Relative Abundance of Alpha-Amylase/Trypsin Inhibitors in Selected Sorghum Cultivars
Abstract
1. Introduction
2. Results and Discussion
2.1. Protein Concentration and SDS PAGE
2.2. Establishment of the LC–MS/MS Method for the Analysis of Sorghum ATIs: Selection of Peptides
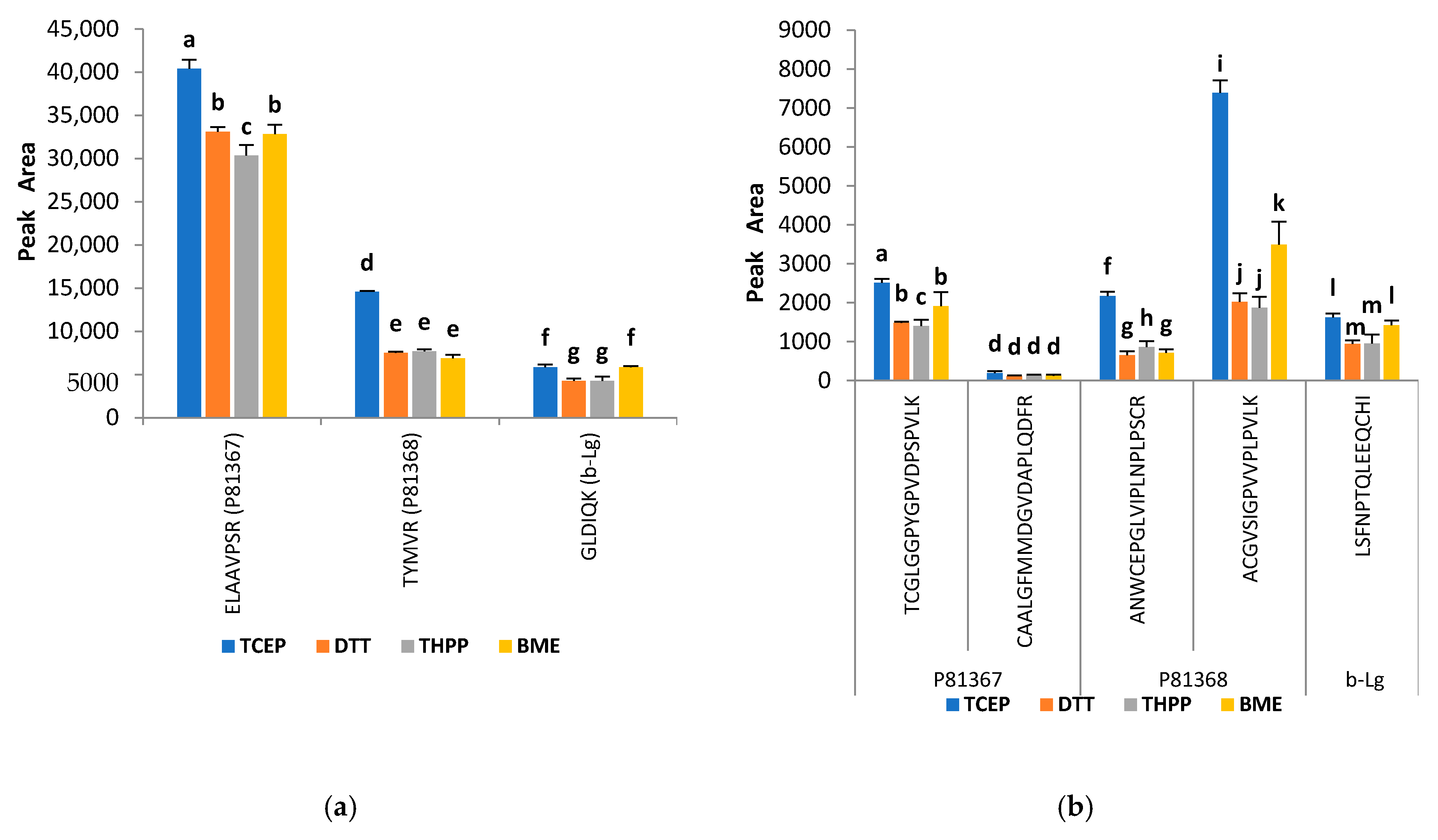
2.3. Improvement of the Trypsin Digestion: Evaluation of Different Reducing and Alkylating Agents
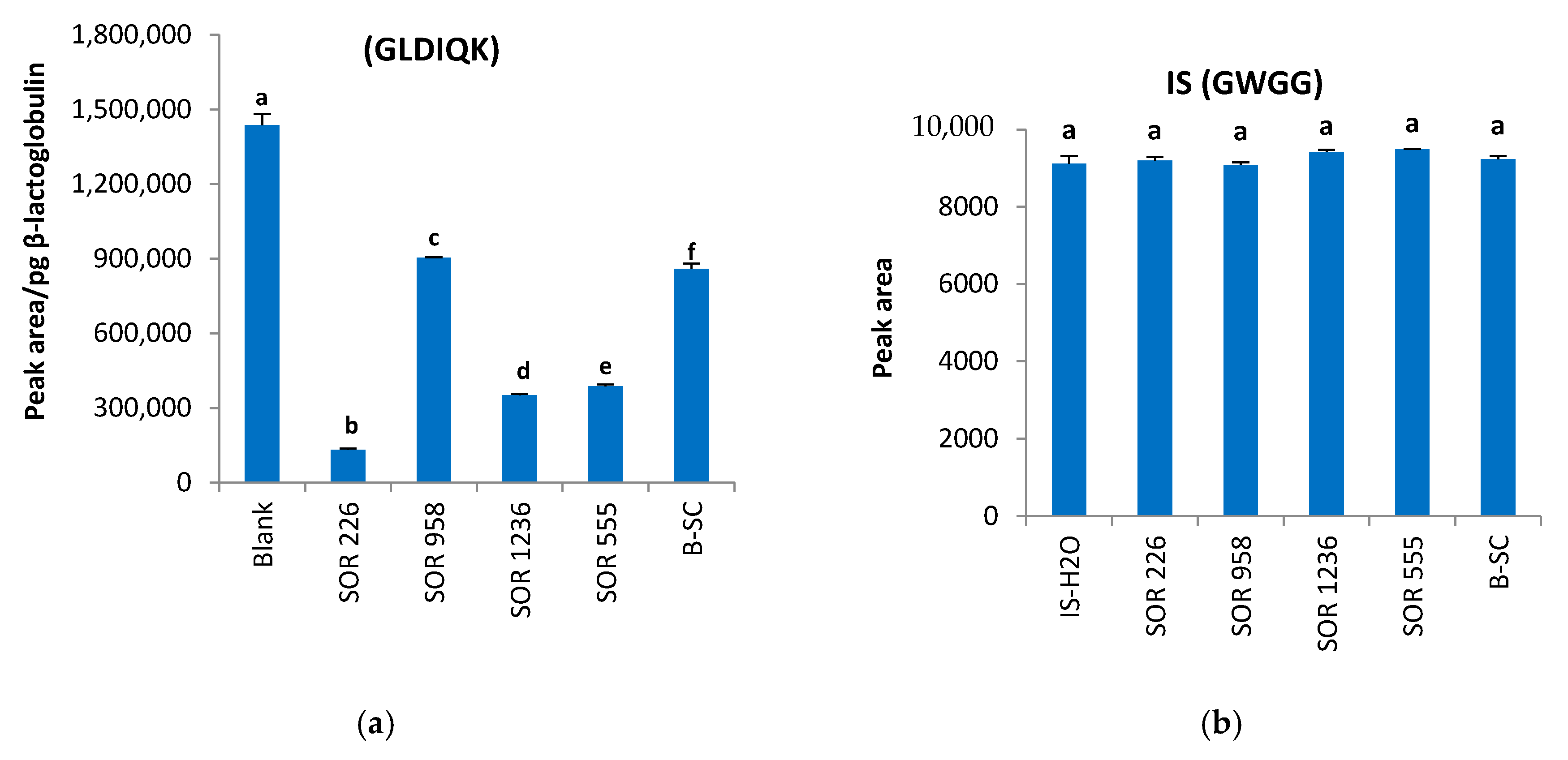
2.4. Matrix Effects and Ion Suppression
2.5. Method Validation
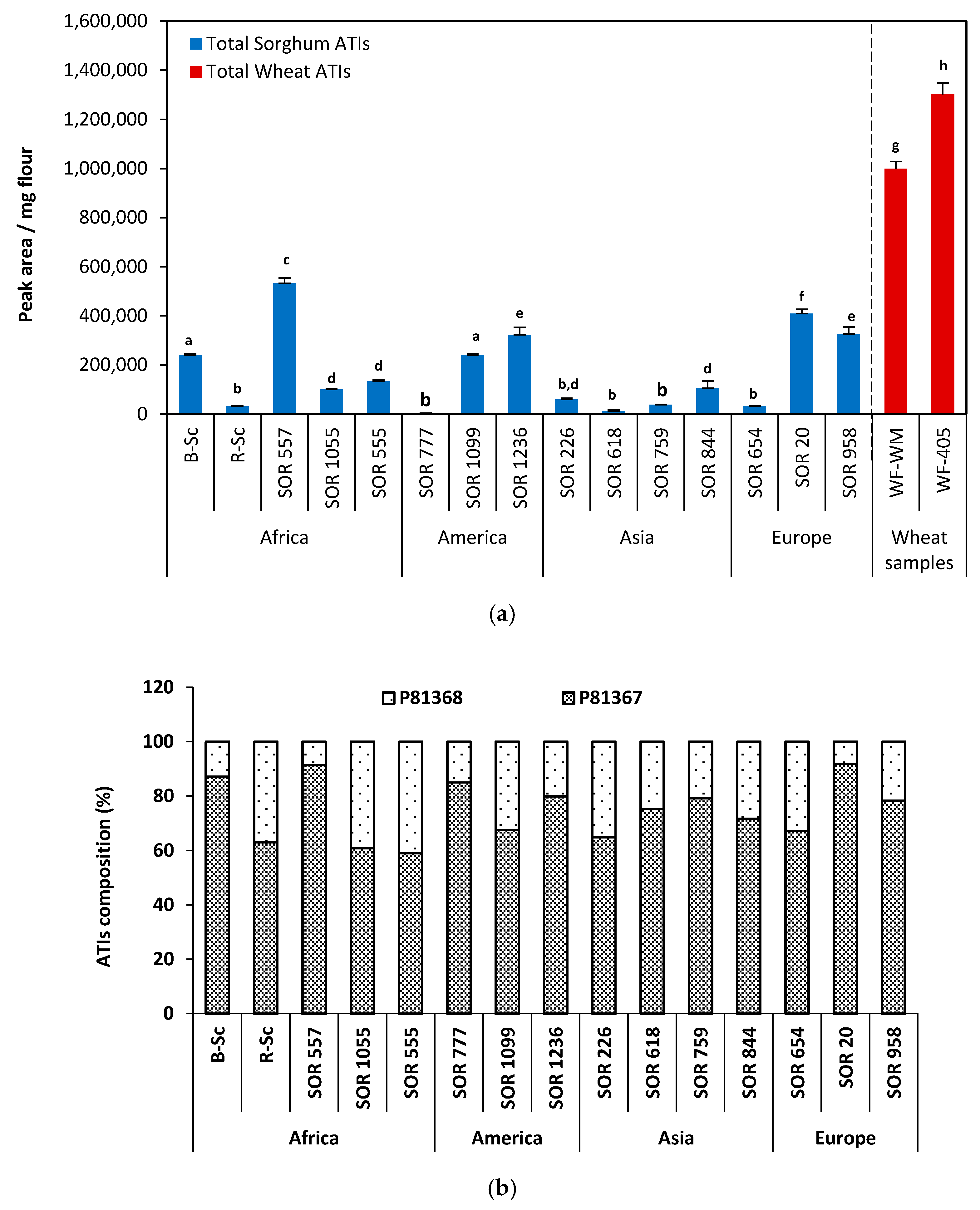
2.6. Relative Quantification of ATIs from the Fifteen Sorghum Samples
3. Materials and Methods
3.1. Materials
3.1.1. Food Material
3.1.2. Standards and Chemicals
3.2. Methods
3.2.1. Protein Extraction and In-Solution Digestion
3.2.2. Solid Phase Extraction
3.2.3. Protein Determination
3.2.4. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS PAGE)
3.2.5. LC–MS/MS Method Development and Validation
