Biophysical Characterization and Anticancer Activities of Photosensitive Phytoanthraquinones Represented by Hypericin and Its Model Compounds
Abstract
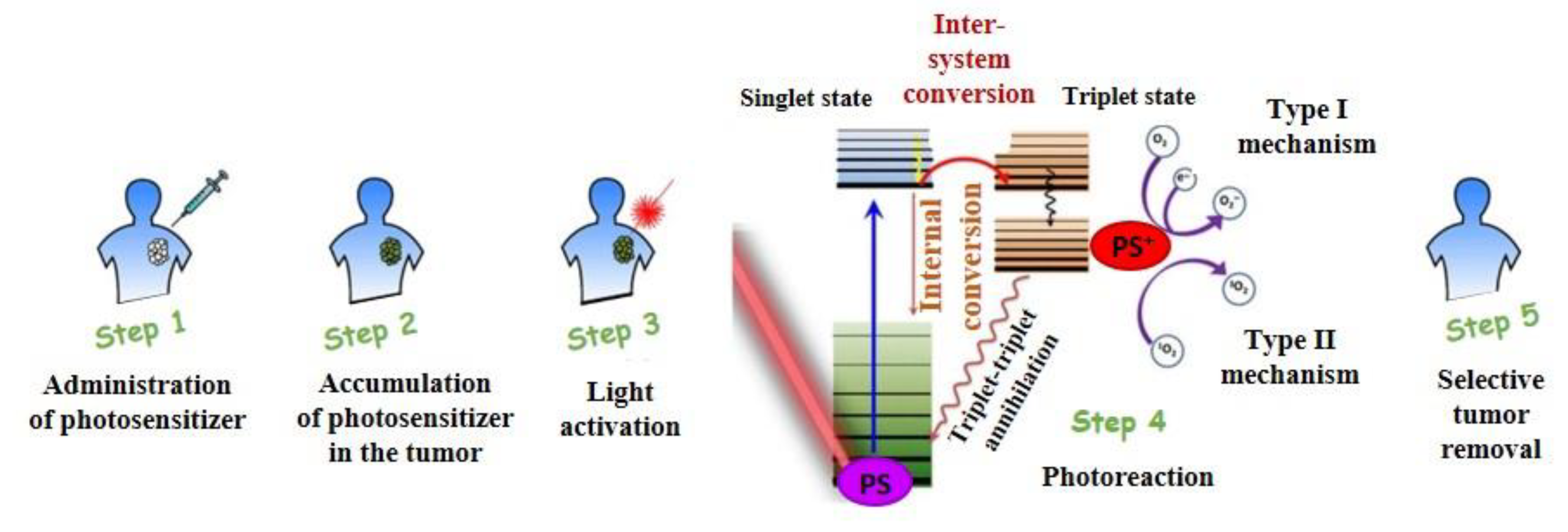
:1. Photodynamic Therapy
1.1. PDT in Cancer Therapy
1.2. Physicochemical Mechanism of PDT
1.3. PDT Applications
2. Plant-Derived Photosensitive Substances
Types of Photosensitive Substances by Hydrophobicity
- Hydrophobic PSs—compounds requiring the presence of transporters, such as liposomes or Cremophor EL, or Tween 80. They have the ability to localize in the inner lipid part of lipoproteins, mainly in LDL and high-density lipoproteins (HDLs), but also in very-low-density lipoproteins (VLDLs). This group includes phthalocyanines (ZnPC, C1A1PC), naphthalocyanines (isoBOSINC), tin-etiopurpurine (SnET2) [84], and hypericin [85].
- Amphiphilic PSs—asymmetric compounds, which can be incorporated into the outer phospholipid and apoprotein layer of lipoprotein particles, e.g., disulfonates (TPPS2a, C1A1PCS2a), lutetium teraphyrin (LuTex), and monoaspartyl chlorine (MACE), which forms a barrier between albumin and HDL [84]. Emodin can be included in this group [86].
- Hydrophilic PSs—drugs that predominantly bind to albumins and globulins, e.g., tetra-sulfone derivates of tetraphenylporfin (TPPS3 and TPPS4) and chloroaluminum phthalocyanine (C1A1PCS3 and C1A1PCS4) [84].
3. Hypericin
4. Emodin
5. Quinizarin
6. Danthron
7. Hypericin and Its Derivatives Interaction with DNA
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Castano, A.P.; Demidova, T.N.; Hamblin, M.R. Mechanisms in photodynamic therapy: Part one—photosensitizers, photochemistry and cellular localization. Photodiagnosis. Photodyn. Ther. 2004, 1, 279–293. [Google Scholar] [CrossRef] [Green Version]
- Deda, D.K.; Araki, K. Nanotechnology, light and chemical action: An effective combination to kill cancer cells. J. Braz. Chem. Soc. 2015, 26, 2448–2470. [Google Scholar] [CrossRef]
- Allen, T.M.; Hansen, C.B.; Demenzes, D.E.L. Pharmacokinetics of long circulating liposomes. Adv. Drug Deliv. Rev. 1995, 16, 267–284. [Google Scholar] [CrossRef]
- Loomis, K.; McNeeley, K.; Bellamkonda, R.V. Nanoparticles with targeting triggered release and imaging functionality for cancer application. Soft. Matter. 2011, 7, 839–856. [Google Scholar] [CrossRef]
- Reddi, E. Role of delivery vehicles for photosensitizers in the photodynamic therapy of tumors. J. Photochem. Photobiol. B 1997, 37, 189–195. [Google Scholar] [CrossRef]
- Huntošová, V.; Buzová, D.; Petrovajová, D.; Kasak, P.; Naďová, Z.; Jancura, D.; Sureau, F.; Miškovský, P. Development of a new LDL-based transport system for hydrophobic/amphiphilic drug delivery to cancer cells. Int. J. Pharm. 2012, 436, 463–471. [Google Scholar] [CrossRef]
- Hally, C.; Delcanale, P.; Nonell, S.; Viappiani, C.; Abbruzzetti, S. Photosensitizing proteins for antibacterial photodynamic inactivation. Transl. Biophotonics 2020, e201900031. [Google Scholar] [CrossRef]
- Ghorbani, J.; Rahban, D.; Aghamiri, S.; Teymouri, A.; Bahador, A. Photosensitizers in antibacterial photodynamic therapy: An overview. Laser Ther. 2018, 27, 293–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buriankova, L.; Buzova, D.; Chorvat, D.; Sureau, F.; Brault, D.; Miskovsky, P.; Jancura, D. Kinetics of hypericin association with low-density lipoproteins. Photochem. Photobiol. 2011, 87, 56–63. [Google Scholar] [CrossRef]
- Lenkavska, L.; Blascakova, L.; Jurasekova, Z.; Macajova, M.; Bilcik, B.; Cavarga, I.; Miskovsky, P.; Huntosova, V. Benefits of hypericin transport and delivery by low- and high-density lipoproteins to cancer cells: From in vitro to ex ovo. Photodiagnosis Photodyn. Ther. 2019, 25, 214–224. [Google Scholar] [CrossRef]
- Konan, Y.N.; Gurny, R.; Allemann, E. State of the art in the delivery of photosensitizers for photodynamic therapy. J. Photochem. Photobiol. B Biol. 2002, 66, 89–106. [Google Scholar] [CrossRef]
- Firestone, R.A. Low-density lipoprotein as a vehicle for targeting antitumor compounds to cancer cells. Bioconjugate Chem. 1994, 5, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Versluis, A.J.; van Geel, P.J.; Oppellar, H.; van Berkel, T.J.; Bijsterbosch, M.K. Receptor-mediated uptake of low-density lipoprotein by B16 melanoma cells in vitro and in vivo in mice. Br. J. Cancer 1996, 4, 525–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rensen, P.C.; de Vrueh, R.L.; Kuiper, J.; Bijsterbosch, M.K.; Biessen, E.A.; van Berkel, T.J. Recombinant lipoproteins: Lipoprotein-like lipid particles for drug targeting. Adv. Drug Deliv. Rev. 2001, 47, 251–276. [Google Scholar] [CrossRef]
- Kader, A.; Pater., A. Loading anticancer drugs into HDL as well as LDL has little affect on properties of complexes and enhances cytotoxicity to human carcinoma cells. J. Control. Release 2002, 80, 29–44. [Google Scholar] [CrossRef]
- Zheng, G.; Chen, J.; Li, H.; Glickson, J.D. Rerouting lipoprotein nanoparticles to selected alternate receptors for the targeted delivery of cancer diagnostic and therapeutic agents. Proc. Natl. Acad. Sci. USA 2005, 102, 17757–17762. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Li, H.; Sunar, U.; Chen, J.; Corbin, I.; Yodh, A.G.; Zheng, G. Naphthalocyanine-reconstituted LDL nanoparticles for in vivo cancer imaging and treatment. Int. J. Nanomed. 2007, 2, 767–774. [Google Scholar]
- Dougherty, T.J.; Gomer, C.J.; Henderson, B.W.; Jori, G.; Kessel, D.; Korbelik, M.; Moan, J.; Peng, Q. Photodynamic therapy. J. Natl. Cancer Inst. 1998, 90, 889–905. [Google Scholar] [CrossRef] [Green Version]
