disulfide formation. J. Biol. Chem. 2017, 292, 6978–6986. [Google Scholar] [CrossRef]
- Woycechowsky, K.J.; Raines, R.T. Native disulfide bond formation in proteins. Curr. Opin. Chem. Biol. 2000, 4, 533–539. [Google Scholar] [CrossRef]
- Bardwell, J.C.; Lee, J.O.; Jander, G.; Martin, N.; Belin, D.; Beckwith, J. A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. USA 1993, 90, 1038–1042. [Google Scholar] [CrossRef]
- Mamathambika, B.S.; Bardwell, J.C. Disulfide-linked protein folding pathways. Annu. Rev. Cell Dev. Biol. 2008, 24, 211–235. [Google Scholar] [CrossRef]
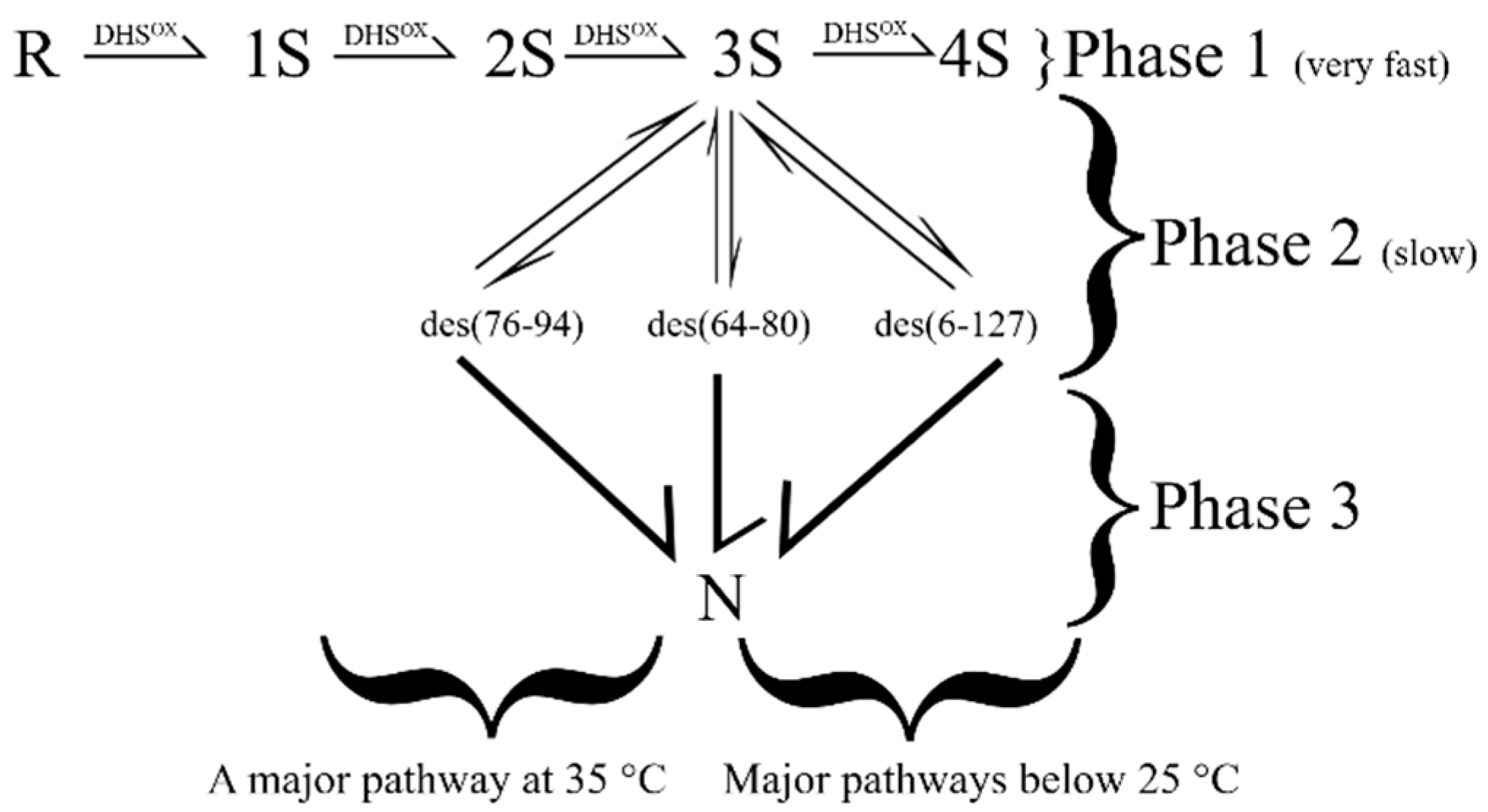
- Arai, K.; Shibagaki, W.; Shinozaki, R.; Iwaoka, M. Reinvestigation of the oxidative folding pathways of hen egg white lysozyme: Switching of the major pathways by temperature control. Int. J. Mol. Sci. 2013, 14, 13194–13212. [Google Scholar] [CrossRef]
- Creighton, T.E.; Goldenberg, D.P. Kinetic role of a meta-stable native-like two-disulphide species in the folding transition of bovine pancreatic trypsin inhibitor. J. Mol. Biol. 1984, 179, 497–526. [Google Scholar] [CrossRef]
- Goldenberg, D.P. Native and non-native intermediates in the BPTI folding pathway. Trends Biochem. Sci. 1992, 17, 257–261. [Google Scholar] [CrossRef]
- Meyer, A.J.; Riemer, J.; Rouhier, N. Oxidative protein folding: State-of-the-art and current avenues of research in plants. New Phytol. 2019, 221, 1230–1246. [Google Scholar] [CrossRef] [PubMed]
- Navon, A.; Ittah, V.; Landsman, P.; Scheraga, H.A.; Haas, E. Distributions of intramolecular distances in the reduced and denatured states of bovine pancreatic ribonuclease A. Folding initiation structures in the C-terminal portions of the reduced protein. Biochemistry 2001, 40, 105–118. [Google Scholar] [CrossRef]
- Meirovitch, H.; Vásquez, M.; Scheraga, H.A. Stability of polypeptide conformational states. II. Folding of a polypeptide chain by the scanning simulation method, and calculation of the free energy of the statistical coil: POLYPEPTIDE CONFORMATIONAL STATES. II. Biopolymers 1988, 27, 1189–1204. [Google Scholar] [CrossRef]
- Smith, L.J.; Fiebig, K.M.; Schwalbe, H.; Dobson, C.M. The concept of a random coil. Residual structure in peptides and denatured proteins. Fold. Des. 1996, 1, R95–R106. [Google Scholar] [CrossRef]
- Toal, S.; Schweitzer-Stenner, R. Local order in the unfolded state: Conformational biases and nearest neighbor interactions. Biomolecules 2014, 4, 725–773. [Google Scholar] [CrossRef]
- Tcherkasskaya, O.; Uversky, V.N. Denatured collapsed states in protein folding: Example of apomyoglobin. Proteins 2001, 44, 244–254. [Google Scholar] [CrossRef]
- Chakravarthi, S.; Jessop, C.E.; Bulleid, N.J. The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EMBO Rep. 2006, 7, 271–275. [Google Scholar] [CrossRef]
- Jessop, C.E.; Bulleid, N.J. Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 2004, 279, 55341–55347. [Google Scholar] [CrossRef]
