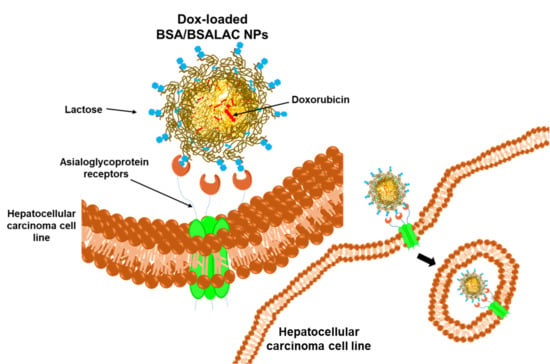
Albumin-Albumin/Lactosylated Core-Shell Nanoparticles: Therapy to Treat Hepatocellular Carcinoma for Controlled Delivery of Doxorubicin
Abstract
1. Introduction
2. Results and Discussion
2.1. Characterization of the Nanoparticles
2.1.1. Size, Zeta Potential and Encapsulation Efficiency of the Nanoparticles
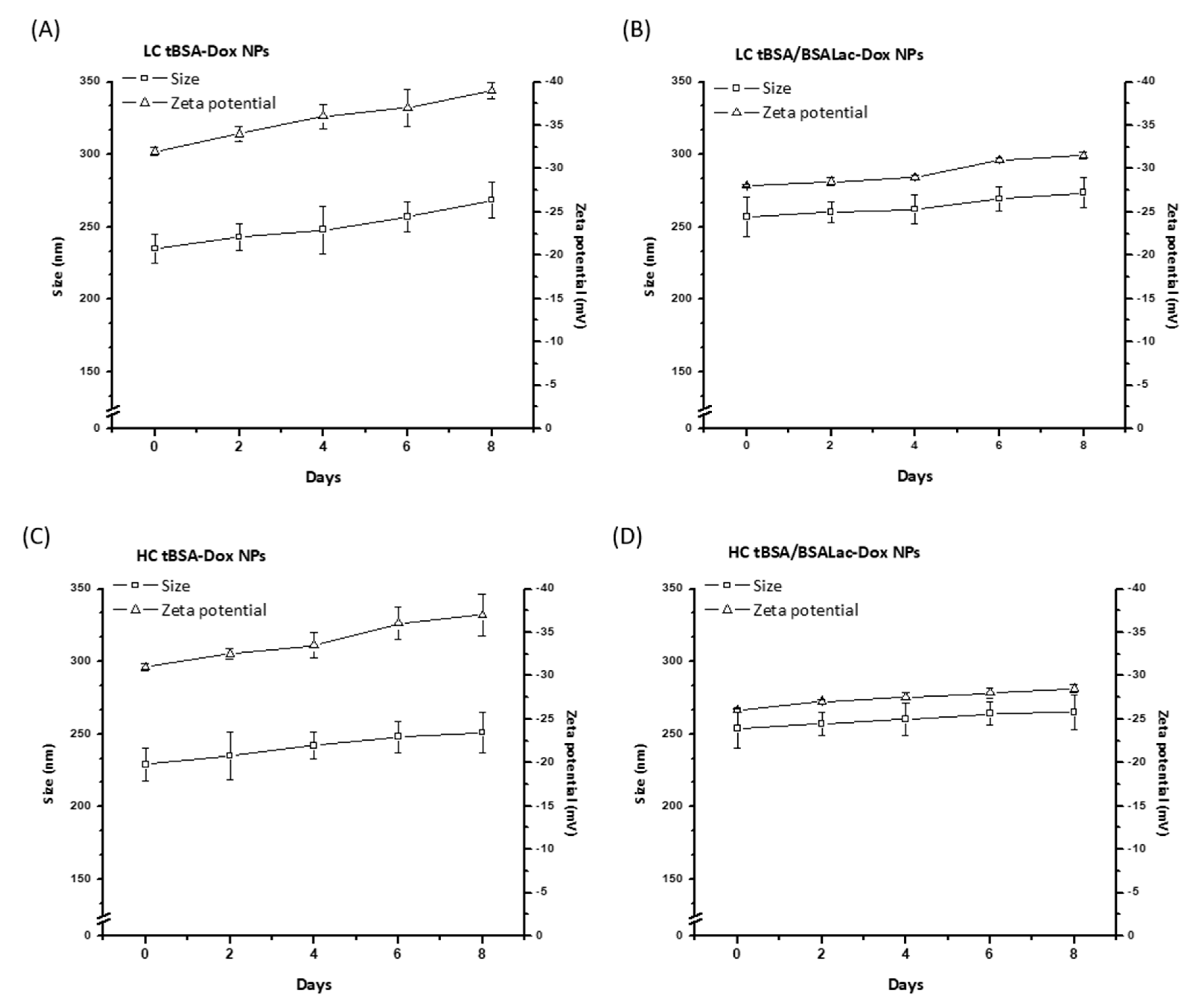
2.1.2. Stability of the Nanoparticles
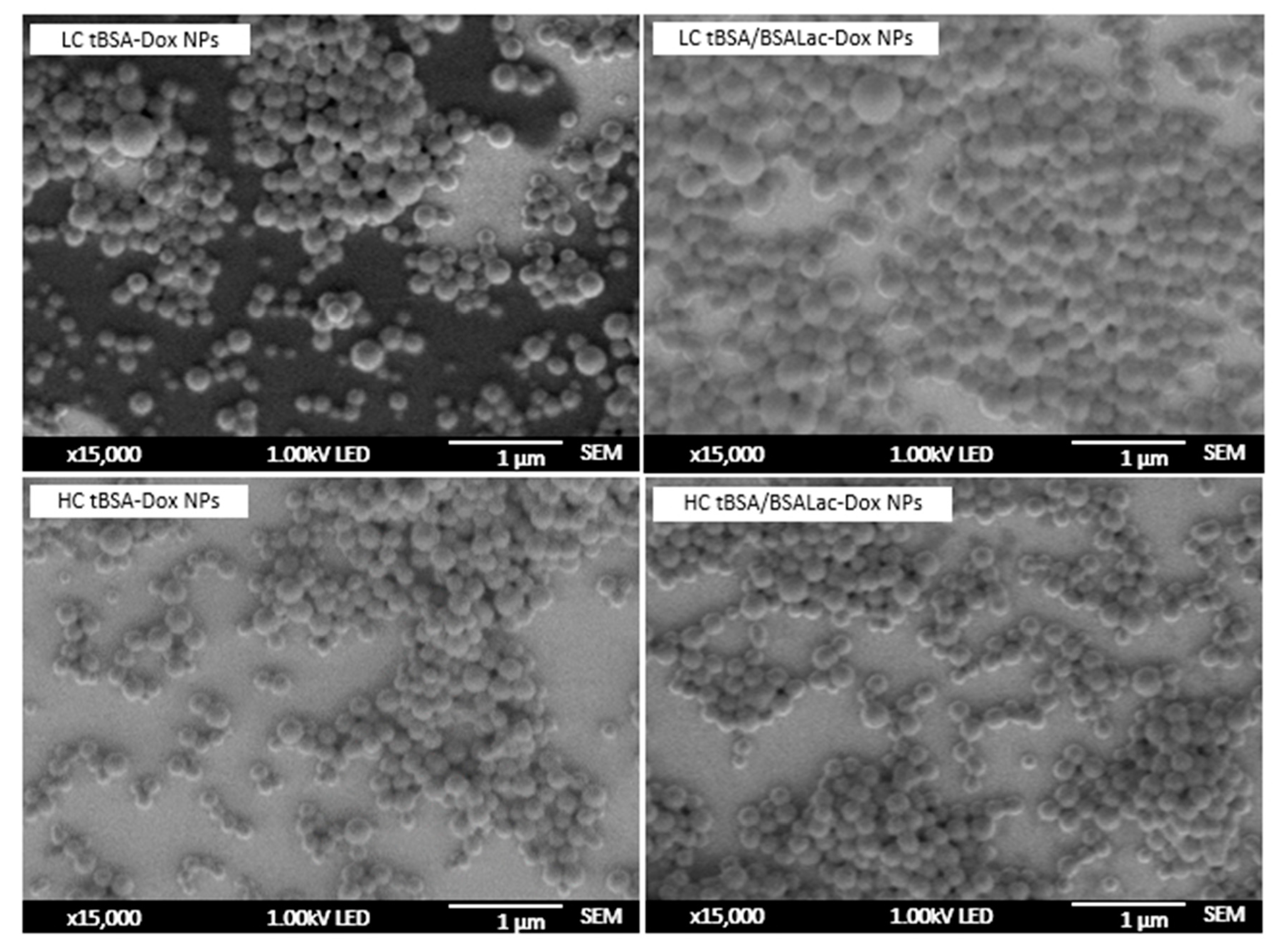
2.1.3. Morphological Characterization of the Nanoparticles
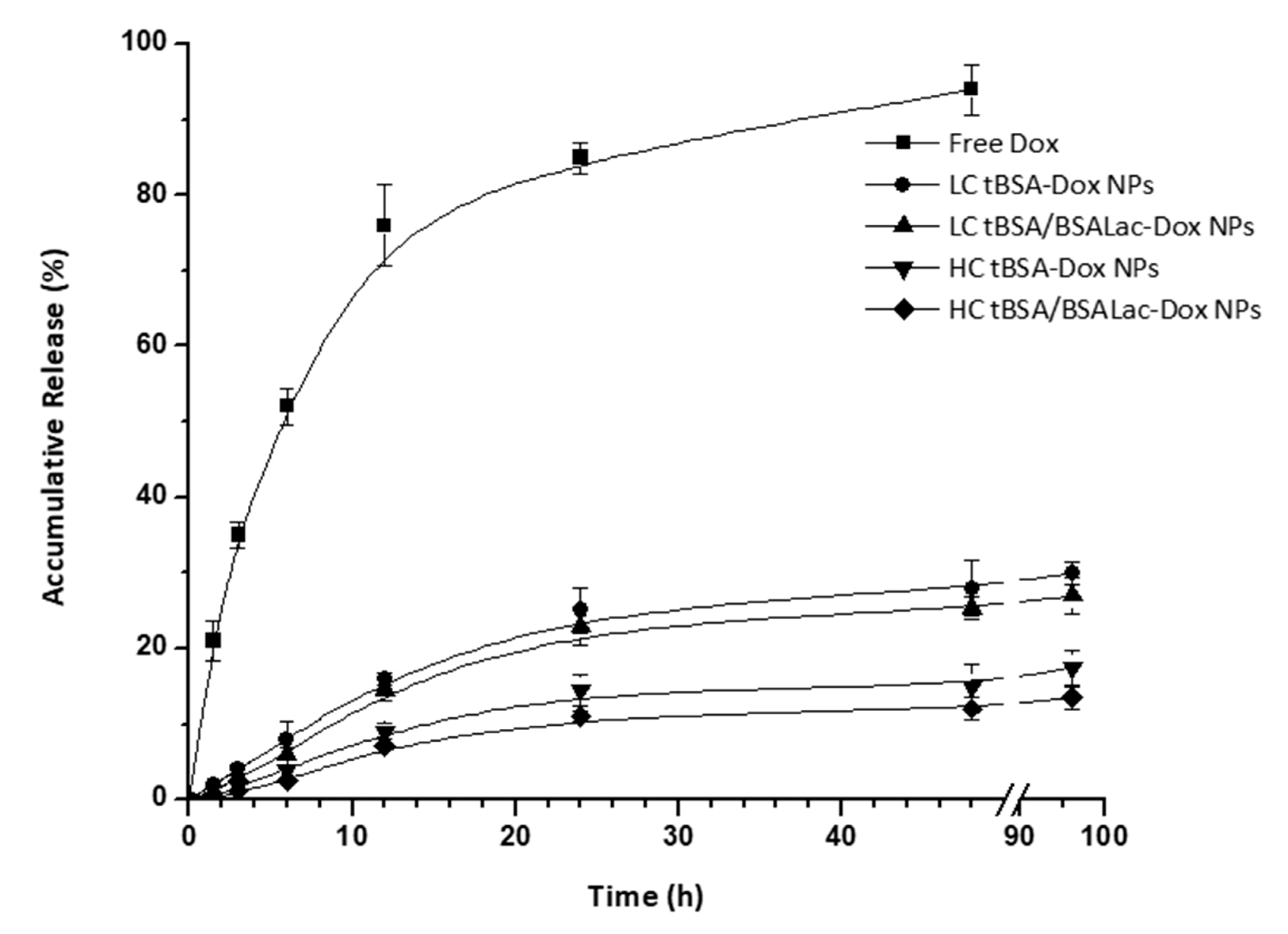
2.2. In Vitro Release Studies
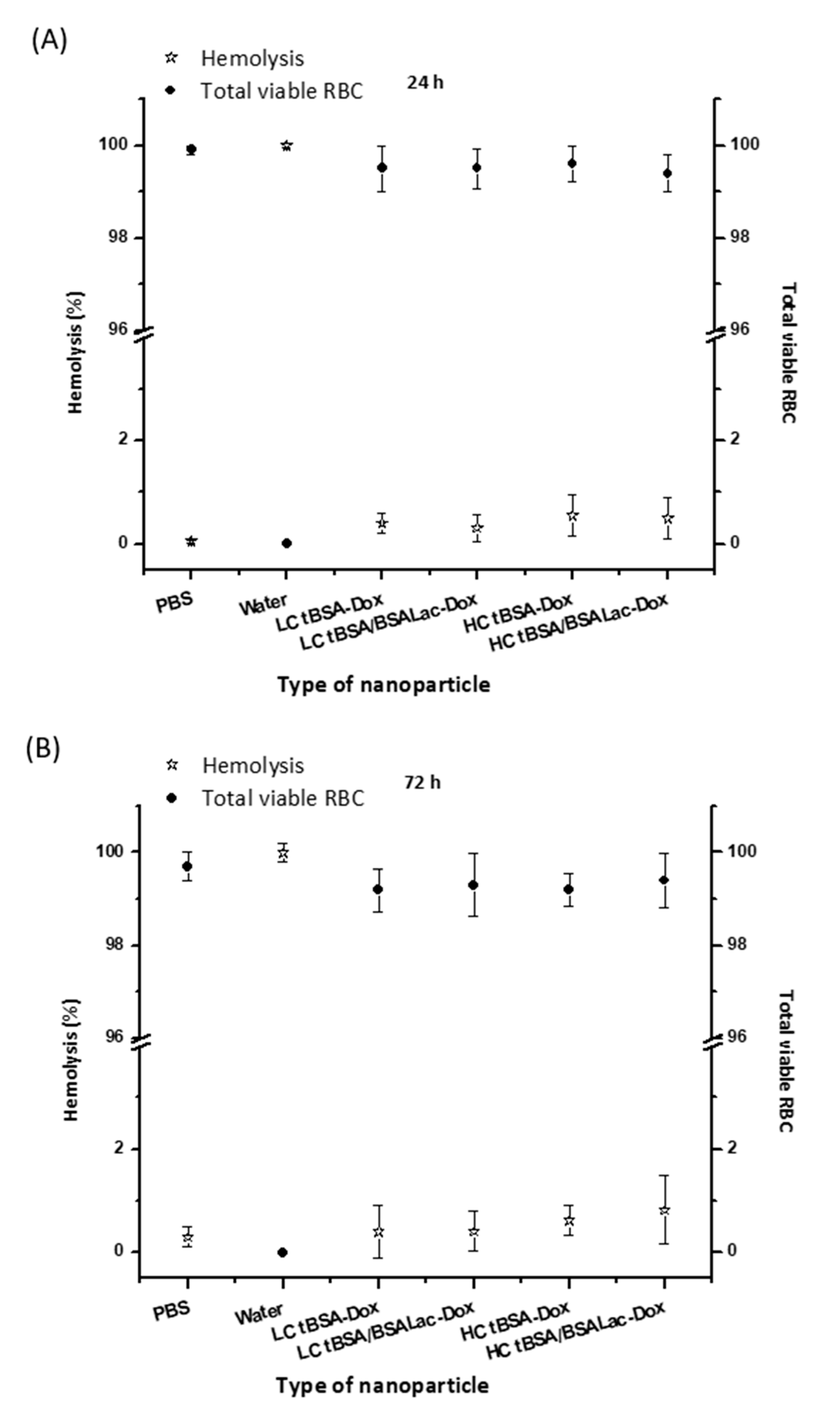
2.3. Hemocompatibility of the Nanoparticles
Hemolysis Assay and RBCs Viability Assay
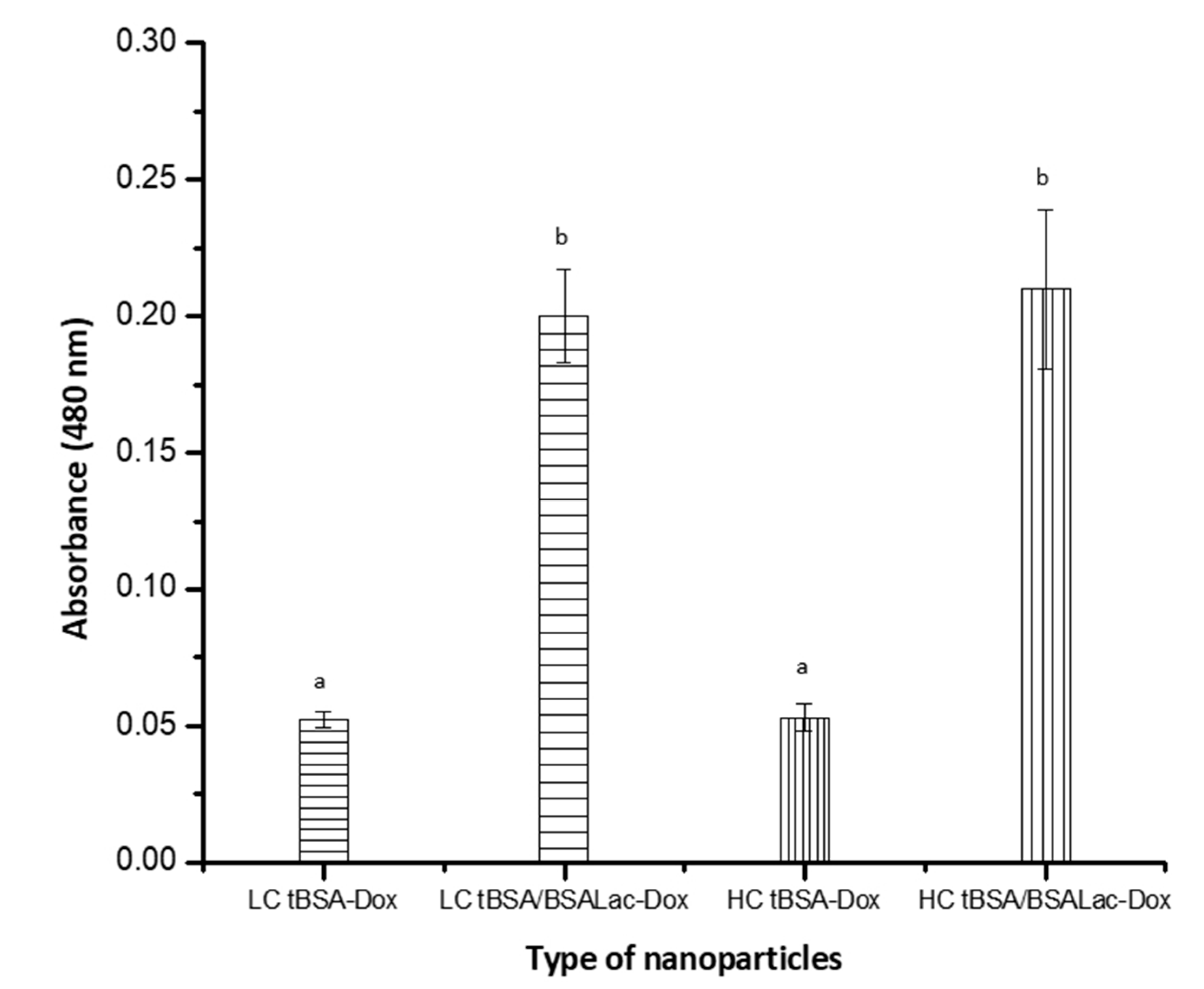
2.4. Specific Biorecognition Assays
Evaluation of the Specific Recognition of Galactose Residues by RCA I
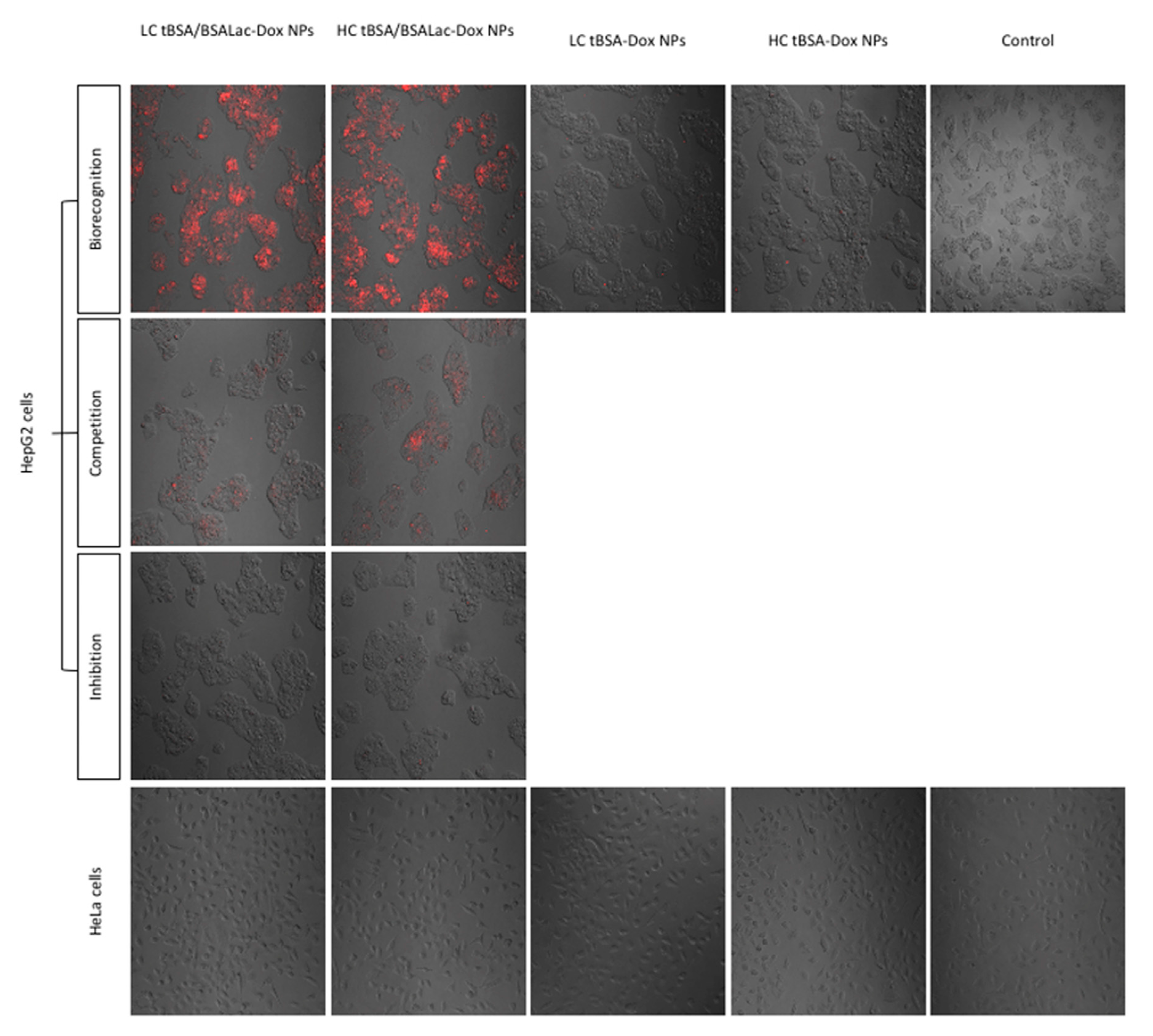
2.5. In Vitro Cytotoxic Activity of the Nanoparticles
Evaluation of Specific Recognition by HepG2 Cells
3. Materials and Methods
3.1. Materials
3.2. Synthesis of Nanoparticles
3.3. Characterization of Nanoparticles
3.3.1. Size, Zeta potential, PDI and Stability of the Nanoparticles
3.3.2. Nanoparticle Morphology
3.4. Encapsulation Efficiency and In Vitro Release Studies
3.5. Hematocompatibily
3.5.1. Hemolysis Assay
3.5.2. Red Blood Cells (RBCs) Viability Assay
3.6. In Vitro Cytotoxic Activity in HepG2 Cells
3.7. Specific Recognition Assays
3.7.1. Enzyme-Linked Lectin Recognition Assays (ELLA)
3.7.2. Evaluation of Specific Recognition by HepG2 Cells
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wallace, K. Doxorubicin-Induced Cardiac Mitochondrionopathy. Pharmacol. Toxicol. 2003, 93, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Cagel, M.; Grotz, E.; Bernabeu, E.; Moretton, M.A.; Chiappetta, D.A. Doxorubicin: Nanotechnological overviews from bench to bedside. Drug Discov. Today 2017, 22, 270–281. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, C.S.; Alam, S.; Aishwarya, R.; Miriyala, S.; Bhuiyan, M.A.N.; Panchatcharam, M.; Pattillo, C.B.; Orr, A.W.; Sadoshima, J.; Hill, J.A.; et al. Doxorubicin-induced cardiomyopathy associated with inhibition of autophagic degradation process and defects in mitochondrial respiration. Sci. Rep. 2019, 9, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Burridge, P.W.; Li, Y.F.; Matsa, E.; Wu, H.; Ong, S.-G.; Sharma, A.; Holmström, A.; Chang, A.C.; Coronado, M.J.; Ebert, A.D.; et al. Human induced pluripotent stem cell–derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat. Med. 2016, 22, 547–556. [Google Scholar] [CrossRef]
- Singal, P.K.; Iliskovic, N. Doxorubicin-Induced Cardiomyopathy. New Engl. J. Med. 1998, 339, 900–905. [Google Scholar] [CrossRef]
- Lipshultz, S.E.; Franco, V.I.; Miller, T.L.; Colan, S.D.; Sallan, S.E. Cardiovascular Disease in Adult Survivors of Childhood Cancer. Annu. Rev. Med. 2015, 66, 161–176. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, X.; Bawa-Khalfe, T.; Lu, L.-S.; Lyu, Y.L.; Liu, L.F.; Yeh, E.T.H. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat. Med. 2012, 18, 1639–1642. [Google Scholar] [CrossRef]
- Octavia, Y.; Tocchetti, C.G.; Gabrielson, K.L.; Janssens, S.; Crijns, H.J.; Moens, A.L. Doxorubicin-induced cardiomyopathy: From molecular mechanisms to therapeutic strategies. J. Mol. Cell. Cardiol. 2012, 52, 1213–1225. [Google Scholar] [CrossRef]
- Giordano, S.H.; Lin, Y.-L.; Kuo, Y.F.; Hortobagyi, G.N.; Goodwin, J.S. Decline in the Use of Anthracyclines for Breast Cancer. J. Clin. Oncol. 2012, 30, 2232–2239. [Google Scholar] [CrossRef]
- Benjanuwattra, J.; Siri-Angkul, N.; Chattipakorn, S.C.; Chattipakorn, N. Doxorubicin and its proarrhythmic effects: A comprehensive review of the evidence from experimental and clinical studies. Pharmacol. Res. 2020, 151, 104542. [Google Scholar] [CrossRef]
- Jiang, B.; Zhang, R.; Zhang, J.; Hou, Y.; Chen, X.; Zhou, M.; Tian, X.; Hao, C.; Fan, K.; Yan, X. GRP78-targeted ferritin nanocaged ultra-high dose of doxorubicin for hepatocellular carcinoma therapy. Theranostics 2019, 9, 2167–2182. [Google Scholar] [CrossRef] [PubMed]
