Compressed Air-Driven Continuous-Flow Thermocycled Digital PCR for HBV Diagnosis in Clinical-Level Serum Sample Based on Single Hot Plate
Abstract
:1. Introduction
2. Methods
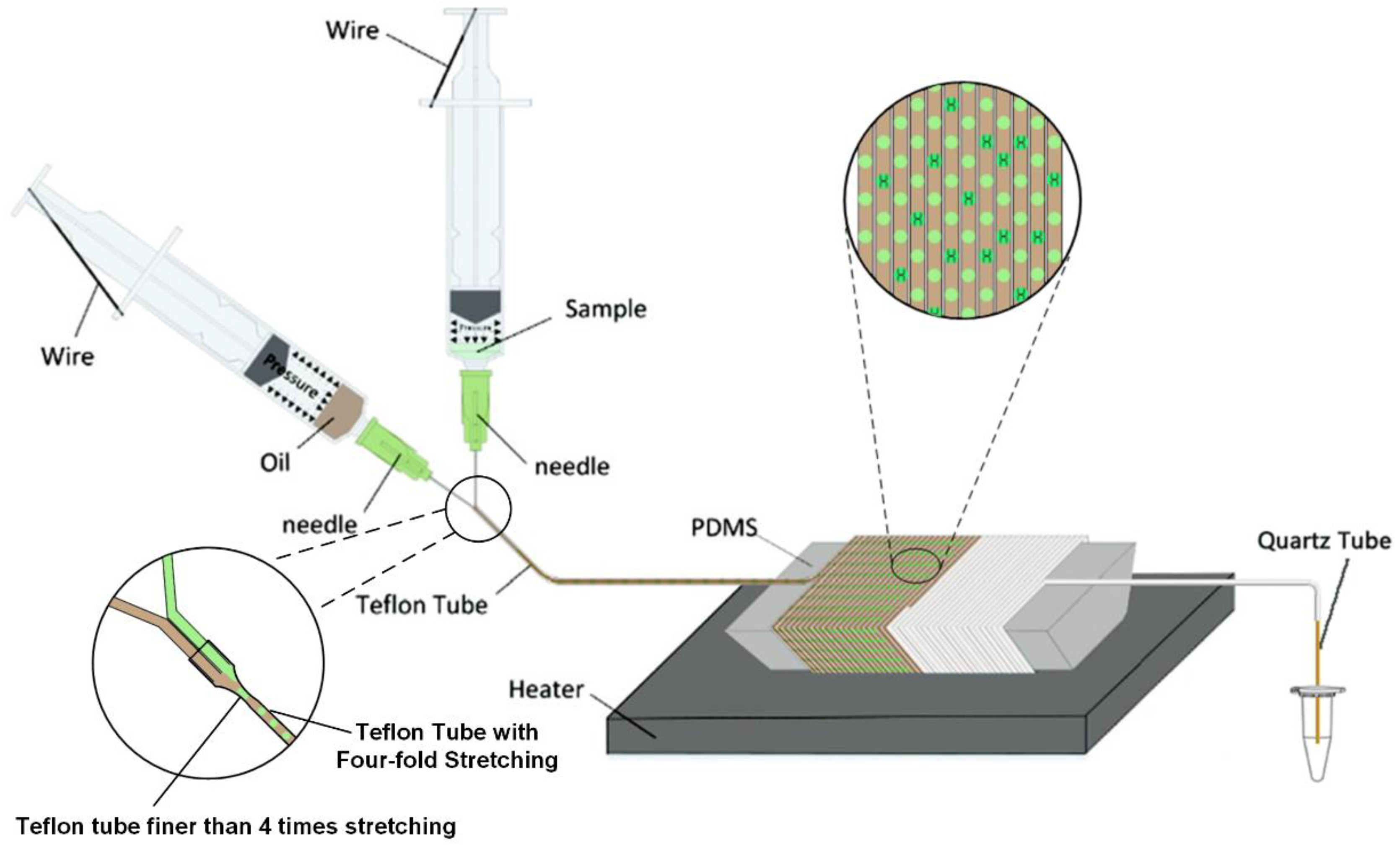
2.1. Composition of Micro Devices
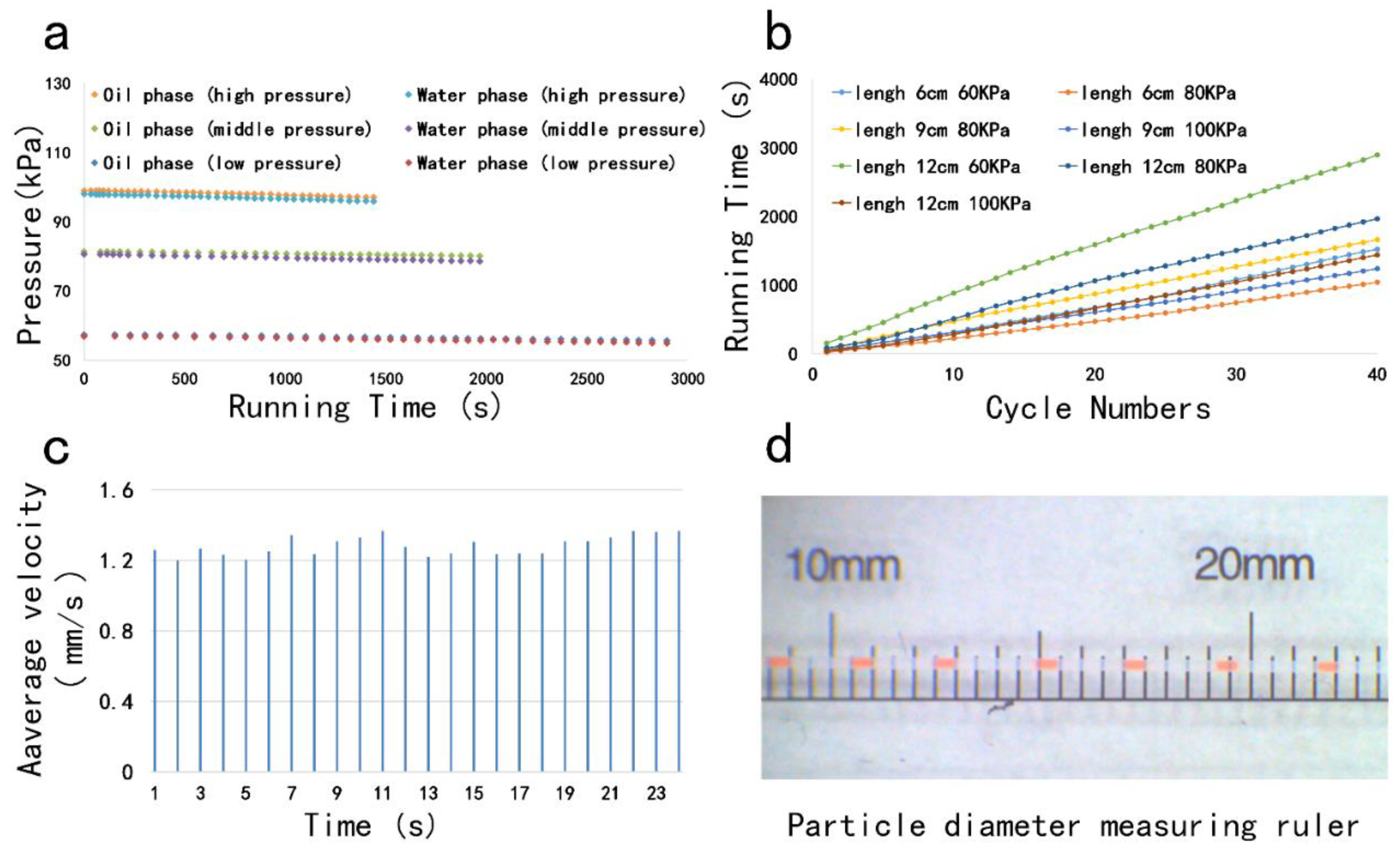
2.2. Droplet Generation
2.3. Reagents
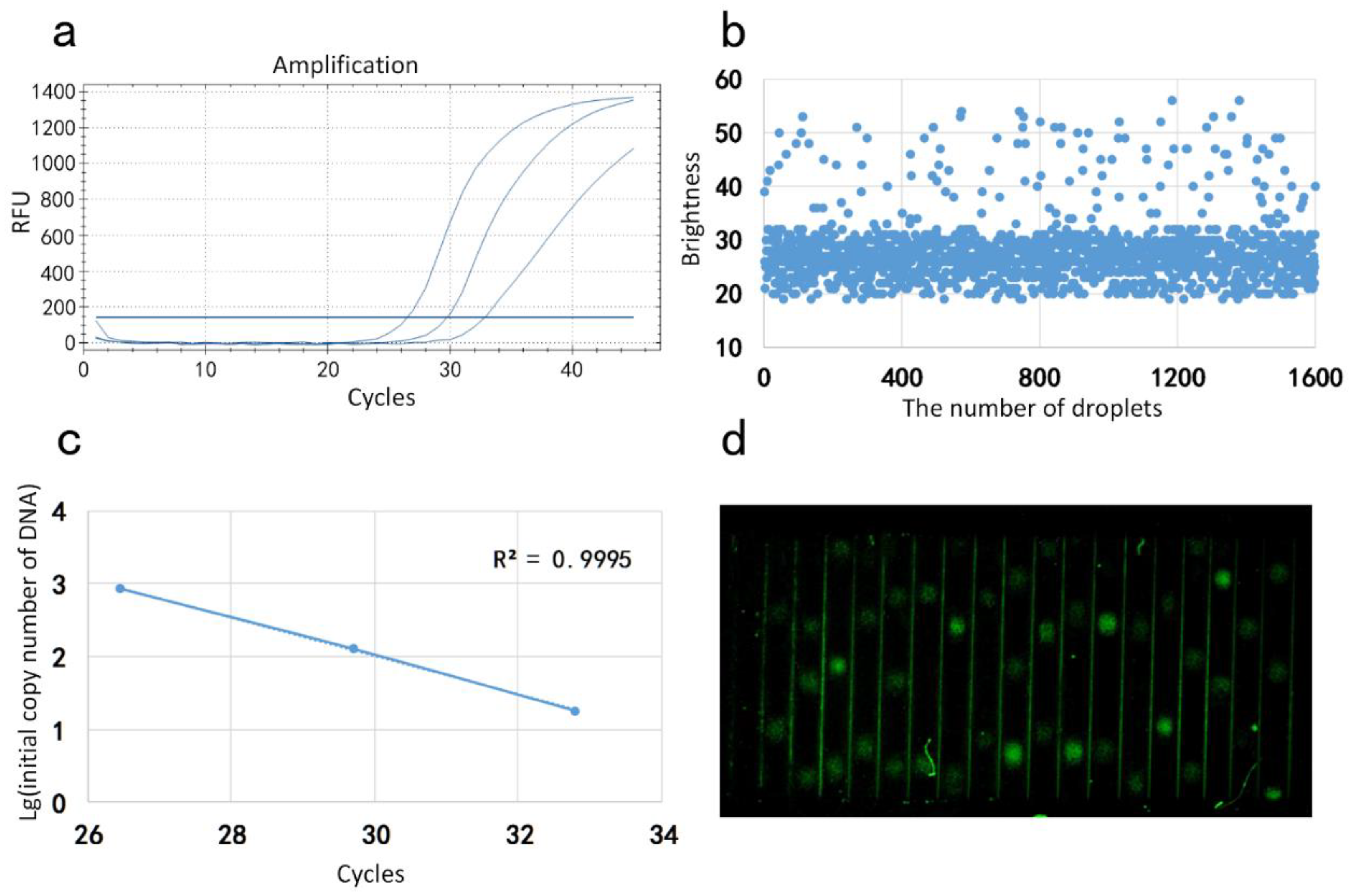
2.4. Image Acquisition and Processing
3. Results and Discussion
Fabrication of Microdevice
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Boni, S.M.; Oyafuso, L.K.; Soler, R.D.; Lindoso, J.A. Efficiency of noninvasive sampling methods (swab) together with Polymerase Chain Reaction (PCR) for diagnosing American Tegumentary Leishmaniasis. Rev. Inst. Med. Trop. Sao Paulo 2017, 59, e38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez Marin, J.E.; Zuluaga, J.D.; Pechene Campo, E.J.; Trivino, J.; de-la-Torre, A. Polymerase chain reaction (PCR) in ocular and ganglionar toxoplasmosis and the effect of therapeutics for prevention of ocular involvement in South American setting. Acta Trop. 2018, 184, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Liu, Q.; Yang, H.; Runyan, R.B.; Eisenberg, C.A.; Xu, M.; Borg, T.K.; Markwald, R.; Wang, Y.; Gao, B.Z. Laser patterning for the study of MSC cardiogenic differentiation at the single-cell level. Light Sci. Appl. 2013, 2, e68. [Google Scholar] [CrossRef]
- Marin, M.J.; Figuero, E.; Herrera, D.; Sanz, M. Quantitative analysis of periodontal pathogens using real-time polymerase chain reaction (PCR). In Oral Biology: Molecular Techniques and Applications; Seymour, G.J., Cullinan, M.P., Heng, N.C.K., Eds.; Springer: New York, NY, USA, 2017; pp. 191–202. [Google Scholar] [CrossRef]
