Impact of C-Terminal Chemistry on Self-Assembled Morphology of Guanosine Containing Nucleopeptides
Abstract
:1. Introduction
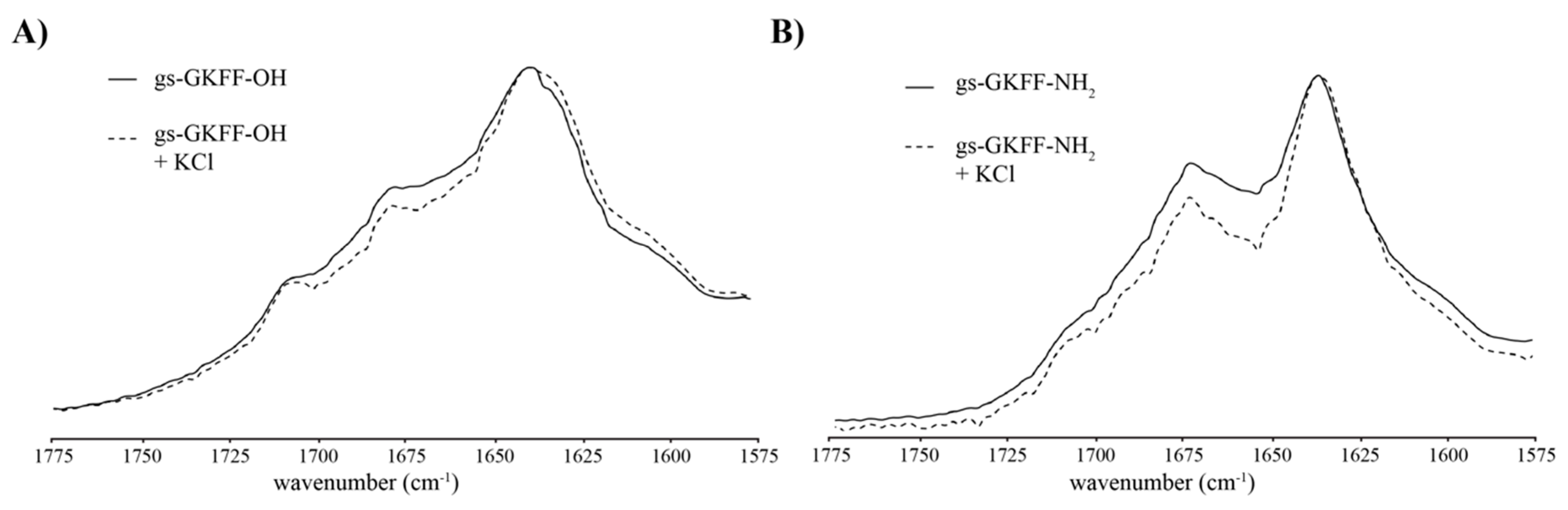
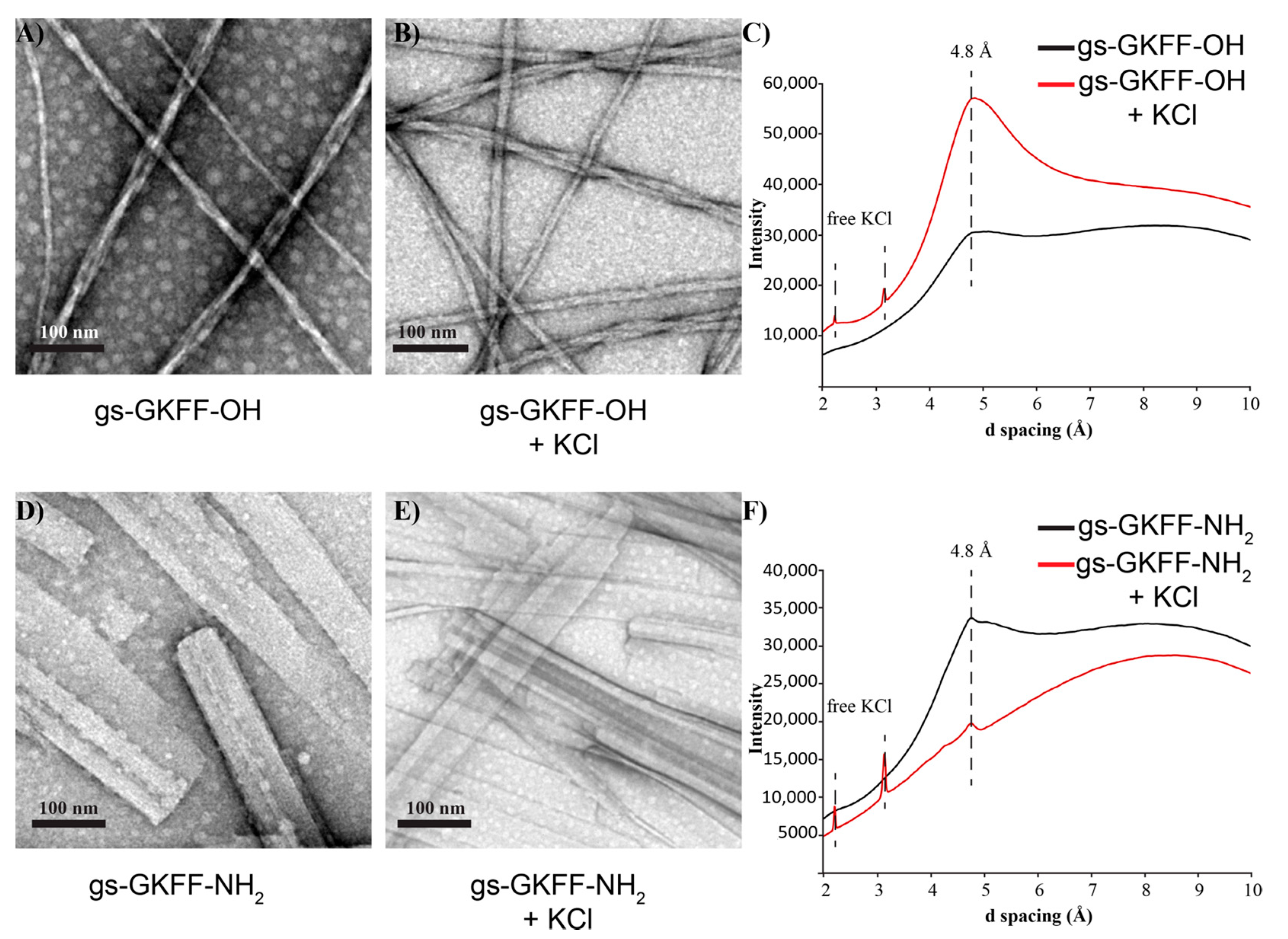
2. Results and Discussion
3. Materials and Methods
3.1. Materials
3.2. Synthesis of 2′,3′-O-Isopropylideneguanosine-5′-Carboxylic Acid
3.3. Synthesis of Nucleopeptide
3.4. Assembly Preparation
3.5. Fourier Transform Infrared Spectroscopy
3.6. TEM Imaging
3.7. Powder X-ray Diffraction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chatterjee, A.; Afrose, S.P.; Ahmed, S.; Venugopal, A.; Das, D. Cross-β amyloid nanotubes for hydrolase-peroxidase cascade reactions. Chem. Commun. 2020, 56, 7869–7872. [Google Scholar] [CrossRef] [PubMed]
- Duncan, K.L.; Ulijn, R.V. Short peptides in minimalistic biocatalyst design. Biocatalysis 2015, 1, 67–81. [Google Scholar] [CrossRef] [Green Version]
- Makhlynets, O.V.; Gosavi, P.M.; Korendovych, I.V. Short self-assembling peptides are able to bind to copper and activate oxygen. Angew. Chem. Int. Ed. 2016, 55, 9017–9020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, Y.; Unsworth, L.D.; Koutsopoulos, S.; Zhang, S. Slow release of molecules in self-assembling peptide nanofiber scaffold. J. Control Release 2006, 115, 18–25. [Google Scholar] [CrossRef]
- Altunbas, A.; Lee, S.J.; Rajasekaran, S.A.; Schneider, J.P.; Pochan, D.J. Encapsulation of curcumin in self-assembling peptide hydrogels as injectable drug delivery vehicles. Biomaterials 2011, 32, 5906–5914. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Ni, R.; Mehta, A.K.; Childers, W.S.; Lakdawala, A.; Pingali, S.V.; Thiyagarajan, P.; Lynn, D.G. Nucleobase-directed amyloid nanotube assembly. J. Am. Chem. Soc. 2008, 130, 16867–16869. [Google Scholar] [CrossRef]
- Ni, R.; Childers, W.S.; Hardcastle, K.I.; Mehta, A.K.; Lynn, D.G. Remodeling cross-β nanotube surfaces with peptide/lipid chimeras. Angew. Chem. Int. Ed. 2012, 51, 6635–6638. [Google Scholar] [CrossRef] [Green Version]
