Inhibiting the NLRP3 Inflammasome
Abstract
:1. Introduction
2. Inflammasomes and Innate Immunity
3. The NLRP3 Inflammasome
4. Structure of NLRP3 and Cryo-EM Structure of NLRP3 Bound to NEK7
5. Role of NEK7 in Inflammasome Activation
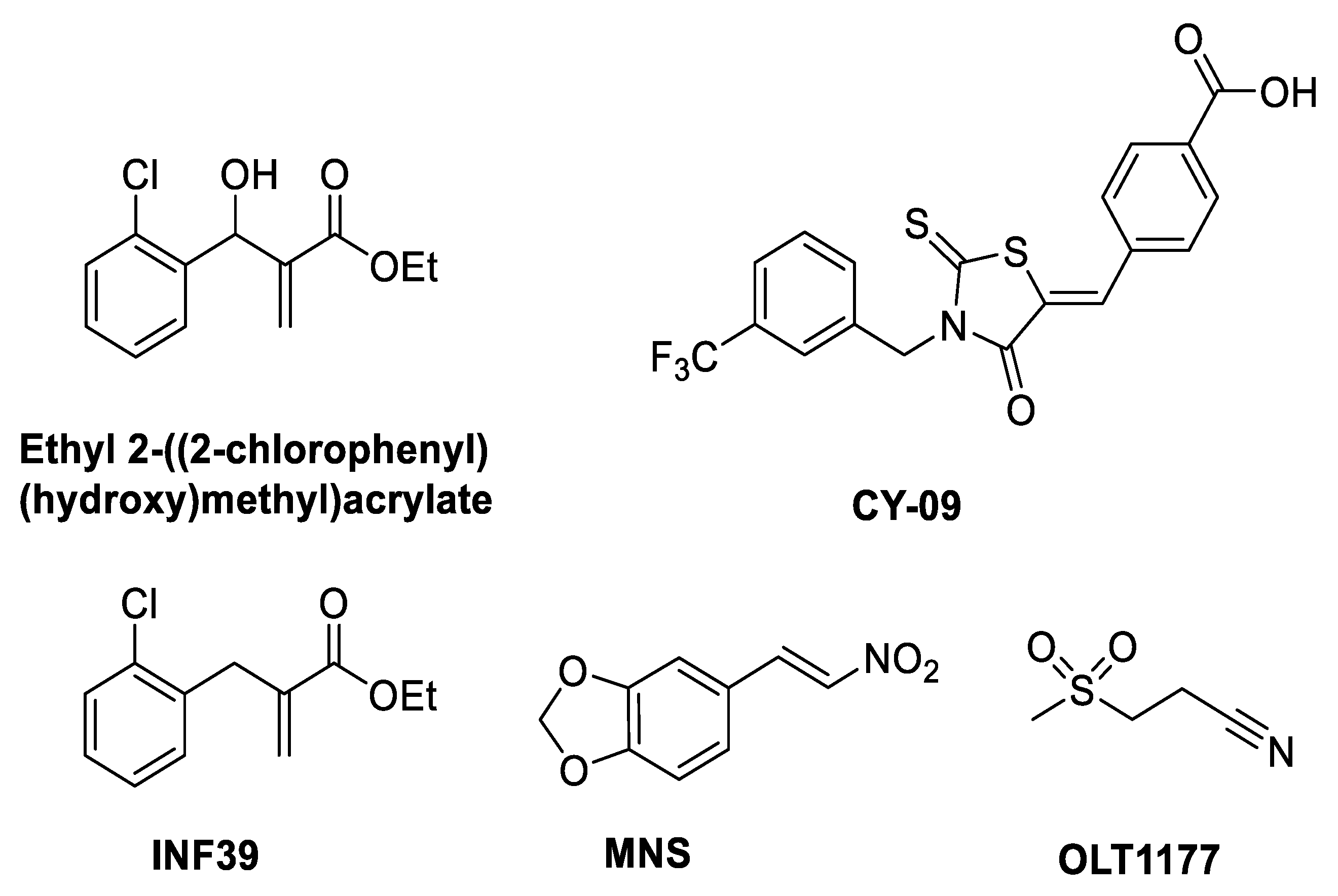
6. Mechanism of Action of NLRP3 Inhibitors: Covalent Modifiers of the NACHT Domain
| Inhibitor | % Inhibition at 10 µM /IC50 | Mechanism of Action |
|---|---|---|
| Ethyl 2-((2-chlorophenyl) (hydroxy)methyl)acrylate | 75.1 ± 2.6% pyroptosis | Inhibits NLRP3 ATPase [26] |
| CY-09 | IC50 = 6 µM | Walker A inhibitor [27] |
| MNS | IC50 = 2 µM | Inhibits NLRP3 ATPase [29] |
| OLT1177 (dapansutrile) | IC50 = 1 nM | ATPase inhibitor blocks NLRP3-ASC interaction [32] |
| INF39 | IC50 = 10 µM | Irreversible NLRP3 inflammasome inhibitor [30] |
| INF58 | IC50 = 74 µM | ATPase inhibitor [31] |
| Oridonin | IC50 = 0.75 µM | ATPase inhibitor blocks NLRP3-NEK7 interaction [28] |
| MCC950/CRID3 | IC50 = 7.5 nM | Walker B inhibitor [33,34,35] |
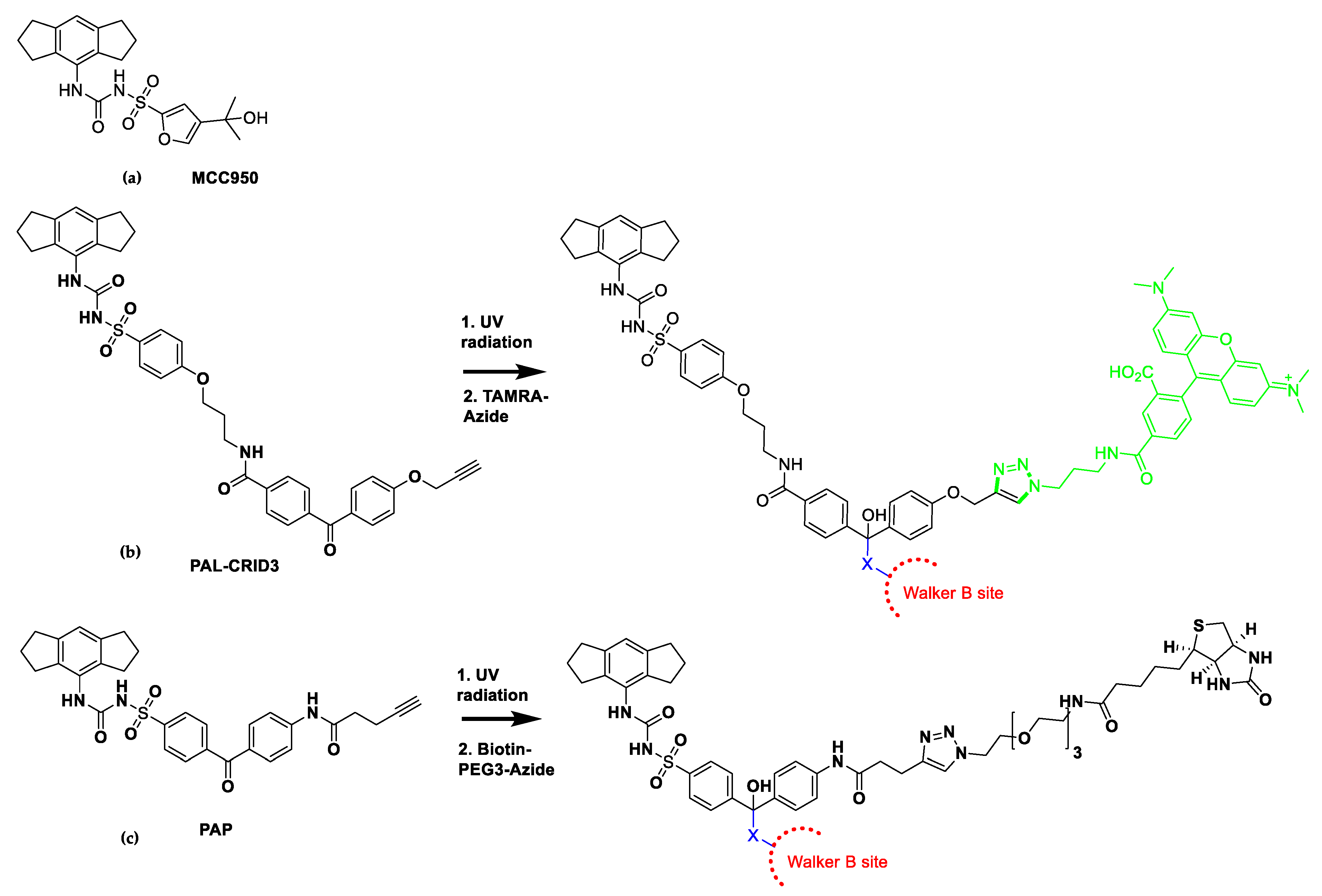
7. Mechanism of Action of NLRP3 Inhibitors: MCC950 Binds to Walker B
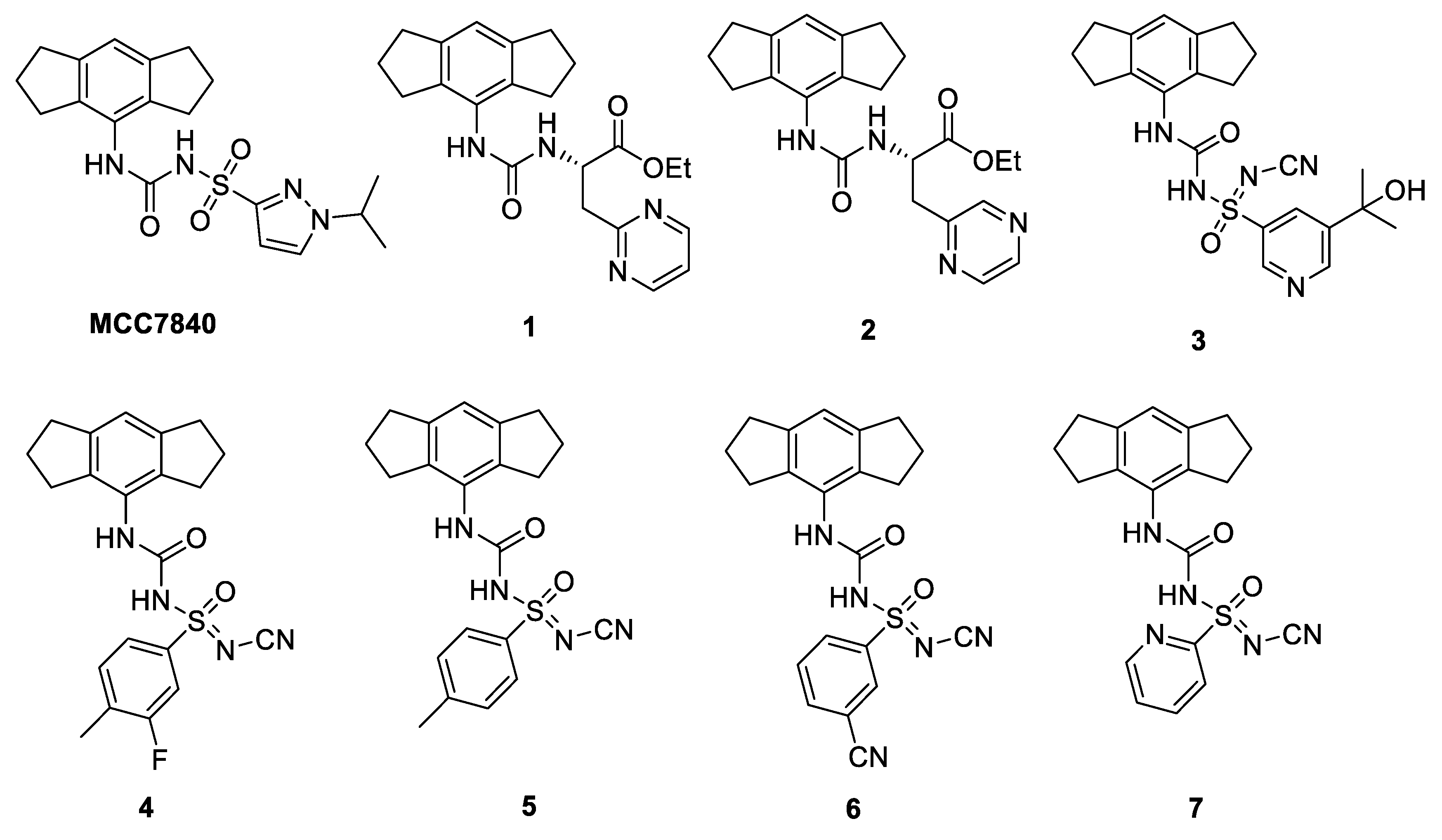
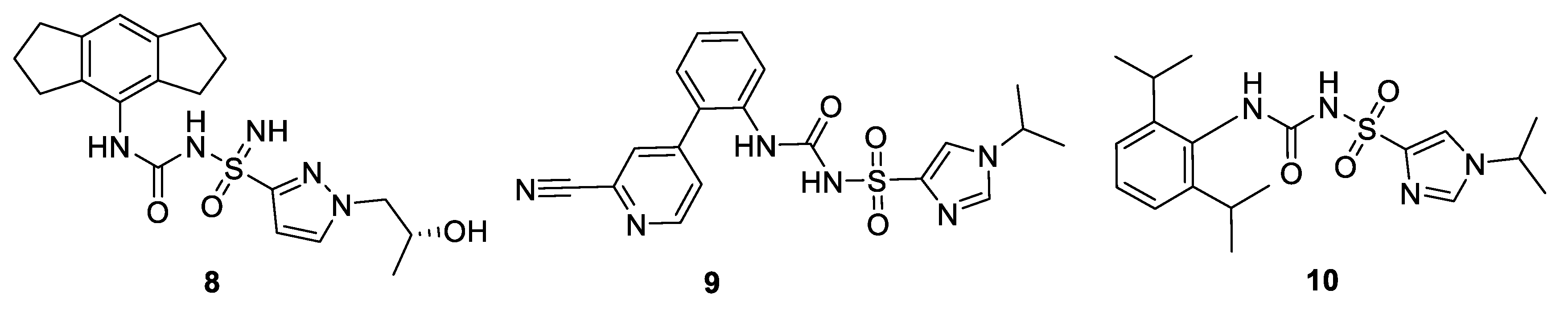
8. Clinical Trials and Patents of NLRP3 Inflammasome Inhibitors in the Pharmaceutical Industry Pipeline
9. Computational Approaches to Design New NLRP3 Inhibitors
9.1. Molecular Dynamics
9.2. Virtual Screening (VS) on ER-β Linked to NLRP3 Inflammasome
9.3. Computational Strategies for the Development of NLRP3 Inhibitors by Direct Binding to NLRP3 Pyrin Domain
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Baldwin, A.G.; Brough, D.; Freeman, S. Inhibiting the Inflammasome: A Chemical Perspective. J. Med. Chem. 2016, 59, 1691–1710. [Google Scholar] [CrossRef]