- Allen, A. The cardiotoxicity of chemotherapeutic drugs. Semin. Oncol. 1992, 19, 529–542. [Google Scholar]
- Chatterjee, K.; Zhang, J.; Honbo, N.; Karliner, J.S. Doxorubicin Cardiomyopathy. Cardiology 2010, 115, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Takemura, G.; Fujiwara, H. Doxorubicin-Induced Cardiomyopathy. Prog. Cardiovasc. Dis. 2007, 49, 330–352. [Google Scholar] [CrossRef]
- Swain, S.; Whaley, F.S.; Ewer, M.S. Congestive heart failure in patients treated with doxorubicin. Cancer 2003, 97, 2869–2879. [Google Scholar] [CrossRef]
- Von Hoff, D.D.; Layard, M.W.; Basa, P.; Davis, H.L.; Von Hoff, A.L.; Rozencweig, M.; Muggia, F.M. Risk Factors for Doxorubicin-lnduced Congestive Heart Failure. Ann. Intern. Med. 1979, 91, 710–717. [Google Scholar] [CrossRef]
- Tan, M.L.; Choong, P.F.; Dass, C.R. Doxorubicin delivery systems based on chitosan for cancer therapy. J. Pharm. Pharmacol. 2009, 61, 131–142. [Google Scholar] [CrossRef]
- Leonetti, B.; Perin, A.; Ambrosi, E.K.; Sponchia, G.; Sgarbossa, P.; Castellin, A.; Riello, P.; Scarso, A. Mesoporous zirconia nanoparticles as drug delivery systems: Drug loading, stability and release. J. Drug Deliv. Sci. Technol. 2020, 64, 102189. [Google Scholar] [CrossRef]
- Mishra, N.; Pant, P.; Porwal, A.; Jaiswal, J.; Aquib, M. Targeted drug delivery: A review. Am. J. Pharm. Tech. Res. 2016, 6, 2249–3387. [Google Scholar]
- Jacob, J.; Haponiuk, J.T.; Thomas, S.; Gopi, S. Biopolymer based nanomaterials in drug delivery systems: A review. Mater. Today Chem. 2018, 9, 43–55. [Google Scholar] [CrossRef]
- Wakaskar, R.R. Passive and active targeting in tumor microenvironment. Int. J. Drug Dev. 2017, 9, 37–41. [Google Scholar]
- Shao, N.; Lu, S.; Wickstrom, E.; Panchapakesan, B. Integrated molecular targeting of IGF1R and HER2 surface receptors and destruction of breast cancer cells using single wall carbon nanotubes. Nanotechnology 2007, 18, 315101. [Google Scholar] [CrossRef]