- Shimodaira, S.; Asano, Y.; Arai, K.; Iwaoka, M. Selenoglutathione diselenide: Unique redox reactions in the GPx-like catalytic cycle and repairing of disulfide bonds in scrambled protein. Biochemistry 2017, 56, 5644–5653. [Google Scholar] [CrossRef] [PubMed]
- Okumura, M.; Shimamoto, S.; Hidaka, Y. Chemical methods for producing disulfide bonds in peptides and proteins to study folding regulation: Chemical methods for producing disulfide bonds. Curr. Protoc. Protein Sci. 2014, 76, 28.7.1–28.7.13. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Rabenstein, D.L. Formation of multiple intramolecular disulfide bonds in peptides using the reagent trans -[Pt(ethylenediamine) 2 Cl 2] 2+. Tetrahedron Lett. 2001, 42, 7203–7206. [Google Scholar] [CrossRef]
- Arai, K.; Ueno, H.; Asano, Y.; Chakrabarty, G.; Shimodaira, S.; Mugesh, G.; Iwaoka, M. Protein folding in the presence of water-soluble cyclic diselenides with novel oxidoreductase and isomerase activities. ChemBioChem 2018, 19, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Pratt, D.A. Radical substitution provides a unique route to disulfides. J. Am. Chem. Soc. 2020, 142, 10284–10290. [Google Scholar] [CrossRef]
- Patel, A.S.; Lees, W.J. Oxidative folding of lysozyme with aromatic dithiols, and aliphatic and aromatic monothiols. Bioorg. Med. Chem. 2012, 20, 1020–1028. [Google Scholar] [CrossRef]
- Pegoraro, S.; Fiori, S.; Cramer, J.; Rudolph-Böhner, S.; Moroder, L. The disulfide-coupled folding pathway of apamin as derived from diselenide-quenched analogs and intermediates. Protein Sci. 1999, 8, 1605–1613. [Google Scholar] [CrossRef]
- Scheraga, H.A.; Konishi, Y.; Rothwarf, D.M.; Mui, P.W. Toward an understanding of the folding of ribonuclease A. Proc. Natl. Acad. Sci. USA 1987, 84, 5740–5744. [Google Scholar] [CrossRef]
- Chang, J.-Y.; Li, L. Pathway of oxidative folding of alpha-lactalbumin: A model for illustrating the diversity of disulfide folding pathways. Biochemistry 2002, 41, 8405–8413. [Google Scholar] [CrossRef]
- Bronsoms, S.; Pantoja-Uceda, D.; Gabrijelcic-Geiger, D.; Sanglas, L.; Aviles, F.X.; Santoro, J.; Sommerhoff, C.P.; Arolas, J.L. Oxidative folding and structural analyses of a Kunitz-related inhibitor and its disulfide intermediates: Functional implications. J. Mol. Biol. 2011, 414, 427–441. [Google Scholar] [CrossRef]
- Creighton, T.E. Disulfide bonds as probes of protein folding pathways. Methods Enzymol. 1986, 131, 83–106. [Google Scholar] [PubMed]
- Kerstin, K.; Reimer, J. Balancing oxidative protein folding: The influence of reducing pathways on disulfide bond formation. Bba Proteins Proteom. 2014, 1844, 1383–1390. [Google Scholar]
- Gahl, R.F.; Scheraga, H.A. Oxidative folding pathway of onconase, a ribonuclease homologue: Insight into oxidative folding mechanisms from a study of two homologues. Biochemistry 2009, 48, 2740–2751. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, C.C.-J.; Chang, J.-Y. Pathway of oxidative folding of secretory leucocyte protease inhibitor: An 8-disulfide protein exhibits a unique mechanism of folding. Biochemistry 2006, 45, 6231–6240. [Google Scholar] [CrossRef]