- Wilson, B.C.; Patterson, M.S. The physics, biophysics and technology of photodynamic therapy. Phys. Med. Biol. 2008, 53, R61–R109. [Google Scholar] [CrossRef]
- Yano, S.; Hirohara, S.; Obata, M.; Hagiya, Y.; Ogura, S.; Ikeda, A.; Kataoka, H.; Tanaka, M.; Joh, T. Current states and future views in photodynamic therapy. J. Photochem. Photobiol. C Photochem. Rev. 2011, 12, 46–67. [Google Scholar] [CrossRef]
- Lim, C.K.; Heo, J.; Shin, S.; Jeong, K.; Seo, Y.H.; Jang, W.D.; Park, C.R.; Park, S.Y.; Kim, S.; Kwon, I.C. Nanophotosensitizers toward advanced photodynamic therapy of cancer. Cancer Lett. 2013, 334, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Bechet, D.; Mordon, S.R.; Guillemin, F.; Barberi-Heyob, M.A. Photodynamic therapy of malignant brain tumours: A complementary approach to conventional therapies. Cancer Treat. Rev. 2014, 40, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Benov, L. Photodynamic therapy: Current status and future direction. Med. Princ. Pract. 2015, 24, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Miguel-Gomez, L.; Vano-Galvan, S.; Perez-Garcia, B.; Carrillo-Gijon, R.; Jaen-Olasolo, P. Treatment of folliculitis decalvans with photodynamic therapy: Results in 10 patients. J. Am. Acad. Dermatol. 2015, 72, 1085–1087. [Google Scholar] [CrossRef]
- Sharman, W.M.; Allen, C.M.; van Lier, J.E. Photodynamic therapeutics: Basic principles and clinical applications. Drug Discov. Today 1999, 4, 507–517. [Google Scholar] [CrossRef]
- Jin, Y.; Zhang, X.; Kang, H.; Du, L.; Li, M. Nanostructures of an amphiphilic zinc phthalocyanine polymer conjugate for photodynamic therapy of psoriasis. Colloids Surf. B 2015, 128, 405–409. [Google Scholar] [CrossRef]
- Sharman, W.M.; van Lier, J.E.; Allen, C.M. Targeted photodynamic theory via receptor mediated delivery systems. Adv. Drug Deliv. Rev. 2004, 56, 53–76. [Google Scholar] [CrossRef]
- Cruess, A.F.; Zlateva, G.; Pleil, A.M.; Wirostko, B. Photodynamic therapy with verteporfin in age-related macular degeneration: A systematic review of efficacy, safety, treatment modifications and pharmacoeconomic properties. Acta Ophthalmol. 2009, 87, 118–132. [Google Scholar] [CrossRef]
- Rockson, S.G.; Lorenz, D.P.; Cheong, W.F.; Woodburn, K.W. Photoangioplasty: An emerging clinical cardiovascular role for photodynamic therapy. Circulation 2000, 102, 591–596. [Google Scholar] [CrossRef] [Green Version]
- Bozzini, G.; Colin, P.; Betrouni, N.; Nevoux, P.; Ouzzane, A.; Puech, P.; Willers, A.; Mordon, S. Photodynamic therapy in urology: What can we do now and where are we heading. Photodiagn. Photodyn. Ther. 2012, 9, 261–273. [Google Scholar] [CrossRef]
- Panzarini, E.; Inguscio, V.; Dini, L. Immunogenic cell death: Can it be exploited in photodynamic therapy for cancer. Biomed. Res. Int. 2013, 2013, 482160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, G.E.; Mennel, S.; Spital, G.; Wachtlin, J.; Jurklies, B.; Heimann, H.; Damato, B.; Meyer, C.H. Different indications of photodynamic therapy in ophthalmology. Klin. Monbl. Augenheilkd. 2009, 226, 725–739. [Google Scholar] [CrossRef] [PubMed]
- Trindade, A.C.; De Figueiredo, J.A.P.; Steier, L.; Weber, J.B.B. Photodynamic therapy in endodontics: A literature review. Photomed. Laser Surg. 2015, 33, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Vohra, F.; Al-Kheraif, A.A.; Qadri, T.; Hassan, M.I.A.; Ahmedef, A.; Warnakulasuriya, S.; Javed, F. Efficacy of photodynamic therapy in the management of oral premalignant lesions. A systematic review. Photodiagn. Photodyn. Ther. 2015, 12, 150–159. [Google Scholar] [CrossRef]
- Karrer, S.; Kohl, E.; Feise, K.; Hiepe-Wegener, D.; Lischner, S.; Philipp-Dormston, W.; Podda, M.; Prager, W.; Walker, T.; Szeimies, R.M. Photodynamic therapy for skin rejuvenation: Review and summary of the literature-results of a consensus conference of an expert group for aesthetic photodynamic therapy. J. Dtsch. Dermatol. Ges. 2013, 11, 137–148. [Google Scholar] [CrossRef]
- Fuhrmann, G.; Serio, A.; Mazo, M.; Nair, R.; Stevens, M.M. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J. Control. Release 2015, 205, 35–44. [Google Scholar] [CrossRef]
- Baptista, M.S.; Wainwright, M. Photodynamic antimicrobial chemotherapy (PACT) for the treatment of malaria, leishmaniasis and trypanosomiasis. Braz. J. Med. Res. 2011, 44, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Goslinski, T.; Piskorz, J. Fluorinated porphyrinoids and their biomedical applications. J. Photochem. Photobiol. C Photochem. Rev. 2011, 12, 304–321. [Google Scholar] [CrossRef]
- Hanakova, A.; Bogdanova, K.; Tomankova, K.; Pizova, K.; Malohlava, J.; Binder, S.; Bajgar, R.; Langova, K.; Kolar, M.; Mosinger, J.; et al. The application of antimicrobial photodynamic therapy on S. aureus and E. coli using porphyrin photosensitizers bound to cyclodextrin. Microbiol. Res. 2014, 169, 163–170. [Google Scholar] [CrossRef]
- Baltazar, L.M.; Ray, A.; Santos, D.A.; Cisalpino, P.S.; Friedman, A.J.; Nosanchuk, J.D. Antimicrobial photodynamic therapy: An effective alternative approach to control fungal injections. Front. Microbiol. 2015, 6, 202. [Google Scholar] [CrossRef] [Green Version]
- Bonnett, R. Progress with heterocyclic photosensitizers for the photodynamic therapy (PDT) of tomours. J. Heterocyclic Chem. 2002, 39, 455. [Google Scholar] [CrossRef]
- Photolitec, LLC. Tumor Specific Imaging Therapy. Photodynamic Therapy. Available online: http://photolitec.org/Tech_PDT.html (accessed on 10 August 2020).