- Available online: https://www.novartis.com/news/media-releases/novartis-adds-clinical-and-preclinical-anti-inflammatory-programs-portfolio-acquisition-ifm-tre (accessed on 17 November 2020).
- Available online: https://www.ifmthera.com/pipeline (accessed on 17 November 2020).
- Available online: https://www.inflazome.com/pipeline (accessed on 17 November 2020).
- Available online: https://www.nodthera.com (accessed on 17 November 2020).
- Available online: http://www.olatec.com (accessed on 17 November 2020).
- Sharif, H.; Wang, L.; Wang, W.L.; Magupalli, V.G.; Andreeva, L.; Qiao, Q.; Hauenstein, A.V.; Wu, Z.; Núñez, G.; Mao, Y.; et al. Structural Mechanism for NEK7-Licensed Activation of NLRP3 Inflammasome. Nature. 2019, 570, 338–343. [Google Scholar] [CrossRef]
- Kumar, V.; Abbas, A.K.; Aster, J.C. Robbins Basic Pathology, 10th ed.; Elsevier: Amsterdam, The Netherlands, 2017; Chapters 3 and 5; pp. 1–935. [Google Scholar]
- Lee, H.E.; Lee, J.Y.; Yang, G.; Kang, H.C.; Cho, Y.-Y.; Lee, H.S.; Lee, J.Y. Inhibition of NLRP3 Inflammasome in Tumor Microenvironment Leads to Suppression of Metastatic Potential of Cancer Cells. Sci. Rep. 2019, 9, 12277. [Google Scholar] [CrossRef] [Green Version]
- Lu, A.; Li, H.; Niu, J.; Wu, S.; Xue, G.; Yao, X.; Guo, Q.; Wan, N.; Abliz, P.; Yang, G.; et al. Hyperactivation of the NLRP3 Inflammasome in Myeloid Cells Leads to Severe Organ Damage in Experimental Lupus. J. Immunol. 2017, 198, 1119–1129. [Google Scholar] [CrossRef]
- Braga, T.T.; Forni, M.F.; Correa-Costa, M.; Ramos, R.N.; Barbuto, J.A.; Branco, P.; Castoldi, A.; Hiyane, M.I.; Davanso, M.R.; Latz, E.; et al. Soluble Uric Acid Activates the NLRP3 Inflammasome. Sci. Rep. 2017, 7, 39884. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Xing, S.; Gong, Z.; Xing, Q. NLRP3 Inflammasomes Show High Expression in Aorta of Patients with Atherosclerosis. Heart Lung Circ. 2013, 22, 746–750. [Google Scholar] [CrossRef] [PubMed]