- Chang, J.Y.; Lu, B.Y.; Lai, P.H. Oxidative folding of hirudin in human serum. Biochem. J. 2006, 394 Pt 1, 249–257. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.H.; Yan, X.; Maier, C.S.; Schimerlik, M.I.; Deinzer, M.L. Conformational analysis of intermediates involved in the in vitro folding pathways of recombinant human macrophage colony stimulating factor beta by sulfhydryl group trapping and hydrogen/deuterium pulsed labeling. Biochemistry 2002, 41, 15495–15504. [Google Scholar] [CrossRef]
- Chandrasekhar, K.; Ke, H.; Wang, N.; Goodwin, T.; Gierasch, L.M.; Gershenson, A.; Hebert, D.N. Cellular folding pathway of a metastable serpin. Proc. Natl. Acad. Sci. USA 2016, 113, 6484–6489. [Google Scholar] [CrossRef]
- Feige, M.J.; Hendershot, L.M. Disulfide bonds in ER protein folding and homeostasis. Curr. Opin. Cell Biol. 2011, 23, 167–175. [Google Scholar] [CrossRef]
- Margittai, E.; Bánhegyi, G. Oxidative folding in the endoplasmic reticulum: Towards a multiple oxidant hypothesis? FEBS Lett. 2010, 584, 2995–2998. [Google Scholar] [CrossRef]
- Narayan, M. Disulfide bonds: Protein folding and subcellular protein trafficking: Disulfide bonds. FEBS J. 2012, 279, 2272–2282. [Google Scholar] [CrossRef]
- Narayan, M. The case of oxidative folding of ribonuclease A: Factors impacting fold maturation of ER-processed proteins. In Folding of Disulfide Proteins; Springer: New York, NY, USA, 2011; pp. 23–42. [Google Scholar]
- Saaranen, M.J.; Ruddock, L.W. Disulfide bond formation in the cytoplasm. Antioxid. Redox Signal. 2013, 19, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Wallis, A.K.; Freedman, R.B. Assisting oxidative protein folding: How do protein disulphide-isomerases couple conformational and chemical processes in protein folding? In Molecular Chaperones; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–34. [Google Scholar]
- Gonzalez, V.; Pal, R.; Narayan, M. The oxidoreductase behavior of protein disulfide isomerase impedes fold maturation of endoplasmic reticulum-processed proteins in the pivotal structure-coupled step of oxidative folding: Implications for subcellular protein trafficking. Biochemistry 2010, 49, 6282–6289. [Google Scholar] [CrossRef] [PubMed]
- Mezghrani, A.; Fassio, A.; Benham, A.; Simmen, T.; Braakman, I.; Sitia, R. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. Embo J. 2001, 20, 6288–6296. [Google Scholar] [CrossRef] [PubMed]
- Irvine, A.G.; Wallis, A.K.; Sanghera, N.; Rowe, M.L.; Ruddock, L.W.; Howard, M.J.; Williamson, R.A.; Blindauer, C.A.; Freedman, R.B. Protein disulfide-isomerase interacts with a substrate protein at all stages along its folding pathway. PLoS ONE 2014, 9, e82511. [Google Scholar] [CrossRef]
- Shin, H.-C.; Song, M.-C.; Scheraga, H.A. Effect of protein disulfide isomerase on the rate-determining steps of the folding of bovine pancreatic ribonuclease A. FEBS Lett. 2002, 521, 77–80. [Google Scholar] [CrossRef]
- Raines, R.T. Ribonuclease A. Chem. Rev. 1998, 98, 1045–1066. [Google Scholar] [CrossRef]