- Loeian, M.S.; Aghaei, S.M.; Farhadi, F.; Rai, V.; Yang, H.W.; Johnson, M.D.; Aqil, F.; Mandadi, M.; Rai, S.N.; Panchapakesan, B. Liquid biopsy using the nanotube-CTC-chip: Capture of invasive CTCs with high purity using preferential adherence in breast cancer patients. Lab Chip 2019, 19, 1899–1915. [Google Scholar] [CrossRef] [PubMed]
- Teran-Saavedra, N.G.; Sarabia-Sainz, J.A.-I.; Silva-Campa, E.; Burgara-Estrella, A.; Guzmán-Partida, A.M.; Montfort, G.R.-C.; Pedroza-Montero, M.; Vázquez-Moreno, L. Lactosylated Albumin Nanoparticles: Potential Drug Nanovehicles with Selective Targeting Toward an In Vitro Model of Hepatocellular Carcinoma. Molecules 2019, 24, 1382. [Google Scholar] [CrossRef]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef]
- Li, Y.; Huang, G.; Diakur, J.; Wiebe, L. Targeted Delivery of Macromolecular Drugs: Asialoglycoprotein Receptor (ASGPR) Expression by Selected Hepatoma Cell Lines used in Antiviral Drug Development. Curr. Drug Deliv. 2008, 5, 299–302. [Google Scholar] [CrossRef]
- Goel, S.; Ferreira, C.A.; Dogra, P.; Yu, B.; Kutyreff, C.J.; Siamof, C.M.; Engle, J.W.; Barnhart, T.E.; Cristini, V.; Wang, Z.; et al. Size-Optimized Ultrasmall Porous Silica Nanoparticles Depict Vasculature-Based Differential Targeting in Triple Negative Breast Cancer. Small 2019, 15, e1903747. [Google Scholar] [CrossRef]
- Bian, Y.; Guo, D. Targeted Therapy for Hepatocellular Carcinoma: Co-Delivery of Sorafenib and Curcumin Using Lactosylated pH-Responsive Nanoparticles. Drug Des. Dev. Ther. 2020, 14, 647–659. [Google Scholar] [CrossRef]
- Valiño, V.; Román, M.F.S.; Ibáñez, R.; Benito, J.M.; Escudero, I.; Ortiz, I. Accurate determination of key surface properties that determine the efficient separation of bovine milk BSA and LF proteins. Sep. Purif. Technol. 2014, 135, 145–157. [Google Scholar] [CrossRef]
- Carrillo-Conde, B.R.; Roychoudhury, R.; Chavez-Santoscoy, A.V.; Narasimhan, B.; Pohl, N.L.B. High-throughput Synthesis of Carbohydrates and Functionalization of Polyanhydride Nanoparticles. J. Vis. Exp. 2012, 65, e3967. [Google Scholar] [CrossRef]
