Purification and Biochemical Characterization of a New Protease Inhibitor from Conyza dioscoridis with Antimicrobial, Antifungal and Cytotoxic Effects
Abstract
:1. Introduction
2. Results
2.1. Extraction of Protease Inhibitor from C. dioscoridis, Solvent Optimization
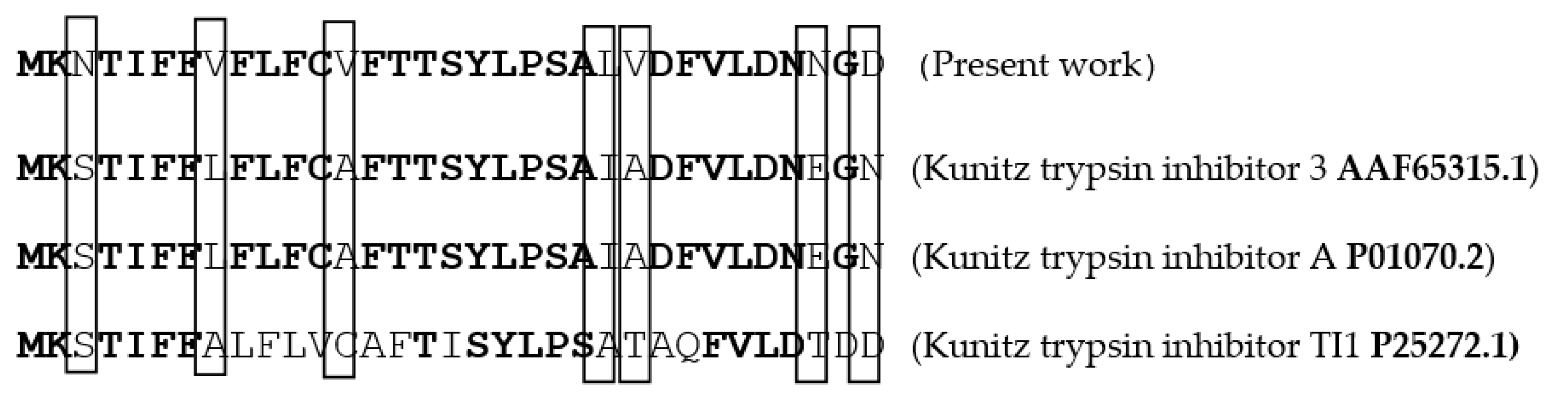
2.2. Purification of Protease Inhibitor from C. dioscoridis (PDInhibitor)
2.3. Biochemical Characterization of the Purified PDInhibitor
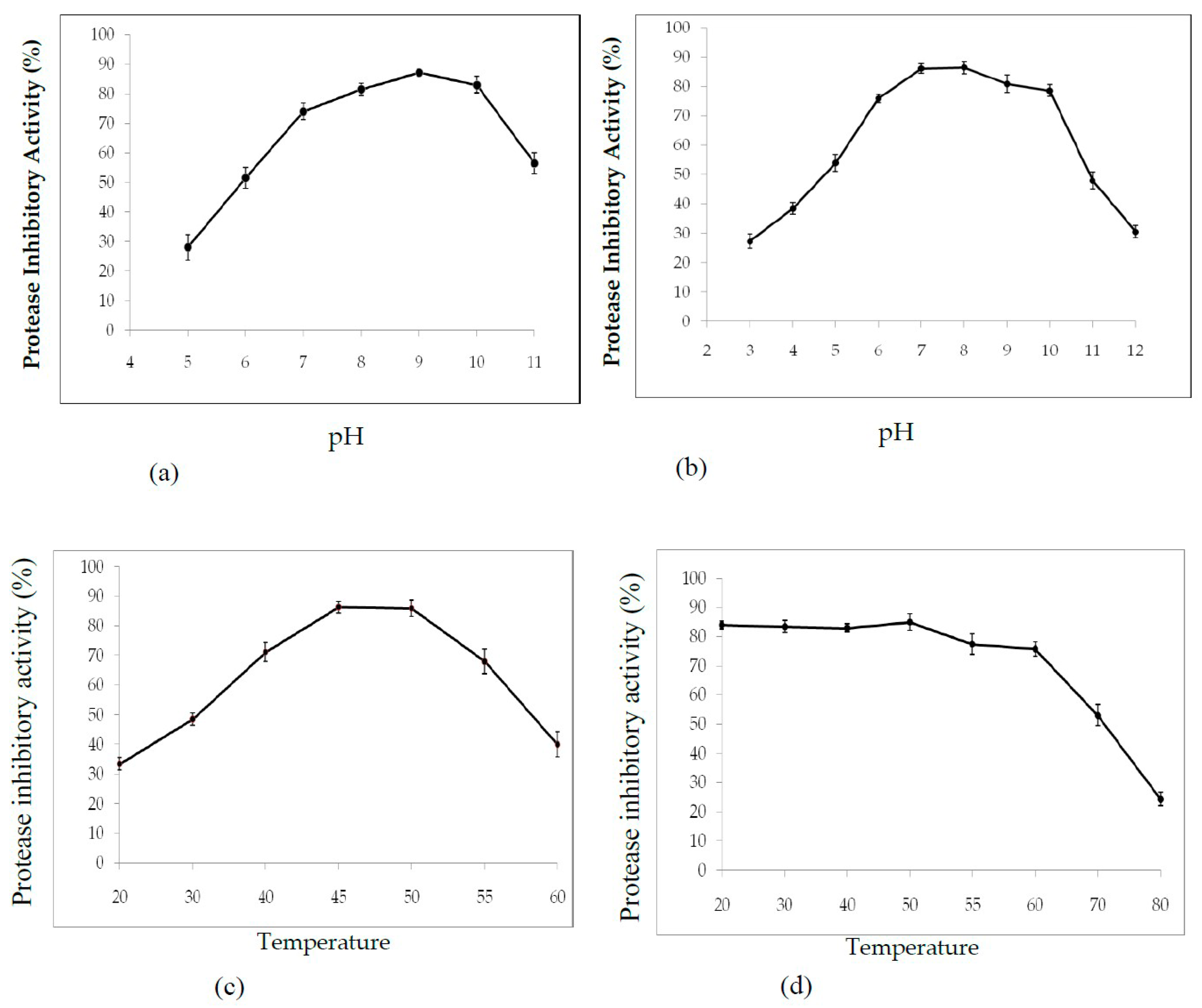
2.3.1. Effect of pH and Temperature on PDInhibitor Activity and Stability
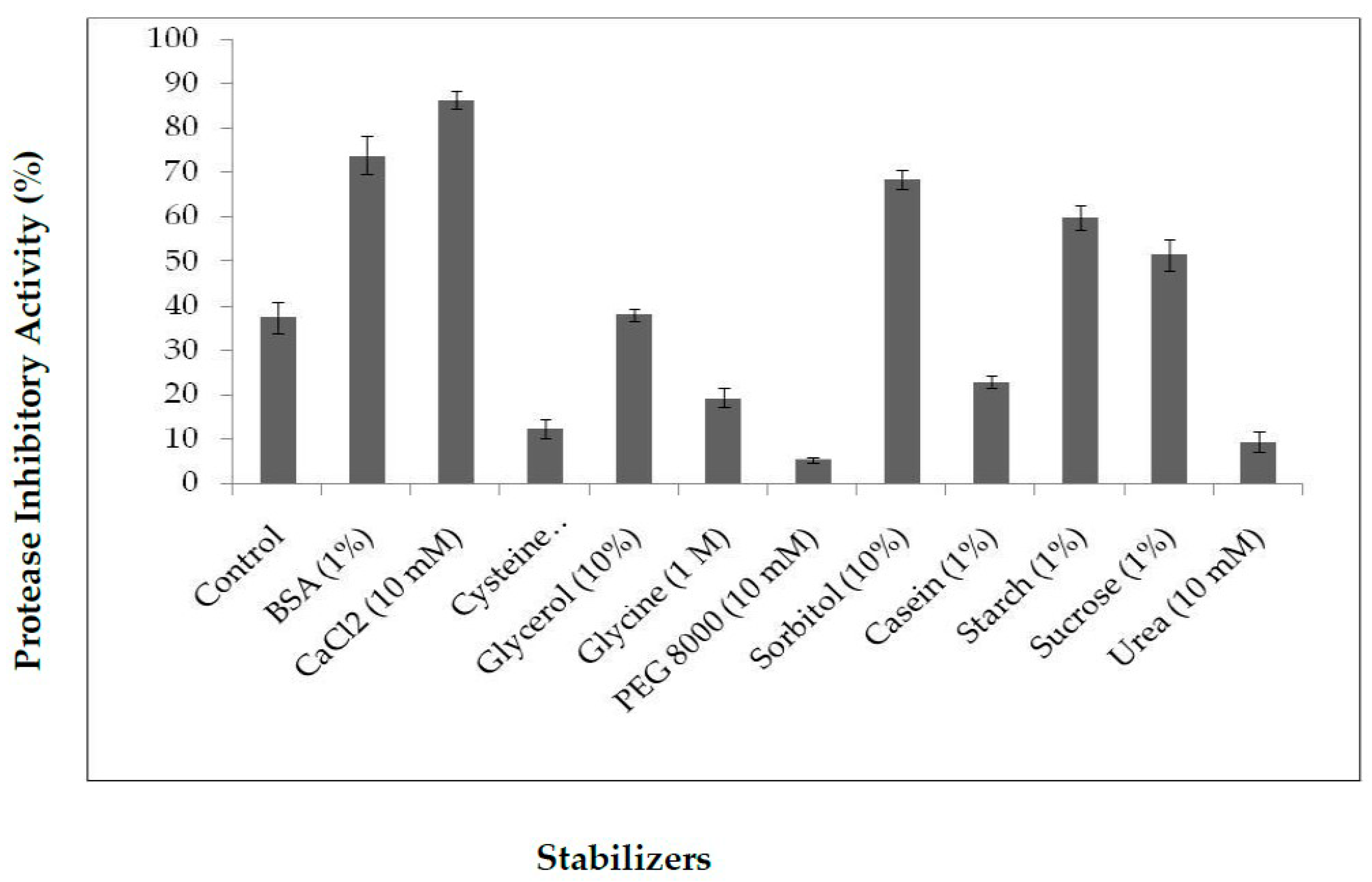
2.3.2. Influence of Stabilizers on PDInhibitor Thermo-Stability
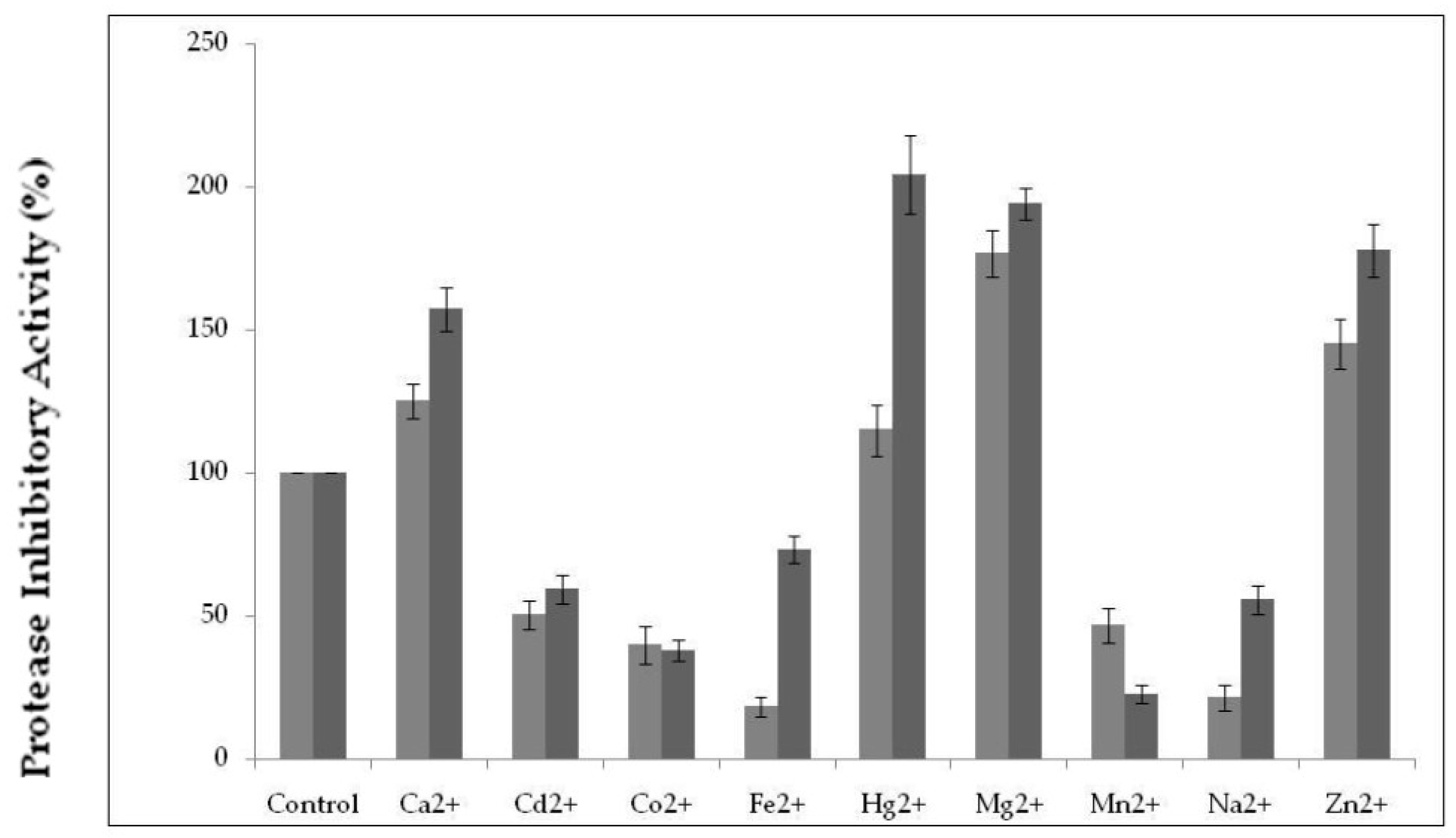
2.3.3. Effect of Various Metal Ions on Protease Inhibitor Activity
2.4. Effect of various Reducing/Oxidizing Agents and Surfactants on PDInhibitor Activity
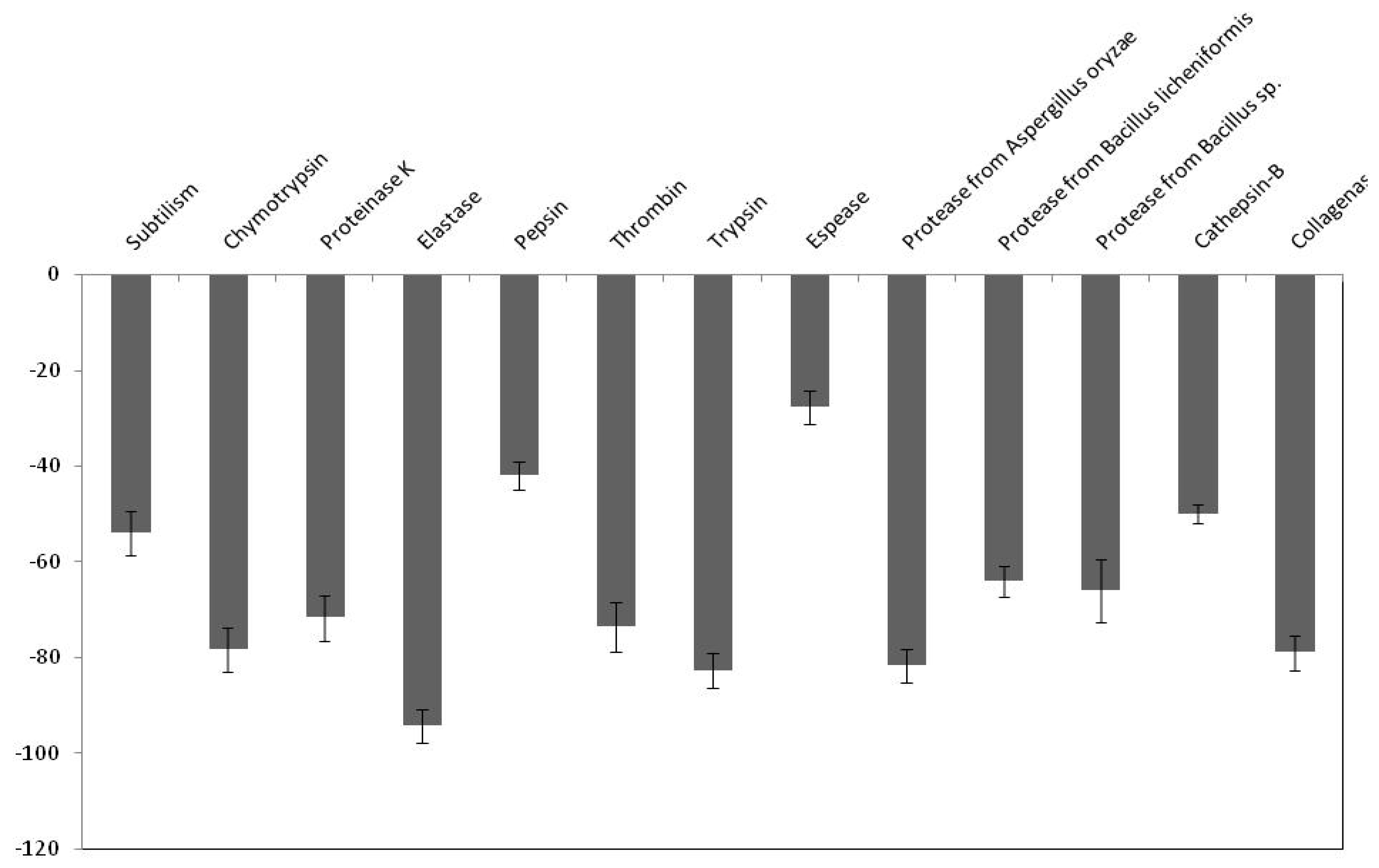
2.5. Effect of PDInhibitor on Available Important Therapeutic and Commercial Proteases
2.6. Antimicrobial Activity of the Purified PDInhibitor
2.7. Cytotoxicity of Protease Inhibitors from C. dioscoridis and from Rhamnus Frangula
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Extraction of Protease Inhibitors
4.2.2. Protease Inhibitor Assays
4.2.3. Purification of Protease Inhibitor from C. dioscoridis (PDInhibitor)
4.2.4. Protein Determination
4.2.5. Amino Acid Sequencing
4.2.6. Effect of pH and Temperature on PDInhibitor Activity and Stability
4.2.7. Influence of Stabilizers on PDInhibitor Thermostability
4.2.8. Influence of Metal Ions on PDInhibitor Activity
4.2.9. Effect of Reducing/Oxidizing Agents and Surfactants on PDInhibitor Activity
4.2.10. Effect of PDInhibitor on Available Important Therapeutic and Commercial Proteases
4.2.11. Antimicrobial Activity of PDInhibitor
4.2.12. Cytotoxicity of Protease Inhibitors