- Vatansever, F.; de Melo, W.C.M.A.; Avci, P.; Vecchio, D.; Sadasivam, M.; Gupta, A.; Chandran, R.; Karimi, M.; Parizotto, N.A.; Yin, R.; et al. Antimicrobial strategies centered around reactive oxygen species—bactericidal antibiotics, photodynamic therapy, and beyond. Microbiol. Rev. 2013, 37, 955–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, B.W.; Dougherty, T.J. How does photodynamic therapy work. Photochem. Photobiol. 1992, 55, 145–157. [Google Scholar] [CrossRef]
- Agostinis, P.; Berg, K.; Cengel, K.A.; Foster, T.H.; Girotti, A.W.; Gollnick, S.O.; Hahn, S.M.; Hamblin, M.R.; Juzeniene, A.; Kessel, D.; et al. Photodynamic therapy of cancer: An update. CA Cancer J. Clin. 2011, 61, 250–281. [Google Scholar] [CrossRef] [PubMed]
- Senapathy, G.J.; George, B.P.; Abrahamse, H. Exloring the role of phytochemicals as potent natural photosensitizers in photodynamic therapy. Anticancer Agents Med. Chem. 2020, 20, 1831–1844. [Google Scholar] [CrossRef] [PubMed]
- Moan, J.; Berg, K. The photodegradation of porphyrins in cells can be used to estimate the lifetime of singlet oxygen. Photochem. Photobiol. 1991, 53, 549–553. [Google Scholar] [CrossRef]
- Hatz, S.; Poulsen, L.; Ogilby, P.R. Time-resolved singlet oxygen phosphorescence measurements from photosensitized experiments in single cells: Effects of oxygen diffusion and oxygen concentration. Photochem. Photobiol. 2008, 84, 1284–1290. [Google Scholar] [CrossRef]
- Nyst, H.J.; Tan, I.B.; Stewart, F.A.; Balm, A.J.M. Is photodynamic therapy a good alternative to surgery and radiotherapy in the treatment of head and neck cancer. Photodiagnosis Photodyn. Ther. 2009, 6, 3–11. [Google Scholar] [CrossRef]
- Schafer, M. High sensitivity of Deinococcus radiodurans to photodynamically-produced singlet oxygen. Int. J. Radiat. Biol. 1998, 74, 249–253. [Google Scholar] [CrossRef]
- Kochevar, I.E.; Lambert, C.R.; Lynch, M.C.; Tedesco, A.C. Comparison of photosensitized plasma membrane damage caused by singlet oxygen and free radicals. Biochim. Biophys. Acta 1996, 1280, 223–230. [Google Scholar] [CrossRef] [Green Version]
- Barr, H.; Tralau, C.J.; Boulos, P.B.; MacRobert, A.J.; Tilly, R.; Bown, S.G. The contrasting mechanisms of colonic collagen damage between photodynamic therapy and thermal injury. Photochem. Photobiol. 1987, 46, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Hopper, C. Photodynamic therapy. A clinical reality in the treatment of cancer. Lancet. Oncol. 2000, 1, 212–219. [Google Scholar] [CrossRef]
- Allison, R.R.; Bagnato, V.S.; Sibata, C.H. Future of oncologic photodynamic therapy. Future Oncol. 2010, 6, 929–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foote, C. Mechanisms of photo-oxygenation. In Porphyrin Localization and Treatment of Tumors, 1st ed.; Doiron, D.R., Gomer, C.J., Eds.; Alan R. Liss: New York, NY, USA, 1984; pp. 3–18. [Google Scholar]
- Boegheim, J.P.; Scholte, H.; Dubbehman, T.M.; Beems, E.; Raap, A.K.; van Steveninck, J. Photodynamic effects of hematoporphyrin-derivative on enzyme activities of murine L929 fibroblasts. J. Photochem. Photobiol. B 1987, 1, 61–73. [Google Scholar] [CrossRef]
- Gibson, S.L.; Hilf, R. Interdependence of fluence, drug, dose and oxygen on hematoporphyrin derivate induced photosensitization of tumor mitochondria. Photochem. Photobiol. 1985, 42, 367–373. [Google Scholar] [CrossRef]
- Dolmans, D.E.; Fukumura, D.; Jain, R.K. Photodynamic therapy for cancer. Nat. Rev. Cancer 2003, 3, 380–387. [Google Scholar] [CrossRef]
- Ochsner, M. Photophysical and photobiological processes in the photodynamic therapy of tumors. J. Photochem. Photobiol. B 1997, 39, 1–18. [Google Scholar] [CrossRef]
- Canti, G.; Lattuada, D.; Morelli, S.; Nicolin, A.; Cubeddu, R.; Taroni, P.; Valentini, G. Efficacy of photodynamic therapy against doxorubicin-resistant murine tumors. Cancer Lett. 1995, 93, 255–259. [Google Scholar] [CrossRef]
- Lofgren, L.A.; Hallgren, S.; Nilsson, E.; Westerborn, A.; Nilsson, C.; Reizenstein, J. photodynamic therapy for recurrent nasopharyngeal cancer. Arch. Otolaryngol. Head Neck Surg. 1995, 121, 997–1002. [Google Scholar] [CrossRef]
- Davies, M.J. Singlet oxygen-mediated damage to proteins and its consequences. Biochem. Biophys. Res. Commun. 2003, 305, 761–770. [Google Scholar] [CrossRef]
- Bicalho, L.S.; Longo, J.P.F.; Pereira, L.O.; Santos, M.F.M.A.; Azevedo, R.B. Photodynamic therapy, a new approach in the treatment of oral cancer. Rev. Univ. Ind. Santander. Salud. 2010, 42, 167–174. [Google Scholar]
- Yoo, J.O.; Lim, Y.C.; Kim, Y.M.; Ha, K.S. Differential cytotoxic responses to low-and high-dose photodynamic therapy in human gastric and bladder cancer cells. J. Cell Biochem. 2011, 112, 3061–3071. [Google Scholar] [CrossRef]
- Moghissi, K.; Dixon, K.; Parson, R.J. A controlled trial of Nd-YAG laser vs photodynamic therapy for odvanced malignant bronchial obstruction. Laser Med. Sci. 1993, 8, 269–273. [Google Scholar] [CrossRef]
- Kato, H.; Okunaka, T.; Shimatani, H. Photodynamic therapy for early state bronchogenic carcinoma. J. Clin. Laser Med. Surg. 1996, 14, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Manoto, S.L.; Abrahamse, H. Effect of a newly synthesized Zn sulfophthalocyanine derivative on cell morphology, viability, proliferation, and cytotoxicity in a human lung cancer cell line (A549). Lasers Med. Sci. 2011, 26, 523–530. [Google Scholar] [CrossRef]
- Jheon, S.; Lee, J.M.; Kim, J.K.; Kim, K.H.; Seo, S.J. Photodynamic therapy for tracheobronchial cancer. Photodiagnosis Photodyn. Ther. 2011, 6, 177. [Google Scholar] [CrossRef]
- Grant, W.E.; MacRobert, A.J.; Bown, S.G.; Hopper, C.; Speight, P.M. Photodynamic therapy of oral cancer: Photosensitization with systemic aminolaevulinic acid. Lancet 1993, 342, 147–148. [Google Scholar] [CrossRef]
- Barr, H.; Shepherd, N.A.; Dix, A.; Roberts, D.J.H.; Tan, W.C.; Krasner, N. Eradication of high-grade dysplasia in columnar-lined (Barrett´s) oesophagus by photodynamic therapy with endogenously generated protoporphyrin IX. Lancet 1996, 348, 584–585. [Google Scholar] [CrossRef]
- Miller, J.D.; Baron, E.D.; Scull, H.; Hsia, A.; Berlin, J.C.; McCormick, T.; Colussi, V.; Kenney, M.E.; Cooper, K.D.; Oleinick, N.L. Photodynamic therapy with the phtalocyanin photosensitizer Pc 4: The case experience with preclinical mechanistic and early clinical-tranlational studies. Toxicol. Appl. Pharmacol. 2007, 224, 290–299. [Google Scholar] [CrossRef] [Green Version]
- Nseyo, U.O.; DeHaven, J.; Dougherty, T.J.; Potter, W.R.; Merrill, D.L.; Lundahl, S.L.; Lamm, D.L. Photodynamic therapy (PDT) in the treatment of patients with resistant superficial bladder cancer: A long-term experience. J. Clin. Laser Med. Surg. 1998, 16, 61–68. [Google Scholar] [CrossRef]