- Cuchillo, C.M.; Nogués, M.V.; Raines, R.T. Bovine pancreatic ribonuclease: Fifty years of the first enzymatic reaction mechanism. Biochemistry 2011, 50, 7835–7841. [Google Scholar] [CrossRef]
- Marshall, G.R.; Feng, J.A.; Kuster, D.J. Back to the future: Ribonuclease A. Biopolymers 2008, 90, 259–277. [Google Scholar] [CrossRef]
- Neira, J.L.; Rico, M. Folding studies on ribonuclease A, a model protein. Fold. Des. 1997, 2, R1–R11. [Google Scholar] [CrossRef]
- Klink, T.A.; Woycechowsky, K.J.; Taylor, K.M.; Raines, R.T. Contribution of disulfide bonds to the conformational stability and catalytic activity of ribonuclease A: Disulfide bonds of ribonuclease A. Eur. J. Biochem. 2000, 267, 566–572. [Google Scholar] [CrossRef]
- Rothwarf, D.M.; Scheraga, H.A. Regeneration of bovine pancreatic ribonuclease A. 1. Steady-state distribution. Biochemistry 1993, 32, 2671–2679. [Google Scholar] [CrossRef]
- Rothwarf, D.M.; Scheraga, H.A. Regeneration of bovine pancreatic ribonuclease A. 2. Kinetics of regeneration. Biochemistry 1993, 32, 2680–2689. [Google Scholar] [CrossRef]
- Rothwarf, D.M.; Scheraga, H.A. Regeneration of bovine pancreatic ribonuclease A. 3. Dependence on the nature of the redox reagent. Biochemistry 1993, 32, 2690–2697. [Google Scholar] [CrossRef]
- Rothwarf, D.M.; Scheraga, H.A. Regeneration of bovine pancreatic ribonuclease A. 4. Temperature dependence of the regeneration rate. Biochemistry 1993, 32, 2698–2703. [Google Scholar] [CrossRef] [PubMed]
- Rothwarf, D.M.; Li, Y.J.; Scheraga, H.A. Regeneration of bovine pancreatic ribonuclease A: Identification of two nativelike three-disulfide intermediates involved in separate pathways. Biochemistry 1998, 37, 3760–3766. [Google Scholar] [CrossRef] [PubMed]
- Rothwarf, D.M.; Li, Y.J.; Scheraga, H.A. Regeneration of bovine pancreatic ribonuclease A: Detailed kinetic analysis of two independent folding pathways. Biochemistry 1998, 37, 3767–3776. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Rothwarf, D.M.; Scheraga, H.A. Nonrandom distribution of the one-disulfide intermediates in the regeneration of ribonuclease A. Biochemistry 1996, 35, 6406–6417. [Google Scholar] [CrossRef]
- Volles, M.J.; Xu, X.; Scheraga, H.A. Distribution of disulfide bonds in the two-disulfide intermediates in the regeneration of bovine pancreatic ribonuclease A: Further insights into the folding process. Biochemistry 1999, 38, 7284–7293. [Google Scholar] [CrossRef]
- Milburn, P.J.; Scheraga, H.A. Local interactions favor the native 8-residue disulfide loop in the oxidation of a fragment corresponding to the sequence Ser-50-Met-79 derived from bovine pancreatic ribonuclease A. J Protein Chem. 1988, 7, 377–398. [Google Scholar] [CrossRef]
- Altmann, K.H.; Scheraga, H.A. Local structure in ribonuclease A. Effect of amino acid substitutions on the preferential formation of the native disulfide loop in synthetic peptides corresponding to residues Cys58-Cys72 of bovine pancreatic ribonuclease A. J. Am. Chem. Soc. 1990, 112, 4926–4931. [Google Scholar] [CrossRef]