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kappelhoff, R.; Puente, X.S.; Wilson, C.H.; Seth, A.; López-Otín, C.; Overall, C.M. Overview of transcriptomic analysis of all human proteases, non-proteolytic homologs and inhibitors: Organ, tissue and ovarian cancer cell line expression profiling of the human protease degradome by the CLIP-CHIP™ DNA microarray. Biochim. Biophys. Acta (BBA) 2017, 1864, 2210–2219. [Google Scholar] [CrossRef] [PubMed]
- Puente, X.S.; Sánchez, L.M.; Overall, C.M.; López-Otín, C. Human and mouse proteases: A comparative genomic approach. Nat. Rev. Genet. 2003, 4, 544–558. [Google Scholar] [CrossRef] [PubMed]
- Eatemadi, A.; Aiyelabegan, H.T.; Negahdari, B.; Mazlomi, M.A.; Negahdari, B.; Daraee, N.; Eatemadi, R.; Sadroddiny, E. Role of protease and protease inhibitors in cancer pathogenesis and treatment. Biomed. Pharmacother. 2017, 86, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Heutinck, K.M.; Berge, I.J.M.T.; Hack, C.E.; Hamann, J.; Rowshani, A.T. Serine proteases of the human immune system in health and disease. Mol. Immunol. 2010, 47, 1943–1955. [Google Scholar] [CrossRef]
- Bond, J.S. Proteases: History, discovery, and roles in health and disease. J. Biol. Chem. 2019, 294, 1643–1651. [Google Scholar] [CrossRef] [Green Version]
- Chew, L.Y.; Toh, G.T.; Ismail, A. Application of Proteases for the Production of Bioactive Peptides. In Enzymes in Food Biotechnology; Kuddus, M., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 247–261. [Google Scholar] [CrossRef]
- Bijina, B.; Chellappan, S.; Krishna, J.G.; Basheer, S.M.; Elyas, K.; Bahkali, A.H.; Chandrasekaran, M. Protease inhibitor from Moringa oleifera with potential for use as therapeutic drug and as seafood preservative. Saudi J. Biol. Sci. 2011, 18, 273–281. [Google Scholar] [CrossRef] [Green Version]