- Okuma, T.A.; Hellberg, R.S. Identification of meat species in pet foods using a real-time polymerase chain reaction (PCR) assay. Food Control 2015, 50, 9–17. [Google Scholar] [CrossRef]
- Rahmati, A.; Brazier, J.S. Typing of Fusobacterium necrophorum Strains Using Polymerase Chain Reaction (PCR) Based Methods. Infect. Epidemiol. Med. 2016, 2, 1–3. [Google Scholar] [CrossRef]
- Subramony, A.; Zachariah, P.; Krones, A.; Whittier, S.; Saiman, L. Impact of multiplex polymerase chain reaction testing for respiratory pathogens on healthcare resource utilization for pediatric inpatients. J. Pediatr. 2016, 173, 196–201.e192. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Jing, F.; Li, G.; Fan, X.; Jia, C.; Zhou, H.; Jin, Q.; Zhao, J. A microfluidic droplet digital PCR for simultaneous detection of pathogenic Escherichia coli O157 and Listeria monocytogenes. Biosens. Bioelectron. 2015, 74, 770–777. [Google Scholar] [CrossRef]
- Cao, L.; Cui, X.; Hu, J.; Li, Z.; Choi, J.R.; Yang, Q.; Lin, M.; Hui, L.Y.; Xu, F. Advances in digital polymerase chain reaction (dPCR) and its emerging biomedical applications. Biosens. Bioelectron. 2017, 90, 459–474. [Google Scholar] [CrossRef]
- Cui, X.; Cao, L.; Huang, Y.; Bai, D.; Huang, S.; Lin, M.; Yang, Q.; Lu, T.J.; Xu, F.; Li, F. In vitro diagnosis of DNA methylation biomarkers with digital PCR in breast tumors. Analyst 2018, 143, 3011–3020. [Google Scholar] [CrossRef]
- Fischer, J.M.; Smith, B.; Waters, A.; Krishnamurthy, J.; Parekh, D.; Thiele, C.; Lieuw, K. Development of a minimal residual disease assay for anaplastic lymphoma kinase mutations using digital droplet polymerase chain reaction. Pediatrics 2018, 142, 609. [Google Scholar]