- Talluri, S.; Falcomer, C.M.; Scheraga, H.A. Energetic and structural basis for the preferential formation of the native disulfide loop involving Cys-65 and Cys-72 in synthetic peptide fragments derived from the sequence of ribonuclease A. J. Am. Chem. Soc. 1993, 115, 3041–3047. [Google Scholar] [CrossRef]
- Welker, E.; Narayan, M.; Volles, M.J.; Scheraga, H.A. Two new structured intermediates in the oxidative folding of RNase A. FEBS Lett. 1999, 460, 477–479. [Google Scholar] [CrossRef]
- Narayan, M.; Welker, E.; Scheraga, H.A. Development of a novel method to study the rate-determining step during protein regeneration: Application to the oxidative folding of RNase A at low temperature reveals BPTI-like kinetic traps. J. Am. Chem. Soc. 2001, 123, 2909–2910. [Google Scholar] [CrossRef]
- Weissman, J.S.; Kim, P.S. Reexamination of the folding of BPTI: Predominance of native intermediates. Science 1991, 253, 1386–1393. [Google Scholar] [CrossRef]
- Creighton, T.E. The disulfide folding pathway of BPTI. Science 1992, 256, 111–114. [Google Scholar] [CrossRef]
- Chang, J.-Y. Distinct folding pathways of two homologous disulfide proteins: Bovine pancreatic trypsin inhibitor and tick anticoagulant peptide. Antioxid. Redox Signal. 2011, 14, 127–135. [Google Scholar] [CrossRef]
- Bulaj, G.; Goldenberg, D.P. Early events in the disulfide-coupled folding of BPTI. Protein Sci. 1999, 8, 1825–1842. [Google Scholar] [CrossRef]
- Weissman, J.S.; Kim, P.S. A kinetic explanation for the rearrangement pathway of BPTI folding. Nat. Struct. Biol. 1995, 2, 1123–1130. [Google Scholar] [CrossRef]
- Mousa, R.; Lansky, S.; Shoham, G.; Metanis, N. BPTI folding revisited: Switching a disulfide into methylene thioacetal reveals a previously hidden path. Chem. Sci. 2018, 9, 4814–4820. [Google Scholar] [CrossRef]
- Metanis, N.; Hilvert, D. Harnessing selenocysteine reactivity for oxidative protein folding. Chem. Sci. 2015, 6, 322–325. [Google Scholar] [CrossRef]
- Roux, P.; Ruoppolo, M.; Chaffotte, A.F.; Goldberg, M.E. Comparison of the kinetics of S-S bond, secondary structure, and active site formation during refolding of reduced denatured hen egg white lysozyme. Protein Sci. 1999, 8, 2751–2760. [Google Scholar] [CrossRef] [PubMed]
- Shioi, S.; Imoto, T.; Ueda, T. Analysis of the early stage of the folding process of reduced lysozyme using all lysozyme variants containing a pair of cysteines. Biochemistry 2004, 43, 5488–5493. [Google Scholar] [CrossRef] [PubMed]
- Muttathukattil, A.N.; Singh, P.C.; Reddy, G. Role of disulfide bonds and topological frustration in the kinetic partitioning of lysozyme folding pathways. J. Phys. Chem. B 2019, 123, 3232–3241. [Google Scholar] [CrossRef]
- Dobson, C.M.; Evans, P.A.; Radford, S.E. Understanding how proteins fold: The lysozyme story so far. Trends Biochem. Sci. 1994, 19, 31–37. [Google Scholar] [CrossRef]