- Puente, X.S. A Genomic Analysis of Rat Proteases and Protease Inhibitors. Genome Res. 2004, 14, 609–622. [Google Scholar] [CrossRef] [Green Version]
- Habib, H.; Majid Fazili, K. Plant protease inhibitors: A defense strategy in plants. Biotechnol. Mol. Biol. Rev. 2007, 2, 68–85. [Google Scholar]
- Clemente, M.; Corigliano, M.G.; Pariani, S.A.; Sánchez-López, E.F.; Sander, V.A.; Ramos-Duarte, V.A. Plant Serine Protease Inhibitors: Biotechnology Application in Agriculture and Molecular Farming. Int. J. Mol. Sci. 2019, 20, 1345. [Google Scholar] [CrossRef] [Green Version]
- Srikanth, S.; Chen, Z. Plant Protease Inhibitors in Therapeutics-Focus on Cancer Therapy. Front. Pharmacol. 2016, 7, 470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drag, M.; Salvesen, G.S. Emerging principles in protease-based drug discovery. Nat. Rev. Drug Discov. 2010, 9, 690–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mannaa, F.A.; El-Shamy, K.A.; El-Shaikh, K.A.; El-Kassaby, M. Efficacy of fish liver oil and propolis as neuroprotective agents in pilocarpine epileptic rats treated with valproate. Pathophysiology 2011, 18, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Zalabani, S.M.; Hetta, M.H.; Ismail, A.S. Anti-inflammatory and Antimicrobial Activity of the Different Conyza dioscoridis L. Desf. Organs. Biosaf. 2013, 2, 106. [Google Scholar] [CrossRef] [Green Version]
- Hamed, M.; Mohamed, M.A.-E.-M.; Ibrahim, M. Phytochemical investigation and cytotoxic characterization of bioactive constituents from Conyza dioscoridis. Planta Medica 2015, 81. [Google Scholar] [CrossRef] [Green Version]
- Balah, M.A. Chemical and biological characterization of Conyza dioscoridis (L.) desf. family (Compositae) in some perennial weeds control. S. Afr. J. Bot. 2016, 103, 268–274. [Google Scholar] [CrossRef]