- Kuwar, R.; Rolfe, A.; Di, L.; Xu, H.; He, L.; Jiang, Y.; Zhang, S.; Sun, D. A Novel Small Molecular NLRP3 Inflammasome Inhibitor Alleviates Neuroinflammatory Response Following Traumatic Brain Injury. J. Neuroinflamm. 2019, 16, 81. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.-Y.; Moriyama, M.; Chang, M.-F.; Ichinohe, T. Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Front. Microbiol. 2019, 10, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafner-Bratkovič, I.; Sušjan, P.; Lainšček, D.; Tapia-Abellán, A.; Cerović, K.; Kadunc, L.; Angosto-Bazarra, D.; Pelegrin, P.; Jerala, R. NLRP3 Lacking the Leucine-Rich Repeat Domain can be Fully Activated via the Canonical Inflammasome Pathway. Nat. Commun. 2018, 9, 5182. [Google Scholar] [CrossRef] [Green Version]
- Lu, A.; Magupalli, V.G.; Ruan, J.; Yin, Q.; Atianand, M.K.; Vos, M.; Schröder, G.F.; Fitzgerald, K.A.; Wu, H.; Egelman, E.H. Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell 2014, 156, 1193–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubois, H.; Sorgeloos, F.; Sarvestani, S.T.; Martens, L.; Saeys, Y.; Mackenzie, J.M.; Lamkanfi, M.; van Loo, G.; Goodfellow, I.; Wullaert, A. NLRP3 Inflammasome Activation and Gasdermin D-Driven Pyroptosis are Immunopathogenic upon Gastrointestinal Norovirus Infection. PLoS Pathog. 2019, 15, e1007709. [Google Scholar] [CrossRef] [PubMed]
- Duncan, J.A.; Bergstralh, D.T.; Wang, Y.; Willingham, S.B.; Ye, Z.; Zimmermann, A.G.; Ting, J.P.