- Sreejith, K.R.; Ooi, C.H.; Jin, J.; Dao, D.V.; Nguyen, N.T. Digital polymerase chain reaction technology—recent advances and future perspectives. Lab Chip 2018, 18, 3717–3732. [Google Scholar] [CrossRef] [PubMed]
- Sreejith, K.R.; Gorgannezhad, L.; Jin, J.; Ooi, C.H.; Stratton, H.; Dao, D.V.; Nguyen, N.T. Liquid marbles as biochemical reactors for the polymerase chain reaction. Lab Chip 2019, 19, 3220–3227. [Google Scholar] [CrossRef]
- Sreejith, K.R.; Gorgannezhad, L.; Jin, J.; Ooi, C.H.; Nguyen, N.T. Core-Shell Beads Made by Composite Liquid Marble Technology as a Versatile Microreactor for Polymerase Chain Reaction. Micromachines 2020, 11, 242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gou, T.; Hu, J.; Wu, W.; Ding, X.; Zhou, S.; Fang, W.; Mu, Y. Smartphone-based mobile digital PCR device for DNA quantitative analysis with high accuracy. Biosens. Bioelectron. 2018, 120, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Hatch, A.C.; Fisher, J.S.; Tovar, A.R.; Hsieh, A.T.; Lin, R.; Pentoney, S.L.; Yang, D.L.; Lee, A.P. 1-Million droplet array with wide-field fluorescence imaging for digital PCR. Lab Chip 2011, 11, 3838–3845. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2011, 84, 1003–1011. [Google Scholar] [CrossRef]
- Song, Q.; Sun, J.; Mu, Y.; Xu, Y.; Zhu, Q.; Jin, Q. A new method for polydimethylsiloxane (PDMS) microfluidic chips to maintain vacuum-driven power using Parylene C. Sens. Actuators B Chem. 2018, 256, 1122–1130. [Google Scholar] [CrossRef]
- Tian, Q.; Yu, B.; Mu, Y.; Xu, Y.; Ma, C.; Zhang, T.; Jin, W.; Jin, Q. An integrated temporary negative pressure assisted microfluidic chip for DNA isolation and digital PCR detection. RSC Adv. 2015, 5, 81889–81896. [Google Scholar] [CrossRef]