- Elshamy, A.E.G.; El Gendy, A.; Farrag, A.H.; Nassar, M.I. Antidiabetic and antioxidant activities of phenolic extracts of Conyza dioscoridis L. shoots. Int. J. Pharm. Pharm. Sci. 2015, 7, 65–72. [Google Scholar]
- El-Meligy, R.M.; Awaad, A.S.; Soliman, G.A.; Kenawy, S.A.; Alqasoumi, S.I. Prophylactic and curative anti-ulcerogenic activity and the possible mechanisms of action of some desert plants. Saudi Pharm. J. 2016, 25, 387–396. [Google Scholar] [CrossRef] [Green Version]
- El-Seedi, H.R.; Azeem, M.; Khalil, N.S.; Sakr, H.H.; Khalifa, S.A.M.; Awang, K.; Saeed, A.; Farag, M.A.; Alajmi, M.F.; Pålsson, K.; et al. Essential oils of aromatic Egyptian plants repel nymphs of the tick Ixodes ricinus (Acari: Ixodidae). Exp. Appl. Acarol. 2017, 73, 139–157. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Jofuku, K.D.; Goldberg, R.B. Kunitz trypsin inhibitor genes are differentially expressed during the soybean life cycle and in transformed tobacco plants. Plant Cell 1989, 1, 1079–1093. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; Schipper, R.D.; Goldberg, R.B. A frameshift mutation prevents Kunitz trypsin inhibitor mRNA accumulation in soybean embryos. Plant Cell 1989, 1, 427–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Bacha, A.; Jemel, I.; Moubayed, N.M.S.; Ben Abdelmalek, I. Purification and characterization of a newly serine protease inhibitor from Rhamnus frangula with potential for use as therapeutic drug. 3 Biotech 2017, 7, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.; Rao, A.A. Reductive unfolding and oxidative refolding of a Bowman–Birk inhibitor from horsegram seeds (Dolichos biflorus): Evidence for ‘hyperreactive’ disulfide bonds and rate-limiting nature of disulfide isomerization in folding. Biochim. Biophys. Acta (BBA) 2002, 1597, 280–291. [Google Scholar] [CrossRef]