- Leon, S.P.; Folkerth, R.D.; Black, P. Microvessel density is a prognostic indicator for patients with astroglial brain tumors. Cancer 1996, 77, 362–372. [Google Scholar] [CrossRef]
- Zhu, T.C.; Finlay, J.C. The role of photodynamic therapy (PDT) physics. Med. Phys. 2008, 35, 3127–3136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- George, B.P.A.; Abrahamse, H. A review on novel breast cancer therapies: Photodynamic therapy and plant derived agent induced cell death mechanisms. Anticancer Agents Med. Chem. 2016, 16, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Kostović, K.; Pastar, Z.; Ceović, R.; Mokos, Z.B.; Buzina, D.S.; Stanimirović, A. Photodynamic therapy in dermatology: Current treatments and implications. Coll. Antropol. 2012, 36, 1477–1481. [Google Scholar] [PubMed]
- Kessel, D. Photosensitization of viral particles. J. Lab. Clin. Med. 1990, 116, 428. [Google Scholar] [PubMed]
- Tardivo, J.P.; Del Giglio, A.; Paschoal, L.H.; Baptista, M.S. New photodynamic therapy protocol to treat AIDS-related Kaposi´s sarcoma. Photomed. Laser Surg. 2006, 24, 528–531. [Google Scholar] [CrossRef] [PubMed]
- Mĺkvy, P. Position and possibilities of photodynamic therapy in oncology. Oncology 2007, 5, 299–301. [Google Scholar]
- Castano, A.P.; Demidova, T.N.; Hamblin, M.R. Mechanisms in photodynamic therapy: Part three—Photosensitizer pharmacokinetics, biodistribution, tumor localization and modes of tumor destruction. Photodiagnosis Photodyn. Ther. 2005, 2, 91–106. [Google Scholar] [CrossRef] [Green Version]
- Muniyandi, K.; George, B.; Parimelazhagan, T.; Abrahamse, H. Role of photoactive phytocompounds in photodynamic therapy of cancer. Molecules 2020, 25, 4102. [Google Scholar] [CrossRef]
- Doherty, R.E.; Sazanovich, I.V.; McKenzie, L.K.; Stasheuski, A.S.; Coyle, R.; Baggaley, E.; Bottomley, S.; Weinstein, J.A.; Bryant, H.E. Photodynamic killing of cancer cells by a platinum(II) complex with cyclometallating ligand. Sci. Rep. 2016, 6, 22668. [Google Scholar] [CrossRef] [Green Version]
- Pouton, C.W.; Wagstaff, K.M.; Roth, D.M.; Moseley, G.W.; Jans, D.A. Targeted delivery to the nucleus. Adv. Drug Deliv. Rev. 2007, 59, 698–717. [Google Scholar] [CrossRef] [PubMed]
- Sobolev, A.S. Novel modular transporters delivering anticancer drugs and foreign DNA to the nuclei of target cancer cells. J. Buon. 2009, 14 (Suppl. 1), S33–S42. [Google Scholar]
- de Melo, W.C.M.A.; Lee, A.N.; Perussi, J.R.; Hamblin, M.R. Electroporation enhances antimicrobial photodynamic therapy mediated by the hydrophobic photosensitizer, hypericin. Photodiagnosis Photodyn. Ther. 2013, 10, 647–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves, D.S.; Pérez-Fons, L.; Estepa, A.; Micol, V. Membrane-related effects underlying the biological activity of the anthraquinones emodin and barbaloin. Biochem. Pharmacol. 2004, 68, 549–561. [Google Scholar] [CrossRef]
- Du, K.; Xia, Q.; Heng, H.; Feng, F. Temozolomide-doxorubicin conjugate as a double intercalating agent and delivery by apoferritin for glioblastoma chemotherapy. ACS Mater. Interfaces 2020, 12, 34599–34609. [Google Scholar] [CrossRef] [PubMed]
- Mandelli, F.; Vignetti, M.; Suciu, S.; Stasi, R.; Petti, M.C.; Meloni, G.; Muus, P.; Marmont, F.; Marie, J.P.; Labar, B.; et al. Daunorubicin versus mitoxantrone versus idarubicin as induction and consolidation chemotherapy for adults with acute myeloid leukemia: The EORTC and GIMEMA groups study AML-10. J. Clin. Oncol. 2009, 27, 5397–5403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wójcik, K.; Zarebski, M.; Cossarizza, A.; Dobrucki, J.W. Daunomycin, an antitumor DNA intercalator, influences histone-DNA interactions. Cancer Biol. Ther. 2013, 14, 823–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Cao, Y.; Yin, X.; Zhu, L.; Chen, Y.; Wang, W.; Hu, J. Design and synthesis of various quinizarin derivatives as potential anticancer agents in acute T lymphoblastic leukemia. Bioorganic Med. Chem. 2019, 27, 1362–1369. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhao, C.; He, R.; Zhou, M.; Liu, Y.; Guo, X.; Wang, M.; Zhu, F.; Qin, R.; Li, X. Danthron suppresses autophagy and sensitizers pancreatic cancer cells to doxorubicin. Toxicol. In Vitro 2019, 54, 345–353. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, M.; Qian, J.; Xu, W.; Wang, J.; Hou, G.; Ji, L.; Suo, A. Sequentially self-assembled polysaccharide-based nanocomplexes for combined chemotherapy and photodynamic therapy of breast cancer. Carbohydr. Polym. 2019, 203, 203–213. [Google Scholar] [CrossRef]
- Lee, H.; Han, J.; Shin, H.; Han, H.; Na, K.; Kim, H. Combination of chemotherapy and photodynamic therapy for cancer treatment with sonoporation effects. J. Control. Release 2018, 283, 190–199. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Liu, D.; Lin, W. self-assembled core-shell nanoparticles for combined chemotherapy and photodynamic therapy of resistant head and neck cancers. ACS Nano 2015, 9, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Gleason, H.A.; Cronquist, A. Manual of Vascular Plants of Northeastern United States and Adjacent Canada, 2nd ed.; New York Botanical Garden: Bronx, New York, NY, USA, 1991; p. 910. [Google Scholar]
- Rahman, A. Bioactive natural products (Part C). In Studies in Natural Products Chemistry, 1st ed.; Rahman, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2000; Volume 22, p. 647. [Google Scholar]
- Miškovský, P. Hypericin—A new antiviral and antitumor photosensitizer: Mechanism of action and interaction with biological macromolecules. Curr. Drug Targets 2002, 3, 55–84. [Google Scholar] [CrossRef] [PubMed]
- Thornett, A. Use of hypericin as antidepressant. Valid measure of antidepressant efficacy in primary care is needed. Brit. Med. J. 2000, 320, 1141. [Google Scholar]
- Butterweck, V.; Winterhoff, H.; Herkenham, M. St John´s wort, hypericin, and imipramine: A comparative analysis of mRNA levels in brain areas involved in HPA axis control following short-term and long-term administration in normal and stressed rats. Mol. Psychiatry 2001, 6, 547–564. [Google Scholar] [CrossRef] [Green Version]
- Eberman, R.; Alth, G.; Kreitner, M.; Kubin, A. Natural products derived from plants as potential drugs for the photodynamic destruction of tumor cells. J. Photochem. Photobiol. B 1996, 36, 95–97. [Google Scholar] [CrossRef]
- Pengelly, A. The Constituents of Medical Plants: An Introduction to the Chemistry and Therapeutics of Herbal Medicine, 2nd ed.; CABI Publishing: Cambridge, UK, 2004; pp. 20–50. [Google Scholar]
- Miroššay, A.; Mirossay, L.; Tóthová, J.; Miškovský, P.; Onderková, H.; Mojžiš, J. Potentiation of hypericin and hypocrellin-induced phototoxicity by omeprazole. Phytomedicine 1999, 6, 311–317. [Google Scholar] [CrossRef]