- Motevalli, S.M.; Eltahan, A.S.; Liu, L.; Magrini, A.; Rosato, N.; Guo, W.; Bottini, M.; Liang, X.-J. Co-encapsulation of curcumin and doxorubicin in albumin nanoparticles blocks the adaptive treatment tolerance of cancer cells. Biophys. Rep. 2019, 5, 19–30. [Google Scholar] [CrossRef]
- Thao, L.Q.; Lee, C.; Kim, B.; Lee, S.; Kim, T.H.; Kim, J.O.; Lee, E.S.; Oh, K.T.; Choi, H.-G.; Yoo, S.D.; et al. Doxorubicin and paclitaxel co-bound lactosylated albumin nanoparticles having targetability to hepatocellular carcinoma. Colloids Surf. B Biointerfaces 2017, 152, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Yao, P.; He, F.; Yu, C.; Huang, C. Nanoparticles with dextran/chitosan shell and BSA/chitosan core—Doxorubicin loading and delivery. Int. J. Pharm. 2010, 393, 177–185. [Google Scholar] [CrossRef]
- Hawkins, M.J.; Soon-Shiong, P.; Desai, N. Protein nanoparticles as drug carriers in clinical medicine. Adv. Drug Deliv. Rev. 2008, 60, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.-S.; Meghani, N. Impact of surface modification in BSA nanoparticles for uptake in cancer cells. Colloids Surf. B Biointerfaces 2016, 145, 653–661. [Google Scholar] [CrossRef]
- De Redín, I.L.; Boiero, C.; Martínez-Ohárriz, M.C.; Agüeros, M.; Ramos, R.; Peñuelas, I.; Allemandi, D.; Llabot, J.M.; Irache, J.M. Human serum albumin nanoparticles for ocular delivery of bevacizumab. Int. J. Pharm. 2018, 541, 214–223. [Google Scholar] [CrossRef]
- Hao, H.; Ma, Q.; Huang, C.; He, F.; Yao, P. Preparation, characterization, and in vivo evaluation of doxorubicin loaded BSA nanoparticles with folic acid modified dextran surface. Int. J. Pharm. 2013, 444, 77–84. [Google Scholar] [CrossRef]
- Li, F.-Q.; Su, H.; Wang, J.; Liu, J.; Zhu, Q.; Fei, Y.-B.; Pan, Y.-H.; Hu, J. Preparation and characterization of sodium ferulate entrapped bovine serum albumin nanoparticles for liver targeting. Int. J. Pharm. 2008, 349, 274–282. [Google Scholar] [CrossRef]