- Silverman, G.A.; Bird, P.I.; Carrell, R.W.; Church, F.C.; Coughlin, P.B.; Gettins, P.G.W.; Irving, J.A.; Lomas, D.A.; Luke, C.J.; Moyer, R.W.; et al. The Serpins Are an Expanding Superfamily of Structurally Similar but Functionally Diverse Proteins. J. Biol. Chem. 2001, 276, 33293–33296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heit, C.; Jackson, B.C.; McAndrews, M.; Seal, R.L.; Thompson, D.C.; A Silverman, G.; Nebert, D.W.; Vasiliou, V. Update of the human and mouse SERPIN gene superfamily. Hum. Genom. 2013, 7, 22. [Google Scholar] [CrossRef] [Green Version]
- Johnson, S.L.; Pellecchia, S.L.J.A.M. Structure- and Fragment-Based Approaches to Protease Inhibition. Curr. Top. Med. Chem. 2006, 6, 317–329. [Google Scholar] [CrossRef]
- Pichare, M.M.; Kachole, M.S. Protease inhibitors of pigeonpea (Cajanus cajan) and its wild relatives. Physiol. Plant. 1996, 98, 845–851. [Google Scholar] [CrossRef]
- Kunitz, M. Isolation of a crystalline protein compound of trypsin and of soybean trypsin-inhibitor. J. Gen. Physiol. 1947, 30, 311–320. [Google Scholar] [CrossRef] [Green Version]
- Creighton, T.E.; Charles, I.G. Sequences of the genes and polypeptide precursors for two bovine protease inhibitors. J. Mol. Biol. 1987, 194, 11–22. [Google Scholar] [CrossRef]
- Mello, G.C.; Oliva, M.L.V.; Sumikawa, J.T.; Machado, O.L.T.; Marangoni, S.; Novello, J.C.; Macedo, M.L.R. Purification and characterization of a new trypsin inhibitor from Dimorphandra mollis seeds. Protein J. 2001, 20, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Pandhare, J.; Zog, K.; Deshpande, V. Differential stabilities of alkaline protease inhibitors from actinomycetes: Effect of various additives on thermostability. Bioresour. Technol. 2002, 84, 165–169. [Google Scholar] [CrossRef]