-Y. Cryopyrin/NALP3 Binds ATP/dATP, is an ATPase, and Requires ATP Binding to Mediate Inflammatory Signaling. Proc. Natl. Acad. Sci. USA 2007, 104, 8041–8046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maharana, J.; Panda, D.; De, S. Deciphering the ATP-Binding Mechanism(s) in NLRP-NACHT 3D Models using Structural Bioinformatics Approaches. PLoS ONE 2018, 13, e0209420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, J.E.; Saraste, M.; Runswick, M.J.; Gay, N.J. Distantly Related Sequences in the Alpha- and Beta-Subunits of ATP Synthase, Myosin, Kinases and other ATP-Requiring Enzymes and a Common Nucleotide Binding Fold. EMBO J. 1982, 8, 945–951. [Google Scholar] [CrossRef]
- Neuwald, A.F.; Aravind, L.; Spouge, J.L.; Koonin, E.V. AAA+: A Class of Chaperone-Like ATPases Associated with the Assembly, Operation, and Disassembly of Protein Complexes. Genome Res. 1999, 9, 27–43. [Google Scholar] [CrossRef]
- Ye, Z.; Lich, J.D.; Moore, C.B.; Duncan, J.A.; Williams, K.L.; Ting, J.P.-Y. ATP Binding by Monarch-1/NLRP12 is Critical for its Inhibitory Function. Mol. Cell. Biol. 2008, 28, 1841–1850. [Google Scholar] [CrossRef] [Green Version]
- Schmid-Burgk, J.L.; Chauhan, D.; Schmidt, T.; Ebert, T.S.; Reinhardt, J.; Endl, E.; Hornung, V. A Genome-wide CRISPR Screen Identifies NEK7 as an Essential Component of NLRP3 Inflammasome Activation. J. Biol. Chem. 2016, 291, 103–109. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Zeng, M.Y.; Yang, D.; Motro, B.; Núñez, G. NEK7 is an Essential Mediator of NLRP3 Activation Downstream of Potassium Efflux. Nature 2016, 530, 354–357. [Google Scholar] [CrossRef] [Green Version]
- Schmacke, N.A.; Gaidt, M.M.; Szymanska, I.; O’Duill, F.; Stafford, C.A.; Chauhan, D.; Fröhlich, A.L.; Nagl, D.; Pinci, F.; Schmid-Burgk, J.L.; et al. Priming Enables a NEK7-Independent Route of NLRP3 Activation. bioRxiv 2019, 799320. [Google Scholar] [CrossRef] [Green Version]