- Cao, Q.; Kim, M.C.; Klapperich, C. Plastic microfluidic chip for continuous-flow polymerase chain reaction: Simulations and experiments. Biotechnol. J. 2011, 6, 177–184. [Google Scholar] [CrossRef]
- Fernández-Carballo, B.L.; McGuiness, I.; McBeth, C.; Kalashnikov, M.; Borrós, S.; Sharon, A.; Sauer-Budge, A.F. Low-cost, real-time, continuous flow PCR system for pathogen detection. Biomed. Microdevices 2016, 18, 34. [Google Scholar] [CrossRef]
- Li, S.; Fozdar, D.Y.; Ali, M.F.; Li, H.; Shao, D.; Vykoukal, D.M.; Vykoukal, J.; Floriano, P.N.; Olsen, M.; McDevitt, J.T.; et al. A continuous-flow polymerase chain reaction microchip with regional velocity control. J. Microelectromech. Syst. 2006, 15, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Schaerli, Y.; Wootton, R.C.; Robinson, T.; Stein, V.; Dunsby, C.; Neil, M.A.; French, P.M.; DeMello, A.J.; Abell, C.; Hollfelder, F. Continuous-flow polymerase chain reaction of single-copy DNA in microfluidic microdroplets. Anal. Chem. 2009, 81, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, H.; Saito, M.; Tsuji, K.; Yamanaka, K.; Tamiya, E. Self-propelled continuous-flow PCR in capillary-driven microfluidic device: Microfluidic behavior and DNA amplification. Sens. Actuators B Chem. 2015, 206, 303–310. [Google Scholar] [CrossRef]
- Chen, J.; Luo, Z.; Li, L.; He, J.; Li, L.; Zhu, J.; Wu, P.; He, L. Capillary-based integrated digital PCR in picoliter droplets. Lab Chip 2018, 18, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Hatch, A.C.; Ray, T.; Lintecum, K.; Youngbull, C. Continuous flow real-time PCR device using multi-channel fluorescence excitation and detection. Lab Chip 2014, 14, 562–568. [Google Scholar] [CrossRef]