- Jamal, S.; Poddar, N.K.; Singh, L.R.; Dar, T.A.; Rishi, V.; Ahmad, F. Relationship between functional activity and protein stability in the presence of all classes of stabilizing osmolytes. FEBS J. 2009, 276, 6024–6032. [Google Scholar] [CrossRef] [PubMed]
- Puntambekar, A.; Dake, M. Protease inhibitor from white cranberry beans (phaseolus vulgaris): Isolation, purification and characterization. Int. J. Pharm. Pharm. Sci. 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.; Travis, J. The oxidative inactivation of human alpha-1-proteinase inhibitor. Further evidence for methionine at the reactive center. J. Biol. Chem. 1979, 254, 4022–4026. [Google Scholar]
- Lima, T.B.; Silva, O.N.; Migliolo, L.; Souza-Filho, C.R.; Gonçalves, E.G.; Vasconcelos, I.M.; Oliveira, J.T.A.; Amaral, A.C.; Franco, O.L. A Kunitz Proteinase Inhibitor from Corms ofXanthosoma blandumwith Bactericidal Activity. J. Nat. Prod. 2011, 74, 969–975. [Google Scholar] [CrossRef]
- Banks, W.A.; Niehoff, M.L.; Brown, R.L.; Chen, Z.-Y.; Cleveland, T.E. Transport of an Antifungal Trypsin Inhibitor Isolated from Corn across the Blood-Brain Barrier. Antimicrob. Agents Chemother. 2002, 46, 2633–2635. [Google Scholar] [CrossRef] [Green Version]
- Turk, B. Targeting proteases: Successes, failures and future prospects. Nat. Rev. Drug Discov. 2006, 5, 785–799. [Google Scholar] [CrossRef]
- Chittepu, V.C.S.R.; Kalhotra, P.; Osorio-Gallardo, T.; Jiménez-Martínez, C.; La Torre, R.R.R.-D.; Gallardo-Velázquez, T.; Osorio-Revilla, G. New Molecular Insights into the Inhibition of Dipeptidyl Peptidase-4 by Natural Cyclic Peptide Oxytocin. Molecules 2019, 24, 3887. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, J.G.; Diniz, P.M.M.; De Paula, C.A.A.; Lobo, Y.A.; Paredes-Gamero, E.J.; Paschoalin, T.; Nogueira-Pedro, A.; Maza, P.K.; Toledo, M.S.; Suzuki, E.; et al. The Impaired Viability of Prostate Cancer Cell Lines by the Recombinant Plant Kallikrein Inhibitor. J. Biol. Chem. 2013, 288, 13641–13654. [Google Scholar] [CrossRef] [Green Version]