- Cavarga, I.; Brezani, P.; Fedorocko, P.; Miskovsky, P.; Bobrov, N.; Longauer, F.; Rybarova, S.; Mirossay, L.; Stubna, J. Photoinduced antitumor effect of hypericin can be enhanced by fractionated dosing. Photomedicine 2005, 12, 680–683. [Google Scholar] [CrossRef]
- Plenagl, N.; Duse, L.; Seitz, B.S.; Goergen, N.; Pinnapireddy, S.R.; Jedelska, J.; Brűβler, J.; Bakowsky, U. Photodynamic therapy—hypericin tetraether liposome conjugates and their antitumor and antiangiogenic activity. J. Drug Deliv. 2019, 26, 23–33. [Google Scholar] [CrossRef] [Green Version]
- Bianchini, P.; Cozzolino, M.; Oneto, M.; Pesce, L.; Pennacchietti, F.; Tognolini, M.; Giorgio, C.; Nonell, S.; Cavanna, L.; Delcanale, P.; et al. Hypericin-apomyoglobin an enhanced photosensitizer complex for the treatment of tumor cells. Biomacromolecules 2019, 20, 2024–2033. [Google Scholar] [CrossRef]
- Fiebich, B.L.; Lieb, A.H. Inhibition of substance P induced cytokine synthesis by St John´s Wort extracts. Pharmacopsychiatry 2001, 34, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Samadi, S.; Khadivzadeh, T.; Emami, A.; Moosavi, N.S.; Tafaghodi, M.; Behnam, H.R. The effect of Hypericum perforatum on the wound healing and scar of cesarean. J. Altern. Complement. Med. 2010, 16, 113–117. [Google Scholar] [CrossRef]
- Hajhashemi, M.; Ghanbari, Z.; Movahedi, M.; Rafieian, M.; Keivani, A.; Haghollahi, F. The effect of Achillea millefolium and Hypericum perforatum ointments on episiotomy wound healing in primiparous women. J. Matern. Fetal Neonatal Med. 2018, 31, 63–69. [Google Scholar] [CrossRef] [PubMed]
- García, I.; Ballesta, S.; Gilaberte, Y.; Rezusta, A.; Pascual, Á. Antimicrobial photodynamic activity of hypericin against methicillin-susceptible and resistant Staphylococcus aureus biofilms. Future Microbiol. 2015, 10, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, V.; Krammer, B.; Plaetzer, K. Antibacterial photodynamic therapy using water-soluble formulations of hypericin or mTHPC is effective in inactivation of Staphylococcus aureus. Photochem. Photobiol. Sci. 2010, 9, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Yow, C.; Tang, H.M.; Chu, E.S.; Huang, Z. Hypericin-mediated photodynamic antimicrobial effect on clinically isolated pathogens. Photochem. Photobiol. 2012, 88, 626–632. [Google Scholar] [CrossRef]
- Rodríguez-Amigo, B.; Delcanale, P.; Rotger, G.; Juárez-Jiménez, J.; Abbruzzetti, S.; Summer, A.; Agut, M.; Luque, F.J.; Nonell, S.; Viappiani, C. The complex of hypericin with β-lactoglobulin has antimicrobial activity with perspective applications in dairy industry. J. Dairy Sci. 2015, 98, 89–94. [Google Scholar] [CrossRef] [Green Version]
- Delcanale, P.; Rodríguez-Amigo, B.; Juárez-Jiménez, J.; Luque, F.J.; Abbruzzetti, S.; Agut, M.; Nonell, S.; Viappiani, C. Tuning the local solvent composition at a drug carrier surface: Effect of dimethyl sulfoxide/water mixture on the photofunctional properties of hyperici-β-lactoglobulin. Mat. Chem. B 2017, 5, 1633–1641. [Google Scholar] [CrossRef] [Green Version]
- Pezzuoli, D.; Cozzolino, M.; Montali, C.; Brancaleon, L.; Bianchini, P.; Zantedeschi, M.; Bonardi, S.; Viappiani, C.; Abbruzzetti, S. Serum albumins are efficient delivery systems for the photosensitizer hypericin in photosensitization-based treatments against Staphylococcus aureus. Food Control. 2018, 94, 254–262. [Google Scholar] [CrossRef]
- Lopez-Bazzocchi, I.; Hudson, J.B.; Towers, G.H. Antiviral activity of photoactive plant pigment hypericin. Photochem. Photobiol. 1991, 54, 95–98. [Google Scholar] [CrossRef]
- Hudson, J.B.; Lopez-Bazzocchi, I.; Towers, G.H. Antiviral activities of hypericin. Antivir. Res. 1991, 15, 101–112. [Google Scholar] [CrossRef]
- Moraleda, G.; Wu, T.T.; Jilbert, A.R.; Aldrich, C.E.; Condreay, L.D.; Larsen, S.H.; Tang, J.C.; Colacino, J.M.; Mason, W.S. Inhibition of duck hepatitis B virus replication by hypericin. Antivir. Res. 1993, 20, 235–247. [Google Scholar] [CrossRef]
- Guedes, R.C.; Eriksson, L.A. Effects of halogen substitution on the photochemical properties of hypericin. J. Photochem. Photobiol. A Chem. 2006, 178, 41–49. [Google Scholar] [CrossRef]
- Meruelo, D.; Lavie, G.; Lavie, D. Therapeutic agents with dramatic antiretroviral activity and little toxicity at effective doses: Aromatic polycyclic diones hypericin and pseudohypericin. Proc. Natl. Acad. Sci. USA 1988, 85, 5230–5234. [Google Scholar] [CrossRef] [Green Version]
- Hudson, J.B.; Imperial, V.; Haugland, R.P.; Diwu, Z. Antiviral activities of photoactive perylenequinones. Photochem. Photobiol. 1997, 65, 352–354. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, C. Raman spectroscopy study on structure of human immunodeficiency virus (HIV) and hypericin-induced photosensitive damage of HIV. Sci. China Ser. C 2005, 48, 117–132. [Google Scholar]
- Gulick, R.M.; McAuliffe, V.; Holden-Wiltse, J.; Crumpacker, C.; Liebes, L.; Stein, D.S.; Meehan, P.; Hussey, S.; Forcht, J.; Valentine, F.T. Phase I studies of hypericin, the active compound in St. John´s Wort, as an antiretroviral agent in HIV-infected adults: AIDS clinical trials group protocols 150 and 258. Ann. Intern. Med. 1999, 130, 510–514. [Google Scholar] [CrossRef]
- Kerb, R.; Reum, T.; Brockmöller, J.; Bauer, S.; Roots, I. No clinically relevant photosensitization after single-dose and steady state in treatment with hypericum extract in man. Eur. J. Clin. Pharmacol. 1995, 49, A156. [Google Scholar]
- Diwu, Z. Novel therapeutic and diagnostic applications of hypocrellins and hypericins. Photochem. Photobiol. 1995, 61, 529–539. [Google Scholar] [CrossRef]
- Wynn, J.L.; Cotton, T.M. Spectroscopic properties of hypericin in solution and at surfaces. J. Phys. Chem. 1995, 99, 4317–4323. [Google Scholar] [CrossRef]
- Yamazaki, T.; Ohta, N.; Yamazaki, I.; Song, P.S. Excited-state properties of hypericin: Electronic spectra and fluorescence decay kinetics. J. Phys. Chem. 1993, 97, 7870–7875. [Google Scholar] [CrossRef]
- Keša, P.; Antalík, M. Determination of pKa constants of hypericin in aqueous solution of the anti-allergic hydrotropic drug Cromolyn disodium salt. Chem. Phys. Lett. 2017, 676, 112–117. [Google Scholar] [CrossRef]
- Redepenning, J.; Tao, N. Measurement of formal potentials for hypericin in dimethylsulfoxide. Photobiol. 1993, 58, 532–535. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Pardini, R.S. Oxygen dependence of hypericin-induced phototoxicity to EMT6 mouse mammary carcinoma cells. Photochem. Photobiol. 1992, 55, 831–837. [Google Scholar] [CrossRef]
- Ehrenberg, B.; Anderson, J.L.; Foote, C.S. Kinetics and yield of singlet oxygen photosensitized by hypericin inorganic and biological media. Photochem. Photobiol. 1998, 68, 135–140. [Google Scholar] [CrossRef]
- Roslaniec, M.; Weitman, H.; Freeman, D.; Mazur, Y.; Ehrenberg, B. Liposome binding constant and singlet oxygen quantum yields of hypericin, tetrahydroxyhelianthrone and their derivatives: Studies in organic solutions and in liposomes. J. Photochem. Photobiol. B Biol. 2000, 57, 149–158. [Google Scholar] [CrossRef]