- Baneshi, M.; Dadfarnia, S.; Shabani, A.M.H.; Sabbagh, S.K.; Haghgoo, S.; Bardania, H. A novel theranostic system of AS1411 aptamer-functionalized albumin nanoparticles loaded on iron oxide and gold nanoparticles for doxorubicin delivery. Int. J. Pharm. 2019, 564, 145–152. [Google Scholar] [CrossRef]
- Yang, R.; Tang, Q.; An, Y.; Miao, F.; Liu, P.; Li, M. Preparation of folic acid-conjugated, doxorubicin-loaded, magnetic bovine serum albumin nanospheres and their antitumor effects in vitro and in vivo. Int. J. Nanomed. 2014, 9, 4231–4243. [Google Scholar] [CrossRef]
- Nosrati, H.; Salehiabar, M.; Manjili, H.K.; Danafar, H.; Davaran, S. Preparation of magnetic albumin nanoparticles via a simple and one-pot desolvation and co-precipitation method for medical and pharmaceutical applications. Int. J. Biol. Macromol. 2018, 108, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Nehate, C.; Alex, M.A.; Kumar, A.; Koul, V. Combinatorial delivery of superparamagnetic iron oxide nanoparticles (γFe2O3) and doxorubicin using folate conjugated redox sensitive multiblock polymeric nanocarriers for enhancing the chemotherapeutic efficacy in cancer cells. Mater. Sci. Eng. C 2017, 75, 1128–1143. [Google Scholar] [CrossRef] [PubMed]
- Jesus, S.; Marques, A.P.; Duarte, A.; Soares, E.; Costa, J.P.; Colaço, M.; Schmutz, M.; Som, C.; Borchard, G.; Wick, P.; et al. Chitosan Nanoparticles: Shedding Light on Immunotoxicity and Hemocompatibility. Front. Bioeng. Biotechnol. 2020, 8, 100. [Google Scholar] [CrossRef] [PubMed]
- Fornaguera, C.; Calderó, G.; Mitjans, M.; Vinardell, M.P.; Solans, C.; Vauthier, C. Interactions of PLGA nanoparticles with blood components: Protein adsorption, coagulation, activation of the complement system and hemolysis studies. Nanoscale 2015, 7, 6045–6058. [Google Scholar] [CrossRef] [PubMed]
- Pedroso-Santana, S.; Sarabia-Saínz, A.; Fleitas-Salazar, N.; Santacruz-Gómez, K.; Acosta-Elías, M.; Pedroza-Montero, M.; Riera, R. Deagglomeration and characterization of detonation nanodiamonds for biomedical applications. J. Appl. Biomed. 2017, 15, 15–21. [Google Scholar] [CrossRef]