- Magee, P.J.; Owusu-Apenten, R.; McCann, M.J.; Gill, C.I.; Rowland, I.R. Chickpea (Cicer arietinum) and Other Plant-Derived Protease Inhibitor Concentrates Inhibit Breast and Prostate Cancer Cell Proliferation In Vitro. Nutr. Cancer 2012, 64, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nat. Cell Biol. 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Schneider, B.A.; Balskus, E.P. Discovery of small molecule protease inhibitors by investigating a widespread human gut bacterial biosynthetic pathway. Tetrahedron 2018, 74, 3215–3230. [Google Scholar] [CrossRef]
- Petersen, L.M.; Tisa, L.S. Molecular Characterization of Protease Activity in Serratia sp. Strain SCBI and Its Importance in Cytotoxicity and Virulence. J. Bacteriol. 2014, 196, 3923–3936. [Google Scholar] [CrossRef] [Green Version]
- Shen, A. Allosteric regulation of protease activity by small molecules. Mol. BioSyst. 2010, 6, 1431–1443. [Google Scholar] [CrossRef] [Green Version]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]




| Extraction Solvent | Protease Inhibitor Activity (%) |
|---|---|
| Phosphate buffer 0.1 M | 83.3 ± 3.05 |
| Distilled water | 47.3 ± 2.5 |
| NaCl 15% | 66,3 ± 4.1 |
| HCl 0.05 M | 35 ± 3.6 |
| NaOH 0.2% | 18.6 ± 3.5 |
| Purification Step | Total a Activity (units) | Protein b (mg) | Specific Activity (PIU/mg) | Activity Recovery (%) | Purification Factor |
|---|---|---|---|---|---|
| Extraction | 21,000 | 1350 | 15.5 | 100 | 1 |
| Ammonium sulfate fractionation(60–90%) | 18,100 | 520 | 34.8 | 86.2 | 2.2 |
| Heat treatment (70 °C, 10 min) | 12,350 | 56 | 220.5 | 58.8 | 14.2 |
| Sephadex G-50 | 5470 | 4.6 | 1189.1 | 26 | 76.7 |