- Gbur, P.; Dedič, R.; Jancura, D.; Miškovský, P.; Hala, J. Time-resolved luminescence and singlet oxygen formation under illumination of hypericin in acetone. J. Lumin. 2008, 128, 765–767. [Google Scholar] [CrossRef]
- Fehr, M.J.; Carpenter, S.L.; Petrich, J.W. The role of oxygen in the photoinduced antiviral activity of hypericin. Bioorg. Med. Chem. Lett. 1994, 4, 1339–1344. [Google Scholar] [CrossRef]
- Carpenter, S.; Fehr, J.M.; Kraus, G.A.; Petrich, J.W. Chemiluminescent activation of the antiviral activity of hypericin: A molecular flashlight. Proc. Natl. Acad. Sci. USA 1994, 91, 12273–12277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verebová, V.; Belej, D.; Joniová, J.; Jurašeková, Z.; Miškovský, P.; Kožár, T.; Horváth, D.; Staničová, J.; Huntošová, V. Deeper insights into the drug defense of glioma cells against hydrophobic molecules. Int. J. Pharm. 2016, 503, 56–67. [Google Scholar] [CrossRef]
- de Witte, P.; Agostinis, P.; Van Lint, J.; Merlevede, W.; Vandenheede, J.R. Inhibition of epidermal growth factor receptor tyrosine kinase activity by hypericin. Biochem. Pharmacol. 1993, 46, 1929–1936. [Google Scholar] [CrossRef]
- Agostinis, P.; Vantieghem, A.; Merlevede, W.; de Witte, P. Hypericin in cancer treatment: More light on the way. Int. J. Biochem. Cell Biol. 2002, 34, 221–241. [Google Scholar] [CrossRef]
- Buytaert, E.; Callewaert, G.; Hendrickx, N.; Scorrano, L.; Hartmann, D.; Missiaen, L.; Vandenheede, J.R.; Heirman, I.; Grooten, J.; Agostinis, P. Role of endoplasmic reticulum depletion and multidomain proapoptic BAX and BAK proteins in shaping cell death after hypericin-mediated photodynamic therapy. Faseb. J. 2006, 20, 756–758. [Google Scholar] [CrossRef]
- Ferrario, A.; von Tiehl, K.; Wong, S.; Luna, M.; Gomer, C.J. Cyclooxygenase-2 inhibitor treatment enhances photodynamic therapy-mediated tumor response. Cancer Res. 2002, 62, 3956–3961. [Google Scholar]
- Delcanale, P.; Pennacchietti, F.; Maestrini, G.; Rodríguez-Amigo, B.; Bianchini, P.; Diaspro, A.; Iagatti, A.; Patrizi, B.; Foggi, P.; Agut, M.; et al. Subdiffraction localization of a nanostructured photosensitizer in bacterial cells. Sci. Rep. 2015, 5, 15564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miškovský, P.; Sureau, F.; Chinsky, L.; Turpin, P.Y. Subcellular distribution of hypericin in human cancer cells. Photochem. Photobiol. 1995, 62, 546–549. [Google Scholar] [CrossRef]
- Miškovský, P.; Chinsky, L.; Wheeler, G.V.; Turpin, P.Y. Hypericin site specific interactions within polynucleotides used as DNA model compounds. J. Biomol. Struct. Dyn. 1995, 13, 547–552. [Google Scholar] [CrossRef]
- Sánchez-Cortés, S.; Miškovský, P.; Jancura, D.; Bertoluzza, A. Specific interactions of antiretroviraly active drug hypericin with DNA as studied by surface-enhanced resonance Raman spectroscopy. J. Phys. Chem. 1996, 100, 1938–1944. [Google Scholar] [CrossRef]
- Das, K.; Smirnov, A.V.; Wen, J.; Miškovský, P.; Petrich, J.W. Photophysics of hypericin and hypocrellin A in complex with subcellular components: Interactions with human serum albumin. Photochem. Photobiol. 1999, 69, 633–645. [Google Scholar] [CrossRef]
- Senthil, V.; Jones, L.R.; Senthil, K.; Grossweiner, L.J. Hypericin photosensitization in aqueous model system. Photochem. Photobiol. 1994, 59, 40–47. [Google Scholar] [CrossRef]
- Mijatovic, S.; Maksimovic-Ivanic, D.; Radovic, J.; Milijkovic, D.; Kaludjerovic, G.N.; Sabo, T.J.; Trajkovic, V. Aloe emodin decreases the ERK-dependent anticancer activity of cisplatin. Cell Mol. Life Sci. 2005, 62, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Qi, Q.H.; Yang, W.X.; Xu, J.; Dong, Z.L. Contractile effects and antracellular Ca2+ signaling induced by emodin in circular smooth muscle cells of rat colon. World J. Gastroenterol. 2003, 9, 1804–1807. [Google Scholar] [CrossRef] [PubMed]
- Janeczko, M.; Masčyk, M.; Kubiński, K.; Golczyk, H. Emodin, a natural inhibitor of protein kinase CK2, suppresses growth, hyphal development, and biofilm formation of Candida Albicans. Yeast 2017, 34, 253–265. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.C.; Lee, C.R.; Lee Chao, P.D.; Chen, C.C.; Chu, S.H. Vasorelaxant effects of emodin, an antraquinone from a Chinese herb. Eur. J. Pharmacol. 1991, 205, 289–294. [Google Scholar]
- Su, Y.T.; Chang, H.L.; Shyue, S.K.; Hsu, S.L. Emodin induces apoptosis in human lung adenocarcinoma cells through a reactive oxygen species-dependent mitochondrial signaling pathway. Biochem. Pharmacol. 2005, 70, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Chukwujelwu, J.C.; Coombes, P.H.; Mulholland, D.A.; van Staden, J. Emodin, an antibacterial anthraquinone from the roots of Cassia occidentalis. S. Afr. J. Bot. 2006, 72, 295–297. [Google Scholar] [CrossRef] [Green Version]
- Srinivas, G.; Babykutty, S.; Sathiadevan, P.P.; Srinivas, P. Molecular mechanism of emodin action: Transition from laxative ingredient to an antitumor agent. Med. Res. Rev. 2007, 27, 591–608. [Google Scholar] [CrossRef] [PubMed]
- Fernand, V.E.; Dinh, D.T.; Washington, S.J.; Fakayode, S.O.; Loss, J.N.; van Ravenswaay, R.O.; Warner, I.M. Determination of pharmacologically active compounds in root extracts of Cassia Alata L. by use of high performance liquid chromatography. Talanta 2008, 74, 896–902. [Google Scholar] [CrossRef] [Green Version]
- Hennebelle, T.; Weniger, B.; Joseph, H.; Sahpaz, S.; Bailleul, F. Senna alata. Fitoterapia 2009, 80, 385–393. [Google Scholar] [CrossRef]
- Andersen, D.O.; Weber, N.D.; Wood, S.G.; Hughes, B.G.; Murray, B.K.; North, J.A. In vitro virucidal activity of selected anthraquinones and anthraquinone derivatives. Antiviral Res. 1991, 16, 185–196. [Google Scholar] [CrossRef]
- Kusari, S.; Lamshöft, M.; Zühlke, S.; Spiteller, M. An endophytic fungus from hypericum perforatum that produces hypericin. J. Nat. Prod. 2008, 71, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Karioti, A.; Bilia, A.R. Hypericins as potential leads for new therapeutics. Int. J. Mol. Sci. 2010, 11, 562–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogdanska, A.; Chmurzyński, L.; Ossowski, T.; Liwo, A.; Jeziorek, D. Protolytic equilibria of dihydroxyanthraquinones in non-aqueous solutions. Anal. Chim. Acta 1999, 402, 339–343. [Google Scholar] [CrossRef]
- Mueller, S.O.; Lutz, W.K.; Stopper, H. Factors affecting the genotoxic potency ranking of natural anthraquinones in mammalian cell culture systems. Mutat. Res. 1998, 414, 125–129. [Google Scholar] [CrossRef]
- Li, J.; Yang, W.; Hu, W.; Wang, J.; Jin, Z.; Wang, X.; Xu, W. Effects of emodin on the activity of K channel in guinea pig taenia coli smooth muscle cells. Acta Pharm. Sin. 1998, 33, 321–325. [Google Scholar]