- Hartgerink, J.D.; Beniash, E.; Stupp, S.I. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science 2001, 294, 1684–1688. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Kuang, Y.; Shi, J.; Gao, Y.; Lin, H.; Xu, B. Multifunctional, biocompatible supramolecular hydrogelators consist only of nucleobase, amino acid, and glycoside. J. Am. Chem. Soc. 2011, 133, 17513–17518. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Du, X.; Gao, Y.; Shi, J.; Kuang, Y.; Xu, B. Supramolecular hydrogels formed by the conjugates of nucleobases, Arg-Gly-Asp (RGD) peptides, and glucosamine. Soft Matter 2012, 8, 7402–7407. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Zhou, J.; Shi, J.; Du, X.; Xu, B. A naphthalene-containing amino acid enables hydrogelation of a conjugate of nucleobase-saccharideamino acids. Chem. Commun. 2014, 50, 1992–1994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Kuang, Y.; Lin, H.; Gao, Y.; Shi, J.; Xu, B. Supramolecular nanofibers and hydrogels of nucleopeptides. Angew. Chem. Int. Ed. 2011, 50, 9365–9369. [Google Scholar] [CrossRef]
- Wuang, H.; Feng, Z.; Qin, Y.; Wang, J.; Xu, B. Nucleopeptide assemblies selectively sequester ATP in cancer cells to increase the efficacy of doxorubicin. Angew. Chem. Int. Ed. 2018, 57, 4931–4935. [Google Scholar] [CrossRef]
- Baek, K.; Noblett, A.D.; Ren, P.; Suggs, L.J. Design and characterization of nucleopeptides for hydrogel self-assembly. ACS Appl. Bio Mater. 2019, 2, 2812–2821. [Google Scholar] [CrossRef]
- Henderson, E.; Hardin, C.C.; Walk, S.K.; Tinoco, I.; Blackburn, E.H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell 1987, 51, 899–908. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, L.; Wang, H.; Luo, Q.; Xu, J.; Liu, J. Reversible organogels triggered by dynamic K+ binding and release. J. Colloid Interface Sci. 2011, 353, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zou, R.; Sun, P.; Wang, Q.; Wu, J. A supramolecular peptide polymer from hydrogen-bond and coordination-driven self-assembly. Polym. Chem. 2018, 9, 69–76. [Google Scholar] [CrossRef]
- Gonnelli, A.; Pieraccini, S.; Baldassarri, E.J.; Funari, S.; Masiero, S.; Ortore, M.G.; Mariani, P. Metallo-respnsive self-assembly of lipophilic guanines in hydrocarbon solvents: A systematic SAXS structural characterization. Nanoscale 2020, 12, 1022–1031. [Google Scholar] [CrossRef]
- Arnal-Herault, C.; Pasc, A.; Michau, M.; Cot, D.; Petit, E.; Barboiu, M. Functional G-quartet macroscopic membrane films. Angew. Chem. Int. Ed. 2007, 46, 8409–8413. [Google Scholar] [CrossRef]
- Görbitz, C.H. Nanotube formation by hydrophobic dipeptides. Chem. Eur. J. 2001, 7, 5153–5159. [Google Scholar] [CrossRef]