- Van den Berg, B.; Chung, E.W.; Robinson, C.V.; Dobson, C.M. Characterisation of the dominant oxidative folding intermediate of hen lysozyme. J. Mol. Biol. 1999, 290, 781–796. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, B.; Chung, E.W.; Robinson, C.V.; Mateo, P.L.; Dobson, C.M. The oxidative refolding of hen lysozyme and its catalysis by protein disulfide isomerase. EMBO J. 1999, 18, 4794–4803. [Google Scholar] [CrossRef] [PubMed]
- Ohkuri, T.; Shioi, S.; Imoto, T.; Ueda, T. Effect of the structure of the denatured state of lysozyme on the aggregation reaction at the early stages of folding from the reduced form. J. Mol. Biol. 2005, 347, 159–168. [Google Scholar] [CrossRef]
- Guez, V.; Roux, P.; Navon, A.; Goldberg, M.E. Role of individual disulfide bonds in hen lysozyme early folding steps. Protein Sci. 2002, 11, 1136–1151. [Google Scholar] [CrossRef]
- Wedemeyer, W.J.; Welker, E.; Scheraga, H.A. Proline cis-trans isomerization and protein folding. Biochemistry 2002, 41, 14637–14644. [Google Scholar] [CrossRef]
- Bhat, R.; Wedemeyer, W.J.; Scheraga, H.A. Proline isomerization in bovine pancreatic ribonuclease A. 2. Folding conditions. Biochemistry 2003, 42, 5722–5728. [Google Scholar] [CrossRef]
- Sendak, R.A.; Rothwarf, D.M.; Wedemeyer, W.J.; Houry, W.A.; Scheraga, H.A. Kinetic and thermodynamic studies of the folding/unfolding of a tryptophan-containing mutant of ribonuclease A. Biochemistry 1996, 35, 12978–12992. [Google Scholar] [CrossRef] [PubMed]
- Houry, W.A.; Rothwarf, D.M.; Scheraga, H.A. The nature of the initial step in the conformational folding of disulphide-intact ribonuclease A. Nat. Struct. Biol. 1995, 2, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Houry, W.A.; Rothwarf, D.M.; Scheraga, H.A. Circular dichroism evidence for the presence of burst-phase intermediates on the conformational folding pathway of ribonuclease A. Biochemistry 1996, 35, 10125–10133. [Google Scholar] [CrossRef] [PubMed]
- Houry, W.A.; Scheraga, H.A. Nature of the unfolded state of ribonuclease A: Effect of cis-trans X-Pro peptide bond isomerization. Biochemistry 1996, 35, 11719–11733. [Google Scholar] [CrossRef] [PubMed]
- Udgaonkar, J.B.; Baldwin, R.L. Early folding intermediate of ribonuclease A. Proc. Natl. Acad. Sci. USA 1990, 87, 8197–81201. [Google Scholar] [CrossRef] [PubMed]
- Anil, B.; Craig-Schapiro, R.; Raleigh, D.P. Design of a hyperstable protein by rational consideration of unfolded state interactions. J. Am. Chem. Soc. 2006, 128, 3144–3145. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J.; Wright, P.E. Unfolded proteins and protein folding studied by NMR. Chem. Rev. 2004, 104, 3607–3622. [Google Scholar] [CrossRef]
- Bowler, B.E. Residual structure in unfolded proteins. Curr. Opin. Struct. Biol. 2012, 22, 4–13. [Google Scholar] [CrossRef][Green Version]
- George, A.A.P.; Heimer, P.; Maaβ, A.; Hamaekers, J.; Hofmann-Apitius Biswas, A.; Imhof, D. Insights into the Folding of Disulfide-Rich μ-Conotoxins. Acs Omega 2018, 3, 12330–12340. [Google Scholar] [CrossRef]