- Ma, T.; Qi, Q.H.; Xu, J.; Dong, Z.L.; Yang, W.X. Signal pathways involved in emodin-induced contraction of smooth muscle cells from rat colon. World J. Gastroenterol. 2004, 10, 1476–1479. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Xiong, Y.; Wang, L.; Lv, B.; Lin, Y. Characteristics of emodin on modulating the contractility of jejunal smooth muscle. Can. J. Physiol. Pharmacol. 2012, 90, 455–462. [Google Scholar] [CrossRef]
- Vargas, F.; Fraile, G.; Velásquez, M.; Correia, H.; Fonseca, G.; Marin, M.; Marcano, E.; Sánchez, Y. Studies on the photostability and phototoxicity of aloe-emodin, emodin and rhein. Pharmazie 2002, 57, 399–404. [Google Scholar]
- Oshida, K.; Hirakata, M.; Maeda, A.; Miyoshi, T.; Miyamoto, Y. Toxicological effect of emodin in mouse testicular gene expression profile. J. Appl. Toxicol. 2011, 31, 790–800. [Google Scholar] [CrossRef]
- Xu, X.; Toselli, A.; Russell, L.D.; Seldin, D.C. Globozospermia in mice lacking the casein kinase II α´catalytic subunit. Nat. Genet. 1999, 23, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Escalier, D.; Silvius, D.; Xu, X. Spermatogenesis of mice lacking CK2alpha´failure of germ cell survival and characteristic modifications of the spermatid nucleus. Mol. Reprod. Dev. 2003, 66, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lin, L.; Ye, B. Electrochemical studies of the interaction of the anticancer herbal drug emodin with DNA. J. Pharm. Biomed. Anal. 2006, 42, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.; Zhang, H.; Qiao, C.; Sun, Y.; Liu, C. Studies of interaction of emodin and DNA in the presence of ethidium bromide by spectroscopic method. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2008, 69, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.T.; Silva, G.; Pungartnik, C.; Brendel, M. Study of DNA-emodin interaction by FTIR and UV-Vis spectroscopy. J. Photochem. Photobiol. B: Biol. 2012, 111, 59–63. [Google Scholar] [CrossRef]
- Li, Y.; Luan, Y.; Qi, X.; Li, M.; Gong, L.; Xue, X.; Wu, X.; Wu, Y.; Chen, M.; Xing, G.; et al. Emodin triggers DNA double-strand breaks by stabilizing topoisomerase II-DNA cleavage complexes and by inhibiting ATP hydrolysis of topoisomerase II. Toxicol. Sci. 2010, 118, 435–443. [Google Scholar] [CrossRef] [Green Version]
- Fabriciova, G.; Sanchez-Cortes, S.; Garcia-Ramos, J.V.; Miskovsky, P. Surface-enhanced Raman spectroscopy study of the interaction of the antitumoral drug emodin with serum albumin. Biopolymers 2004, 74, 125–130. [Google Scholar] [CrossRef] [Green Version]
- Bi, S.Y.; Song, D.O.; Kan, Y.H.; Xu, D.; Tian, Y.; Zhou, X.; Zhang, H.Q. Spectroscopic characterization of effective components anthraquinones in Chinese medicinal herbs binding with serum albumins. Spectrochim. Acta A: Biomol. Spectrosc. 2005, 62, 203–212. [Google Scholar] [CrossRef]
- Koistinen, K.M.; Soininen, P.; Venalainen, T.A.; Hayrinen, J.; Laatikainen, R.; Perakyla, M.; Tervahauta, A.I.; Karenlampi, S.O. Birch PR-10c interacts with several biologically important ligands. Phytochemistry 2005, 66, 2524–2533. [Google Scholar] [CrossRef]
- Sevilla, P.; Rivas, J.M.; García-Blanco, F.; García-Ramos, J.V.; Sánchez-Cortés, S. Identification of the antitumoral drug emodin binding sites in bovine serum albumin by spectroscopic methods. BBA-Proteins Proteom. 2007, 1774, 1359–1369. [Google Scholar] [CrossRef]
- Pecere, T.; Gazzola, M.V.; Mucignat, C.; Parolin, C.; Dalla, V.F.; Cavaggioni, A.; Basso, G.; Diaspro, A.; Salvato, B.; Carli, M.; et al. Aloe-emodin is a new type of anticancer agent with selective activity against neuroectodermal tumors. Cancer Res. 2000, 60, 2800–2804. [Google Scholar]
- Chun-Guan, W.; Jun-Qing, Y.; Bei-Zhong, L.; Dan-Ting, J.; Chong, W.; Liang, Z.; Dan, Z.; Yan, W. Anti-tumor activity of emodin against human chronic myelocytic leukemia K562 cell lines in vitro and in vivo. Eur. J. Pharmacol. 2010, 627, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Kuo, T.C.; Yang, J.S.; Lin, M.W.; Hsu, S.C.; Lin, J.J.; Lin, H.J.; Hsia, T.C.; Liao, L.; Yang, M.D.; Fan, M.J.; et al. Emodin has cytotoxic and pretoctive effect in rat c6 glioma cells: Roles of Mdr1a and nuclear factor κB in cell survival. J. Pharmacol. Exp. Ther. 2009, 330, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.C.; Su, Y.J.; Lin, S.T.; Jhan, J.Y.; Ciou, S.C.; Cheng, C.M.; Lin, Y.W. Suppression of ERCC1 and Rad51 expression through ER1/2 inactivation is essential in emodin-mediated cytotoxicity in human non-small cell lung cancer cells. Biochem. Pharmacol. 2010, 79, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Chemicalland21. Quinizarin. Available online: http://www.chemicalland21.com/specialtychem/finechem/QUINIZARIN.htm (accessed on 7 September 2020).
- Quinti, L.; Allen, N.S.; Edge, M.; Murphy, B.P.; Perotti, A. A study of the strongly fluorescent species formed by the interaction of the dye 1,4-dihydroxyanthraquinone (quinizarin) with A(III). J. Photochem. Photobiol. A Chem. 2003, 155, 79–91. [Google Scholar] [CrossRef]
- Yohida, M. Chemistry and hair dyes application of dihydroxy derivates of anthraquinone and naphtoquinone. Prog. Org. Coat. 1997, 31, 63–72. [Google Scholar] [CrossRef]
- Brinkworth, R.I.; Fairle, D.P. Hydroxyquinones are competitive nonpeptide inhibitors of HIV-1 proteinase. Biochim. Biophys. Acta 1995, 1253, 5–8. [Google Scholar] [CrossRef]
- Brown, J.P. A review of the genetic effects of naturally occurring flavonoids, anthraquinones and related compounds. Mutat. Res. 1980, 75, 243–277. [Google Scholar] [CrossRef]
- Longo, V.; Amato, G.; Salvetti, A.; Gervasi, P.G. Heterogenous effect of anthraquinones on drug-metabolizing enzymes in the liver and small intestine of rat. Chem. Biol. Interact. 2000, 126, 63–77. [Google Scholar] [CrossRef]
- Carter, T.P.; Gillispie, G.D.; Connoll, M.A. Intramolecular hydrogen bonding in substituted anthraquinones by laser-induced fluorescence. 1. 1,4-dihydroxyanthraquinone (quinizarin). J. Phys. Chem. 1982, 86, 192–196. [Google Scholar] [CrossRef]
- Smulevich, G.; Angeloni, L.; Giovannardi, S.; Marzocchi, M.P. Resonance Raman and polarized light infrared spectra of 1,4-dihydroxyanthraquinone, vibrational studies of the ground and excited electronic states. Chem. Phys. 1982, 65, 313–322. [Google Scholar] [CrossRef]
- Nigam, G.; Deppisch, B. Redetermination of the structure of 1,4-dihydroxyanthraquinone (C14H8O4). Z. Kristallogr. 1980, 151, 185–191. [Google Scholar] [CrossRef]
- Fabriciová, G.; Garcia-Ramos, J.V.; Miškovský, P.; Sanchez-Cortes, S. Absorption and acidic behavior of anthraquinone drugs quinizarin and danthron on Ag nanoparticles studied by Raman spectroscopy. Vib. Spectrosc. 2004, 34, 273–281. [Google Scholar] [CrossRef]