- Reches, M.; Gazit, E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, T.O.; Chirgadze, D.Y.; Levin, A.; Adler-Abramovich, L.; Gazit, E.; Knowles, T.P.; Buell, A.K. Expanding the solvent chemical space for self-assembly of dipeptide nanostructures. ACS Nano 2014, 8, 1243–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, J.T. G-quartets 40 years later: From 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004, 43, 668–698. [Google Scholar] [CrossRef] [PubMed]
- Gottarelli, G.; Spada, G.P. The stepwise supramolecular organization of guanosine derivatives. Chem. Rec. 2004, 4, 39–49. [Google Scholar] [CrossRef]
- Lena, S.; Masiero, S.; Pieraccini, S.; Spad, G.P. Guanosine hydrogen-bonded scaffolds: A new way to control the bottom-up realization of well-defined nanoarchitectures. Chem. Eur. J. 2009, 15, 7792–7806. [Google Scholar] [CrossRef]
- Dash, J.; Saha, P. Functional architectures derived from guanine quartets. Org. Biomol. Chem. 2016, 14, 2157–2163. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [Green Version]
- Xiao, S.; Davis, J.T. G4-quartet hydrogels from 5′hydrazino-guanosine for the non-covalent and covalent remediation of contaminants from water. Faraday Discuss. 2018, 209, 97–112. [Google Scholar] [CrossRef]
- Setnička, V.; Urbanová, M.; Volka, K.; Nampally, S.; Lehn, J. Investigation of guanosine-quartet assemblies by vibrational and electronic circular dichroism spectroscopy, a novel approach for studying supramolecular entities. Chem. Eur. J. 2006, 12, 8735–8743. [Google Scholar] [CrossRef]
- Singh, V.; Snigdha, K.; Singh, C.; Sinha, N.; Thakur, A.K. Understanding the slef-assembly f Fmoc-phenylalanine to hydrogel formation. Soft Matter 2015, 11, 5353–5364. [Google Scholar] [CrossRef]
- Jian, Z.; Hejing, W. The physical meanings of 5 basic parameters for an X-ray diffraction peak and their application. Chin. J. Geochem. 2003, 22, 38–44. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, X.; Huang, R.; Qi, W.; Wang, Y.; Su, R.; He, Z. Electrostatic and aromatic interaction-directed supramolecular self-assembly of a designed Fmoc-tripeptide into helical nanoribbons. Langmuir 2015, 31, 2885–2894. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Liu, K.; Mo, S.; Mao, Y.; Yi, T. From G-quartets to G-ribbon gel by concentration and sonication control. Org. Biomol. Chem. 2013, 11, 1525–1532. [Google Scholar] [CrossRef] [PubMed]