- Cocco, M.; Garella, D.; Di Stilo, A.; Borretto, E.; Stevanato, L.; Giorgis, M.; Marini, E.; Fantozzi, R.; Miglio, G.; Bertinaria, M. Electrophilic Warhead-Based Design of Compounds Preventing NLRP3 Inflammasome-Dependent Pyroptosis. J. Med. Chem. 2014, 57, 10366–10382. [Google Scholar] [CrossRef]
- Jiang, H.; He, H.; Chen, Y.; Huang, W.; Cheng, J.; Ye, J.; Wang, A.; Tao, J.; Wang, C.; Liu, Q.; et al. Identification of a Selective and Direct NLRP3 Inhibitor to Treat Inflammatory Disorders. J. Exp. Med. 2017, 214, 3219–3238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.; Jiang, H.; Chen, Y.; Ye, J.; Wang, A.; Wang, C.; Liu, Q.; Liang, G.; Deng, X.; Jiang, W.; et al. Oridonin is a Covalent NLRP3 Inhibitor with Strong Anti-Inflammasome Activity. Nat. Commun. 2018, 9, 2550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Varadarajan, S.; Muñoz-Planillo, R.; Burberry, A.; Nakamura, Y.; Núñez, G. 3,4-Methylenedioxy-β-nitrostyrene Inhibits NLRP3 Inflammasome Activation by Blocking Assembly of the Inflammasome. J. Biol. Chem. 2014, 289, 1142–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cocco, M.; Pellegrini, C.; Martínez-Banaclocha, H.; Giorgis, M.; Marini, E.; Costale, A.; Miglio, G.; Fornai, M.; Antonioli, L.; López-Castejón, G.; et al. Development of an Acrylate Derivative Targeting the NLRP3 Inflammasome for the Treatment of Inflammatory Bowel Disease. J. Med. Chem. 2017, 60, 3656–3671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cocco, M.; Miglio, G.; Giorgis, M.; Garella, D.; Marini, E.; Costale, A.; Regazzoni, L.; Vistoli, G.; Orioli, M.; Massulaha-Ahmed, R.; et al. Design, Synthesis, and Evaluation of Acrylamide Derivatives as Direct NLRP3 Inflammasome Inhibitors. ChemMedChem 2016, 11, 1790–1803. [Google Scholar] [CrossRef]