- Metanis, N.; Foletti, C.; Beld, J.; Hilvert, D. Selenoglutathione-mediated rescue of kinetically trapped intermediates in oxidative protein folding. Isr. J. Chem. 2011, 51, 953–959. [Google Scholar] [CrossRef]
- Arolas, J.L.; Aviles, F.X.; Chang, J.Y.; Ventura, S. Folding of small disulfide-rich proteins: Clarifying the puzzle. Trends Biochem. Sci. 2006, 31, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Ventura, S. Folding of Disulfide Proteins; Springer Science: Berlin/Heidelberg, Germany, 2011; Volume 14. [Google Scholar]
- Chakravarty, S.; Varadarajan, R. Elucidation of determinants of protein stability through genome sequence analysis. FEBS Lett. 2000, 470, 65–69. [Google Scholar] [CrossRef]
- Patel, S.; Indu, S.; Ramakrishnan, C.; Varadarajan, R. Protein disulfide analysis and design. In Biomolecular Forms and Functions; World Scientific/Indian Inst of Science: Bangalore, India, 2013; pp. 296–311. [Google Scholar]
- Dani, V.S.; Ramakrishnan, C.; Varadarajan, R. MODIP revisited: Re-evaluation and refinement of an automated procedure for modeling of disulfide bonds in proteins. Protein Eng. 2003, 16, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.; Raleigh, D. Protein aggregation. Protein Sci. 2018, 27, 1149–1150. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Abedini, A.; Raleigh, D.P. Aggregation of islet amyloid polypeptide: From physical chemistry to cell biology. Curr. Opin. Struct. Biol. 2013, 23, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Moriarty, D.F.; Raleigh, D.P. Effects of sequential proline substitutions on amyloid formation by human amylin20-29. Biochemistry 1999, 38, 1811–1818. [Google Scholar] [CrossRef]
- Mossuto, M.F. Disulfide bonding in neurodegenerative misfolding diseases. Int. J. Cell Biol. 2013, 2013, 318319. [Google Scholar] [CrossRef]
- Mossuto, M.F.; Bolognesi, B.; Guixer, B.; Dhulesia, A.; Agostini, F.; Kumita, J.R.; Tartaglia, G.G.; Dumoulin, M.; Dobson, C.M.; Salvatella, X. Disulfide bonds reduce the toxicity of the amyloid fibrils formed by an extracellular protein. Angew. Chem. Weinheim. Bergstr. Ger. 2011, 123, 7186–7189. [Google Scholar]
- Furukawa, Y.; Kaneko, K.; Nukina, N. Tau protein assembles into isoform- and disulfide-dependent polymorphic fibrils with distinct structural properties. J. Biol. Chem. 2011, 286, 27236–27246. [Google Scholar] [CrossRef]
- Römisch, K. Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 2005, 21, 435–456. [Google Scholar] [CrossRef]
- Römisch, K. A cure for traffic jams: Small molecule chaperones in the endoplasmic reticulum: Small molecule chaperones in the ER. Traffic (Cph. Den.) 2004, 5, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-X.; Pang, X. Electrostatic interactions in protein structure, folding, binding, and condensation. Chem. Rev. 2018, 118, 1691–1741. [Google Scholar] [CrossRef] [PubMed]
- Kosuri, P.; Alegre-Cebollada, J.; Feng, J.; Kaplan, A.; Inglés-Prieto, A.; Badilla, C.L.; Fernández, J.M. Protein folding drives disulfide formation. Cell 2012, 151, 794–806. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.-M.; Lü, S.Q.; Liu, Z.-P.; Zhang, J.; Gao, B.-X.; Yao, Z.-Y.; Wu, Y. Conformational folding and disulfide bonding drive distinct stages of protein structure formation. Sci. Rep. 2018, 8, 1494. [Google Scholar] [CrossRef]