- Gao, H.; Yang, Z.; Zhang, S.; Cao, S.; Shen, S.; Pang, Z.; Jiang, X. Ligand modified nanoparticles increases cell uptake, alters endocytosis and elevates glioma distribution and internalization. Sci. Rep. 2013, 3, srep02534. [Google Scholar] [CrossRef]
- Diaz-Galvez, K.R.; Teran-Saavedra, N.G.; Burgara-Estrella, A.; Fernández-Quiroz, D.; Silva-Campa, E.; Acosta-Elias, M.; Sarabia-Sainz, H.M.; Pedroza-Montero, M.; Sarabia-Sainz, J.A.-I. Specific capture of glycosylated graphene oxide by an asialoglycoprotein receptor: A strategic approach for liver-targeting. RSC Adv. 2019, 9, 9899–9906. [Google Scholar] [CrossRef]
- Quan, G.; Pan, X.; Wang, Z.; Wu, Q.; Li, G.; Dian, L.; Chen, B.; Wu, C. Lactosaminated mesoporous silica nanoparticles for asialoglycoprotein receptor targeted anticancer drug delivery. J. Nanobiotechnol. 2015, 13, 1–12. [Google Scholar] [CrossRef]
- He, Q.; Gao, Y.; Zhang, L.; Zhang, Z.; Gao, F.; Ji, X.; Li, Y.; Shi, J. A pH-responsive mesoporous silica nanoparticles-based multi-drug delivery system for overcoming multi-drug resistance. Biomaterials 2011, 32, 7711–7720. [Google Scholar] [CrossRef]
- Sarabia-Sainz, J.A.-I.; Sarabia-Sainz, H.M.; Montfort, G.R.-C.; Mata-Haro, V.; Guzmán-Partida, A.M.; Guzmán-Zamudio, R.; Garcia-Soto, M.J.; Vázquez-Moreno, L. K88 Fimbrial Adhesin Targeting of Microspheres Containing Gentamicin Made with Albumin Glycated with Lactose. Int. J. Mol. Sci. 2015, 16, 22425–22437. [Google Scholar] [CrossRef]


| Nanoparticles | ||||
|---|---|---|---|---|
| LC tBSA-Dox | LC tBSA/BSALac-Dox | HC tBSA-Dox | HC tBSA/BSALac-Dox | |
| Size (nm) | 235 ± 10 | 257 ± 14 | 229 ± 11 | 254 ± 14 |
| PDI | 0.17 ± 0.0 | 0.14 ± 0.1 | 0.18 ± 0.0 | 0.13 ± 0.1 |
| Zeta potential (mV) | −32.0 ± 0.5 | −28.0 ± 0.1 | −31.0 ± 0.5 | −26.0 ± 0.2 |
| E.E. % | 71.8 ± 1.3 | 73.4 ± 0.8 | 89 ± 2 | 91 ± 2 |
| HepG2 Cells | |||
|---|---|---|---|
| Nanoparticles with Dox | IC50 (µg/mL) | Nanoparticles without Dox | IC50 (µg/mL) |
| LC tBSA-Dox | 1.05 ± 0.13 a | LC tBSA | ND |