- Zhang, W.; Li, N.; Koga, D.; Zhang, Y.; Zeng, H.; Nakajima, H.; Lin, J.M.; Uchiyama, K. Inkjet printing based droplet generation for integrated online digital polymerase chain reaction. Analytical chemistry. Anal. Chem. 2018, 90, 5329–5334. [Google Scholar] [CrossRef]
- Bardin, D.; Lee, A.P. Low-cost experimentation for the study of droplet microfluidics. Lab Chip 2014, 14, 3978–3986. [Google Scholar] [CrossRef] [Green Version]
- Baroud, C.N.; Gallaire, F.; Dangla, R. Dynamics of microfluidic droplets. Lab Chip 2010, 10, 2032–2045. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Chen, T.; Chen, Z.; Fei, P.; Yu, Z.; Pang, Y.; Huang, Y. Squeeze-chip: A finger-controlled microfluidic flow network device and its application to biochemical assays. Lab Chip 2012, 12, 1587–1590. [Google Scholar] [CrossRef]
- Seemann, R.; Brinkmann, M.; Pfohl, T.; Herminghaus, S. Droplet based microfluidics. Reports on progress in physics. Rep. Prog. Phys. 2011, 75, 016601. [Google Scholar] [CrossRef]
- Teh, S.Y.; Lin, R.; Hung, L.H.; Lee, A.P. Droplet microfluidics. Lab Chip 2008, 8, 198–220. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Yao, S. A facile on-demand droplet microfluidic system for lab-on-a-chip applications. Microfluid. Nanofluid. 2013, 16, 667–675. [Google Scholar] [CrossRef]
- Sasaki, N.; Shinjo, M.; Hirakawa, S.; Nishinaka, M.; Tanaka, Y.; Mawatari, K.; Kitamori, T.; Sato, K. A palmtop-sized microfluidic cell culture system driven by a miniaturized infusion pump. Electrophoresis 2012, 33, 1729–1735. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Li, Y.; Wu, D.; Wu, W. A handheld continuous-flow real-time fluorescence qPCR system with a PVC microreactor. Analyst 2020, 145, 2767–2773. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, Y.; Wang, K.; Wu, W. Passive Micropump for Highly Stable, Long-Termed, and Large Volume of Droplet Generation/Transport Inside 3D Microchannels Capable of Surfactant-Free and Droplet-Based Thermocycled Reverse Transcription-Polymerase Chain Reactions Based on a Single Thermostatic Heater. Anal. Chem. 2018, 90, 11925–11932. [Google Scholar] [PubMed]
Sample Availability: Samples of the compounds HBV device are available from the authors. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K.; Li, B.; Wu, W. Compressed Air-Driven Continuous-Flow Thermocycled Digital PCR for HBV Diagnosis in Clinical-Level Serum Sample Based on Single Hot Plate. Molecules 2020, 25, 5646. https://doi.org/10.3390/molecules25235646
Wang K, Li B, Wu W. Compressed Air-Driven Continuous-Flow Thermocycled Digital PCR for HBV Diagnosis in Clinical-Level Serum Sample Based on Single Hot Plate. Molecules. 2020; 25(23):5646. https://doi.org/10.3390/molecules25235646
Chicago/Turabian StyleWang, Kangning, Bin Li, and Wenming Wu. 2020. "Compressed Air-Driven Continuous-Flow Thermocycled Digital PCR for HBV Diagnosis in Clinical-Level Serum Sample Based on Single Hot Plate" Molecules 25, no. 23: 5646. https://doi.org/10.3390/molecules25235646
APA StyleWang, K., Li, B., & Wu, W. (2020). Compressed Air-Driven Continuous-Flow Thermocycled Digital PCR for HBV Diagnosis in Clinical-Level Serum Sample Based on Single Hot Plate. Molecules, 25(23), 5646. https://doi.org/10.3390/molecules25235646