- Marchetti, C.; Swartzwelter, B.; Gamboni, F.; Neff, C.P.; Richter, K.; Azam, T.; Carta, S.; Tengesdal, I.; Nemkov, T.; D’Alessandro, A.; et al. OLT1177, a β-Sulfonyl Nitrile Compound, Safe in Humans, Inhibits the NLRP3 Inflammasome and Reverses the Metabolic Cost of Inflammation. Proc. Natl. Acad. Sci. USA 2018, 115, 1530–1539. [Google Scholar] [CrossRef] [Green Version]
- Coll, R.C.; Robertson, A.A.B.; Chae, J.J.; Higgins, S.C.; Muñoz-Planillo, R.; Inserra, M.C.; Vetter, I.; Dungan, L.S.; Monks, B.G.; Stutz, A.; et al. A Small-Molecule Inhibitor of the NLRP3 Inflammasome for the Treatment of Inflammatory Diseases. Nat. Med. 2015, 21, 248–255. [Google Scholar] [CrossRef] [Green Version]
- Vande Walle, L.; Stowe, I.B.; Šácha, P.; Lee, B.L.; Demon, D.; Fossoul, A.; Van Hauwermeiren, F.; Saavedra, P.H.V.; Šimon, P.; Šubrt, V.; et al. MCC950/CRID3 Potently Targets the NACHT Domain of Wild-Type NLRP3 but not Disease-Associated Mutants for Inflammasome Inhibition. PLoS Biol. 2019, 17, e3000354. [Google Scholar] [CrossRef] [Green Version]
- Coll, R.C.; Hill, J.R.; Day, C.J.; Zamoshnikova, A.; Boucher, D.; Massey, N.L.; Chitty, J.L.; Fraser, J.A.; Jennings, M.P.; Robertson, A.A.B.; et al. MCC950 Directly Targets the NLRP3 ATP-Hydrolysis Motif for Inflammasome Inhibition. Nat. Chem. Biol. 2019, 15, 556–559. [Google Scholar] [CrossRef]
- Tapia-Abellán, A.; Angosto-Bazarra, D.; Martínez-Banaclocha, H.; Torre-Minguela, C.D.; Cerón-Carrasco, J.P.; Pérez-Sánchez, H.; Arostegui, J.I.; Pelegrin, P. MCC950 Closes the Active Conformation of NLRP3 to an Inactive State. Nat. Chem. Biol. 2019, 15, 560–564. [Google Scholar] [CrossRef]
- Coss, R. Could an NLRP3 Inhibitor Be the One Drug to Conquer Common Diseases? C EN Glob. Enterp. 2020, 98, 26–31. [Google Scholar] [CrossRef]
- Available online: https://www.zyversa.com (accessed on 17 November 2020).