- Epp, J.B.; Widlanski, T.S. Facile preparation of nucleoside-5′-carboxylic acids. J. Org. Chem. 1999, 64, 293–295. [Google Scholar] [CrossRef]
- Harraz, D.M.; Davis, J.T. A self-assembled peroxidase from 5′-GMP and heme. Chem. Commun. 2018, 54, 1587–1590. [Google Scholar] [CrossRef]
- Bai, J.; Sun, X.; Wang, H.; Li, C.; Qiao, R. Guanosine-based self-assembly as an enantioselective catalyst scaffold. J. Org. Chem. 2020, 85, 2010–2018. [Google Scholar] [CrossRef]
- Banwell, E.F.; Piette, B.M.; Taormina, A.; Heddle, J.G. Reciprocal nucleopeptides as the ancestral Darwinian self-replicator. Mol. Biol. Evol. 2018, 35, 404–416. [Google Scholar] [CrossRef] [Green Version]
- Van der Gulik, P.T.; Speijer, D. How amino acids and peptides shaped the RNA world. Life 2015, 5, 230–246. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.E.; Mowles, A.K.; Mehta, A.K.; Lynn, D.G. Looked at life from both sides now. Life 2014, 4, 887–902. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the nucleopeptides are not available from the authors. |

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boback, K.; Bacchi, K.; O’Neill, S.; Brown, S.; Dorsainvil, J.; Smith-Carpenter, J.E. Impact of C-Terminal Chemistry on Self-Assembled Morphology of Guanosine Containing Nucleopeptides. Molecules 2020, 25, 5493. https://doi.org/10.3390/molecules25235493
Boback K, Bacchi K, O’Neill S, Brown S, Dorsainvil J, Smith-Carpenter JE. Impact of C-Terminal Chemistry on Self-Assembled Morphology of Guanosine Containing Nucleopeptides. Molecules. 2020; 25(23):5493. https://doi.org/10.3390/molecules25235493
Chicago/Turabian StyleBoback, Katherine, Katherine Bacchi, Sarah O’Neill, Samantha Brown, Jovelt Dorsainvil, and Jillian E. Smith-Carpenter. 2020. "Impact of C-Terminal Chemistry on Self-Assembled Morphology of Guanosine Containing Nucleopeptides" Molecules 25, no. 23: 5493. https://doi.org/10.3390/molecules25235493
APA StyleBoback, K., Bacchi, K., O’Neill, S., Brown, S., Dorsainvil, J., & Smith-Carpenter, J. E. (2020). Impact of C-Terminal Chemistry on Self-Assembled Morphology of Guanosine Containing Nucleopeptides. Molecules, 25(23), 5493. https://doi.org/10.3390/molecules25235493