| LC tBSA/BSALac-Dox | 0.7 ± 0.09 a | LC tBSA/BSALac | ND |
| HC tBSA-Dox | 1.04 ± 0.15 a | HC tBSA | ND |
| HC tBSA/BSALac-Dox | 0.59 ± 0.07 a | HC tBSA/BSALac | ND |
| Free Dox * | 1.90 ± 0.42 b | ||
| Type of Nanoparticles | Amount of Glutaraldehyde | BSA-Lac Shell |
|---|---|---|
| LC tBSA-Dox (Control) | 5 μL | No |
| LC tBSA/BSALac-Dox | 5 μL | Yes |
| HC tBSA-Dox (Control) | 10 μL | No |
| HC tBSA/BSALac-Dox | 10 μL | Yes |
Sample Availability: Samples of the compounds are not available from the authors. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teran-Saavedra, N.G.; Sarabia-Sainz, J.A.; Velázquez-Contreras, E.F.; Ramos-Clamont Montfort, G.; Pedroza-Montero, M.; Vazquez-Moreno, L. Albumin-Albumin/Lactosylated Core-Shell Nanoparticles: Therapy to Treat Hepatocellular Carcinoma for Controlled Delivery of Doxorubicin. Molecules 2020, 25, 5432. https://doi.org/10.3390/molecules25225432
Teran-Saavedra NG, Sarabia-Sainz JA, Velázquez-Contreras EF, Ramos-Clamont Montfort G, Pedroza-Montero M, Vazquez-Moreno L. Albumin-Albumin/Lactosylated Core-Shell Nanoparticles: Therapy to Treat Hepatocellular Carcinoma for Controlled Delivery of Doxorubicin. Molecules. 2020; 25(22):5432. https://doi.org/10.3390/molecules25225432
Chicago/Turabian StyleTeran-Saavedra, Nayelli Guadalupe, Jose Andrei Sarabia-Sainz, Enrique Fernando Velázquez-Contreras, Gabriela Ramos-Clamont Montfort, Martín Pedroza-Montero, and Luz Vazquez-Moreno. 2020. "Albumin-Albumin/Lactosylated Core-Shell Nanoparticles: Therapy to Treat Hepatocellular Carcinoma for Controlled Delivery of Doxorubicin" Molecules 25, no. 22: 5432. https://doi.org/10.3390/molecules25225432
APA StyleTeran-Saavedra, N. G., Sarabia-Sainz, J. A., Velázquez-Contreras, E. F., Ramos-Clamont Montfort, G., Pedroza-Montero, M., & Vazquez-Moreno, L. (2020). Albumin-Albumin/Lactosylated Core-Shell Nanoparticles: Therapy to Treat Hepatocellular Carcinoma for Controlled Delivery of Doxorubicin. Molecules, 25(22), 5432. https://doi.org/10.3390/molecules25225432