- Sánchez-Fernández, A.; Skouras, D.B.; Dinarello, C.A.; López-Vales, R. OLT1177 (Dapansutrile), a Selective NLRP3 Inflammasome Inhibitor, Ameliorates Experimental Autoimmune Encephalomyelitis Pathogenesis. Front. Immunol. 2019, 10, 2578. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.businesswire.com/news/home/20201027005190/en/Olatec-Therapeutics-Enrolls-its-First-Patients-in-a-Phase-2-Clinical-Trial-in-COVID-19-with-its-Selective-NLRP3-Inhibitor-Oral-Dapansutrile (accessed on 17 November 2020).
- Bock, M.G.; Watt, A.P.; Porter, R.A.; Harrison, D.; Boutard, N.F.P.; Levenets, O.; Fabritius, C.-H.R.Y.; Topolnicki, G.W. Selective Inhibitors of NLRP3 Inflammasome. WO/2019/025467, 7 February 2019. [Google Scholar]
- Available online: https://www.nodthera.com/nodthera-announces-close-of-55-million-series-b-financing/ (accessed on 17 November 2020).
- Harrison, D.; Boutard, N.; Brzozka, K.; Bugaj, M.; Chmielewski, S.; Cierpich, A.; Doedens, J.R.; Fabritius, C.-H.R.Y.; Gabel, C.A.; Galezowski, M.; et al. Discovery of a Series of Ester-substituted NLRP3 Inflammasome Inhibitors. Bioorg. Med. Chem. Lett. 2020, 30, 127560. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.; Watt, A.P.; Boutard, N.; Fabritius, C.-H.; Galezowski, M.; Kowalczyk, P.; Levents, O.; Woyciechowski, J. Chemical Compounds. WO/2018/167468, 20 September 2018. [Google Scholar]
- Available online: https://www.businesswire.com/news/home/20200226006027/en/Inflazome’s-Somalix-Demonstrates-Positive-Safety-Tolerability-Pharmacodynamic (accessed on 17 November 2020).
- Available online: https://www.businesswire.com/news/home/20200326005102/en/Inzomelid-completes-Phase-studies-shows-positive-results (accessed on 17 November 2020).