| Detergents | ||
|---|---|---|
| Surfactant | Concentration (%) | Residual Activity (%) |
| Tween-20 | 1 | 80 ± 4.2 |
| Tween-80 | 1 | 64.5 ± 3.5 |
| Triton-X100 | 1 | 63.9 ± 4.1 |
| SDS | 1 | 141 ± 8.4 |
| NaTDC | 1 | 193.5 ± 6.3 |
| Oxidizing agents | ||
| H2O2 | 1 | 58.35 ± 3.3 |
| 2 | 40.75 ± 3.1 | |
| 3 | 26.15 ± 3.3 | |
| 4 | 18.45 ± 2.1 | |
| 5 | 8.1 ± 1.2 | |
| NaOCl | 1 | 64 ± 4.2 |
| 2 | 37.2 ± 3.1 | |
| 3 | 21.2 ± 2.2 | |
| 4 | 10.15 ± 1.6 | |
| 5 | 6.05 ± 0.9 | |
| DMSO | 1 | 89.7 ± 3.8 |
| 2 | 83.15 ± 3 | |
| 3 | 74.6 ± 3.6 | |
| 4 | 64.9 ± 2.6 | |
| 5 | 50.5 ± 3.5 | |
| Reducing agents | ||
| DTT | 0.2 | 107 ± 4.2 |
| 0.4 | 133 ± 5.6 | |
| 0.6 | 144 ± 4.2 | |
| 0.8 | 157.5 ± 3.5 | |
| 1 | 160 ± 4.2 | |
| βME | 0.2 | 103 ± 2.8 |
| 0.4 | 122 ± 4.2 | |
| 0.6 | 139.5 ± 3.5 | |
| 0.8 | 151.5 ± 3.5 | |
| 1 | 167 ± 2.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karray, A.; Alonazi, M.; Smaoui, S.; Michaud, P.; Soliman, D.; Ben Bacha, A. Purification and Biochemical Characterization of a New Protease Inhibitor from Conyza dioscoridis with Antimicrobial, Antifungal and Cytotoxic Effects. Molecules 2020, 25, 5452. https://doi.org/10.3390/molecules25225452
Karray A, Alonazi M, Smaoui S, Michaud P, Soliman D, Ben Bacha A. Purification and Biochemical Characterization of a New Protease Inhibitor from Conyza dioscoridis with Antimicrobial, Antifungal and Cytotoxic Effects. Molecules. 2020; 25(22):5452. https://doi.org/10.3390/molecules25225452
Chicago/Turabian StyleKarray, Aida, Mona Alonazi, Slim Smaoui, Philippe Michaud, Dina Soliman, and Abir Ben Bacha. 2020. "Purification and Biochemical Characterization of a New Protease Inhibitor from Conyza dioscoridis with Antimicrobial, Antifungal and Cytotoxic Effects" Molecules 25, no. 22: 5452. https://doi.org/10.3390/molecules25225452