- Bondy, G.S.; Armstrong, C.L.; Dawson, B.A.; Héroux-Metcalf, C.; Neville, G.A.; Rogers, C.G. Toxicity of structurally related anthraquinones and anthrones to mammalian-cell in vitro. Toxicol. In Vitro 1994, 8, 329–335. [Google Scholar] [CrossRef]
- Verebová, V.; Adamčík, J.; Danko, P.; Podhradský, D.; Miškovský, P.; Staničová, J. Anthraquinones quinizarine and danthron unwind negatively supercoiled DNA and lengthen linear DNA. Biochem. Biophys. Res. Commun. 2014, 444, 50–55. [Google Scholar] [CrossRef] [PubMed]
- IARC. Dantron (chrysazin; 1,8-dihydroxyanthraquinone). In Pharmaceutical Drugs; International Agency for Research on Cancer: Lyon, France, 1990; pp. 265–275. [Google Scholar]
- Mueller, S.O.; Stopper, H.; Dekant, W. biotransformation of the anthraquinones emodin and chrysophanol by cytochrome P450 enzymes. Bioactivation to genotoxic metabolites. Drug Metab. Dispos. 1998, 26, 540–546. [Google Scholar]
- Müller, S.O.; Eckert, I.; Lutz, W.K.; Stopper, H. Genotoxicity of the laxative drug components emodin, aloe-emodin and danthron in mammalian cells: Topoisomerase II mediated. Mutat. Res. Genet. Toxicol. 1996, 371, 165–173. [Google Scholar] [CrossRef]
- Downes, C.S.; Mullinger, A.M.; Johnson, R.T. Inhibitors of DNA topoisomerase II prevent chromatid separation in mammalians cells but do not prevent exit from mitosis. Proc. Natl. Acad. Sci. USA 1991, 88, 8895–8899. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.R.; Caron, P.R.; Kim, R.A. The role of DNA topoisomerases in recombination and genomic stability: A double-edged sword. Cell 1990, 62, 403–406. [Google Scholar] [CrossRef]
- Neimeikaité-Čéniené, A.; Sergediené, E.; Nivinskas, H.; Čénas, N. Cytotoxicity of natural hydroxyanthraquinones: Role of oxidative stress. Z. Naturforsch. C 2002, 57, 822–827. [Google Scholar] [CrossRef]
- Lieberman, D.F.; Fink, R.C.; Schaefer, F.L.; Mulcahy, R.J.; Stark, A. Mutagenicity of anthraquinone and hydroxylated anthraquinones in the Ames/Salmonella microsome system. Appl. Environ. Microbiol. 1982, 43, 1354–1359. [Google Scholar] [CrossRef] [Green Version]
- Ferreiro, M.L.; Rodriguez-Otero, J. Ab inition study of the intramolecular proton transfer in dihydroxyanthraquinones. J. Mol. Struct. 2001, 542, 63–77. [Google Scholar] [CrossRef]
- Ansari, S.S.; Khan, R.H.; Naqvi, S. Probing the intermolecular interactions into serum albumin and anthraquinone systems: A spectroscopic and docking approach. J. Biomol. Struct. Dyn. 2018, 36, 3362–3375. [Google Scholar] [CrossRef] [PubMed]
- Jutkova, A.; Chorvat, D.; Miskovsky, P.; Jancura, D.; Datta, S. Encapsulation of anticancer drug curcumin and co-loading with photosensitizer hypericin into lipoproteins investigated by fluorescence resonance energy transfer. Int. J. Pharm. 2019, 564, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Kozsup, M.; Dömötör, O.; Nagy, S.; Farkas, E.; Enyedy, E.A.; Buglyó, P. Synthesis, characterization and albumin binding capabilities of quinizarin containing ternary cobalt(III) complexes. J. Inorg. Biochem. 2020, 204, 110963. [Google Scholar] [CrossRef]
- Crlikova, H.; Kostrhunova, H.; Pracharova, J.; Kozsup, M.; Nagy, S.; Buglyó, P.; Brabec, V.; Kasparkova, J. Antiproliferative, DNA binding, and cleavage properties of dinuclear Co(III) complexes containing the bioactive quinizarin ligand. J. Biol. Inorg. Chem. 2020, 25, 339–350. [Google Scholar] [CrossRef]
- Kočišová, E.; Chinsky, L.; Miškovský, P. Sequence specific interaction of the photoactive drug hypericin depends on the structural arrangement and the stability of the structure containing its specific 5´AG3´target: A resonance Raman spectroscopy study. J. Biomol. Struct. Dyn. 1999, 17, 51–59. [Google Scholar] [CrossRef]
- Staničová, J.; Verebová, V.; Strejčková, A. Potential anticancer agent hypericin and its model compound emodin: Interaction with DNA. Čes. Slov. Farm. 2016, 65, 28–31. [Google Scholar]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef] [Green Version]
- Aller, S.G.; Yu, J.; Ward, A.; Weng, Y.; Chittaboina, S.; Zhou, R.P.; Harrell, P.M.; Trinh, Y.T.; Zhang, Q.H.; Urbatsch, I.L.; et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 2009, 323, 1718–1722. [Google Scholar] [CrossRef] [Green Version]
- Regina, A.; Demeule, M.; Laplante, A.; Jodoin, J.; Dagenais, C.; Berthelet, F.; Moghrabi, A.; Beliveau, R. Multidrug resistance in brain tumors: Roles of the blood-brain barrier. Cancer Metastasis Rev. 2001, 20, 13–25. [Google Scholar] [CrossRef]
- Szaflarski, W.; Sujka-Kordowska, P.; Januchowski, R.; Wojtowicz, K.; Andrzejewska, M.; Nowicki, M.; Zabel, M. Nuclear localization of P-glycoprotein is responsible for protection of the nucleus from doxorubicin in the resistant LoVo cell line. Biomed. Pharmacother. 2013, 67, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Weyergang, A.; Berstad, M.E.; Bull-Hansen, B.; Olsen, C.E.; Selbo, P.K.; Berg, K. Photochemical activation of drug for the treatment of therapy-resistant cancers. Photochem. Photobiol. Sci. 2015, 14, 1465–1475. [Google Scholar] [CrossRef] [PubMed]




| Photosensitizer | Hypericin | Emodin | Quinizarin | Danthron | |
|---|---|---|---|---|---|
| Hydrophobicity | high | amphiphilic with mild hydrophobicity | low | Low | |
| AlogP | 5.040 | 2.568 | 2.324 | 2.324 | |
| Absorption maximum in DMSO λmax (nm) | 560 600 | 440 | 475 | 430 | |
| Fluorescence maximum λexc. (nm) | 603 | 520 | 540 | 575 | |
| 560 | 440 | 475 | 430 | ||
| Dissociated form (nm) | PS0 | 560 | 440 | 475 | 430 |
| PS1- | 600 [127] | 480 | 560 | 475 | |
| PS2- | 650 [127] | 525 | 595 | 500 | |
| PS4- | 630 [127] | ||||
| PS6- | 640 [127] | ||||
| Dissociated constant | pKa1 | 2 [127] | 7.2 | 11.3 | 10.5 |
| pKa2 | 7.8 [127] | 10.6 | 12.7 | 12.9 | |
| pKa3 | 11.5 [127] | ||||
| pKa4 | 13 [127] | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verebová, V.; Beneš, J.; Staničová, J. Biophysical Characterization and Anticancer Activities of Photosensitive Phytoanthraquinones Represented by Hypericin and Its Model Compounds. Molecules 2020, 25, 5666. https://doi.org/10.3390/molecules25235666
Verebová V, Beneš J, Staničová J. Biophysical Characterization and Anticancer Activities of Photosensitive Phytoanthraquinones Represented by Hypericin and Its Model Compounds. Molecules. 2020; 25(23):5666. https://doi.org/10.3390/molecules25235666
Chicago/Turabian StyleVerebová, Valéria, Jiří Beneš, and Jana Staničová. 2020. "Biophysical Characterization and Anticancer Activities of Photosensitive Phytoanthraquinones Represented by Hypericin and Its Model Compounds" Molecules 25, no. 23: 5666. https://doi.org/10.3390/molecules25235666
APA StyleVerebová, V., Beneš, J., & Staničová, J. (2020). Biophysical Characterization and Anticancer Activities of Photosensitive Phytoanthraquinones Represented by Hypericin and Its Model Compounds. Molecules, 25(23), 5666. https://doi.org/10.3390/molecules25235666