- O’Neill, L.; Coll, R.; Cooper, M.; Robertson, A.; Schroder, K. Sulfonylureas and Related Compounds and Use of Same. WO/2016/131098, 25 August 2016. [Google Scholar]
- Sharma, R.; Iyer, P.; Agarwal, S. Novel Substituted Sulfoximine Compounds. WO/2018/225018, 13 December 2018. [Google Scholar]
- Agarwal, S.; Sasane, S.; Shah, H.A.; Pethani, J.P.; Deshmukh, P.; Vyas, V.; Iyer, P.; Bhavsar, H.; Viswanathan, K.; Bandyopadhyay, D.; et al. Discovery of N-Cyano-sulfoximineurea Derivatives as Potent and Orally Bioavailable NLRP3 Inflammasome Inhibitors. ACS Med. Chem. Lett. 2020, 11, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.; Miller, D.; Macleod, A.; Van Wiltenberg, J.; Thom, S.; ST-Gallay, S.; Shannon, J. Sulfonylureas and Sulfonylthioureas as NLRP3 Inhibitors. WO/2019/008029, 10 January 2019. [Google Scholar]
- Mekni, N.; De Rosa, M.; Cipollina, C.; Gulotta, M.R.; De Simone, G.; Lombino, J.; Padova, A.; Perricone, U. In Silico Insights towards the Identification of NLRP3 Druggable Hot Spots. Int. J. Mol. Sci. 2019, 20, 4974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Zhao, W.; Ying, H.; Zhang, Y.; Du, J.; Chen, S.; Li, J.; Shen, B. Estradiol Inhibits NLRP3 Inflammasome in Fibroblast-Like Synoviocytes Activated by Lipopolysaccharide and Adenosine Triphosphate. Int. J. Rheum. Dis. 2018, 21, 2002–2010. [Google Scholar] [CrossRef]
- Souza, P.C.T.; Textor, L.C.; Melo, D.C.; Nascimento, A.S.; Skaf, M.S.; Polikarpov, I. An Alternative Conformation of ERβ Bound to Estradiol Reveals H12 in a Stable Antagonist Position. Sci. Rep. 2017, 7, 3509. [Google Scholar] [CrossRef]
- Abdullaha, M.; Ali, M.; Kour, D.; Kumar, A.; Bharate, S.B. Discovery of Benzo[cd]indol-2-one and Benzylidene-Thiazolidine-2,4-Dione as New Classes of NLRP3 Inflammasome Inhibitors via ER-β Structure Based Virtual Screening. Bioorg. Chem. 2020, 95, 103500. [Google Scholar] [CrossRef]
- Bae, J.Y.; Park, H.H. Crystal Structure of NALP3 Protein Pyrin Domain (PYD) and its Implications in Inflammasome Assembly. J. Biol. Chem. 2011, 286, 39528–39536. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Lee, H.E.; Moon, S.-J.; Ko, K.M.; Koh, J.H.; Seok, J.K.; Min, J.-K.; Heo, T.-H.; Kang, H.C.; Cho, Y.-Y.; et al. Direct Binding to NLRP3 Pyrin Domain as a Novel Strategy to Prevent NLRP3-Driven Inflammation and Gouty Arthritis. Arthritis Rheumatol. 2020, 72, 1192–1202. [Google Scholar] [CrossRef]
- Huang, Y.; Jiang, H.; Chen, Y.; Wang, X.; Yang, Y.; Tao, J.; Deng, X.; Liang, G.; Zhang, H.; Jiang, W.; et al. Tranilast Directly Targets NLRP3 to Treat Inflammasome-Driven Diseases. EMBO Mol. Med. 2018, 10, e8689. [Google Scholar] [CrossRef] [PubMed]
- Seoane, P.I.; Lee, B.; Hoyle, C.; Yu, S.; Lopez-Castejon, G.; Lowe, M.; Brough, D. The NLRP3-Inflammasome as a Sensor of Organelle Dysfunction. J. Cell. Biol. 2020, 219, e202006194. [Google Scholar] [CrossRef]
- Shim, D.-W.; Lee, K.-H. Posttranslational Regulation of the NLR Family Pyrin Domain-Containing 3 Inflammasome. Front. Immunol. 2018, 9, 1054. [Google Scholar] [CrossRef] [Green Version]
- Green, J.; Swanton, T.; Morris, L.; El-Sharkawy, L.; Cook, J.; Yu, S.; Beswick, J.; Adamson, A.; Humphreys, N.; Bryce, R.; et al. LRRC8A is Essential for Hypotonicity-, but not for DAMP-Induced NLRP3 Inflammasome Activation. eLife 2020, 9, e59704. [Google Scholar] [CrossRef] [PubMed]
- Swanton, T.; Beswick, J.A.; Hammadi, H.; Morris, L.; Williams, D.; de Cesco, S.; El-Sharkawy, L.; Yu, S.; Green, J.; Davis, J.B.; et al. Selective Inhibition of the K+ Efflux Sensitive NLRP3 Pathway by Cl− Channel Modulation. Chem. Sci. 2020, 11, 11720–11728. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Sharkawy, L.Y.; Brough, D.; Freeman, S. Inhibiting the NLRP3 Inflammasome. Molecules 2020, 25, 5533. https://doi.org/10.3390/molecules25235533
El-Sharkawy LY, Brough D, Freeman S. Inhibiting the NLRP3 Inflammasome. Molecules. 2020; 25(23):5533. https://doi.org/10.3390/molecules25235533
Chicago/Turabian StyleEl-Sharkawy, Lina Y., David Brough, and Sally Freeman. 2020. "Inhibiting the NLRP3 Inflammasome" Molecules 25, no. 23: 5533. https://doi.org/10.3390/molecules25235533