- Esperante, S.A.; Covaleda, G.; Trejo, S.A.; Bronsoms, S.; Aviles, F.X.; Ventura, S. Plasticity in the oxidative folding pathway of the high affinity Nerita Versicolor carboxypeptidase inhibitor (NvCI). Sci. Rep. 2017, 7, 5457. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, Y. Overview of the regulation of disulfide bond formation in Peptide and protein folding. Curr. Protoc. Protein Sci. 2014, 76, 28.6.1–28.6.6. [Google Scholar] [CrossRef]
- Okada, S.; Matsusaki, M.; Arai, K.; Hidaka, Y.; Inaba, K.; Okumura, M.; Muraoka, T. Coupling effects of thiol and urea-type groups for promotion of oxidative protein folding. Chem. Commun. 2019, 55, 759–762. [Google Scholar] [CrossRef]
- Carmack, M. Chirality of the disulfide in the prion proteins. J. Chem. Inf. Comput. Sci. 2004, 44, 286–288. [Google Scholar] [CrossRef]
- Takeda, M.; Miyanoiri, Y.; Terauchi, T.; Kainosho, M. (13)C-NMR studies on disulfide bond isomerization in bovine pancreatic trypsin inhibitor (BPTI). J. Biomol. Nmr 2016, 66, 37–53. [Google Scholar] [CrossRef]
- Ramanujam, V.; Shen, Y.; Ying, J.; Mobli, M. Residual dipolar couplings for resolving cysteine bridges in disulfide-rich peptides. Front. Chem. 2019, 7, 889. [Google Scholar] [CrossRef]
- Silvers, R.; Sziegat, F.; Tachibana, H.; Segawa, S.I.; Whittaker, S.; Günther, U.L.; Gabel, F.; Huang, J.R.; Blackledge, M.; Wirmer-Bartoschek, J.; et al. Modulation of structure and dynamics by disulfide bond formation in unfolded states. J. Am. Chem. Soc. 2012, 134, 6846–6854. [Google Scholar] [CrossRef]
- Schulte, L.; Mao, J.; Reitz, J.; Sreeramulu, S.; Kudlinzki, D.; Hodirnau, V.V.; Meier-Credo, J.; Saxena, K.; Buhr, F.; Langer, J.D.; et al. Cysteine oxidation and disulfide formation in the ribosomal exit tunnel. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Ceroni, A.; Passerini, A.; Vullo, A.; Frasconi, P. Disulfind: A disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res. 2006, 34, W177–W181. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narayan, M. Revisiting the Formation of a Native Disulfide Bond: Consequences for Protein Regeneration and Beyond. Molecules 2020, 25, 5337. https://doi.org/10.3390/molecules25225337
Narayan M. Revisiting the Formation of a Native Disulfide Bond: Consequences for Protein Regeneration and Beyond. Molecules. 2020; 25(22):5337. https://doi.org/10.3390/molecules25225337
Chicago/Turabian StyleNarayan, Mahesh. 2020. "Revisiting the Formation of a Native Disulfide Bond: Consequences for Protein Regeneration and Beyond" Molecules 25, no. 22: 5337. https://doi.org/10.3390/molecules25225337
APA StyleNarayan, M. (2020). Revisiting the Formation of a Native Disulfide Bond: Consequences for Protein Regeneration and Beyond. Molecules, 25(22), 5337. https://doi.org/10.3390/molecules25225337