Method Development
Method Validation
3.2.6. Analysis of Wheat Samples
3.2.7. Quantitative Assessment and Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Olalde-Portugal, V.; Cabrera-Ponce, J.L.; Gastelum-Arellanez, A.; Guerrero-Rangel, A.; Winkler, R.; Valdés-Rodríguez, S. Proteomic analysis and interactions network in leaves of mycorrhizal and nonmycorrhizal sorghum plants under water deficit. PeerJ 2020, 8, e8991. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Ng, K.; Zhang, P.; Warner, R.D.; Shen, S.; Tang, H.Y.; Liang, Z.; Fang, Z. In Vitro alpha-Glucosidase and alpha-Amylase Inhibitory Activities of Free and Bound Phenolic Extracts from the Bran and Kernel Fractions of Five Sorghum Grain Genotypes. Foods 2020, 9, 1301. [Google Scholar] [CrossRef] [PubMed]
- Pontieri, P.; Mamone, G.; De Caro, S.; Tuinstra, M.R.; Roemer, E.; Okot, J.; De Vita, P.; Ficco, D.B.M.; Alifano, P.; Pignone, D.; et al. Sorghum, a Healthy and Gluten-free Food for Celiac Patients As Demonstrated by Genome, Biochemical, and Immunochemical Analyses. J. Agric. Food Chem. 2013, 61, 2565–2571. [Google Scholar] [CrossRef] [PubMed]
- Tuinstra, M.R. Food-grade sorghum varieties and production considerations: A review. J. Plant Interact. 2008, 3, 69–72. [Google Scholar] [CrossRef]
- Wu, G.; Johnson, S.K.; Bornman, J.F.; Bennett, S.J.; Clarke, M.W.; Singh, V.; Fang, Z. Growth temperature and genotype both play important roles in sorghum grain phenolic composition. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Pezzali, J.G.; Suprabha-Raj, A.; Siliveru, K.; Aldrich, C.G. Characterization of white and red sorghum flour and their potential use for production of extrudate crisps. PLoS ONE 2020, 15, e0234940. [Google Scholar] [CrossRef]
- Schober, T.J.; Bean, S.R.; Boyle, D.L. Gluten-Free Sorghum Bread Improved by Sourdough Fermentation: Biochemical, Rheological, and Microstructural Background. J. Agric. Food Chem. 2007, 55, 5137–5146. [Google Scholar] [CrossRef]
- Irondi, E.A.; Adegoke, B.M.; Effion, E.S.; Oyewo, S.O.; Alamu, E.O.; Boligon, A.A. Enzymes inhibitory property, antioxidant activity and phenolics profile of raw and roasted red sorghum grains in vitro. Food Sci. Hum. Wellness 2019, 8, 142–148. [Google Scholar] [CrossRef]
- Dicko, M.H.; Gruppen, H.; Traoré, A.S.; Voragen, A.G.J.; Van Berkel, W.J.H. Sorghum grain as human food in Africa: Relevance of content of starch and amylase activities. Afr. J. Biotechnol. 2006, 5, 384–395. [Google Scholar]
- Mtelisi, D.N.; Xu, F.; Zhao, R. The efficacy of sorghum flour addition on dough rheological properties and bread quality: A short review. Grain Oil Sci. Technol. 2020. [Google Scholar] [CrossRef]
- Oria, M.P.; Hamaker, B.R.; Shull, J.M. Resistance of Sorghum .alpha.-, .beta.-, and .gamma.-Kafirins to Pepsin Digestion. J. Agric. Food Chem. 1995, 43, 2148–2153. [Google Scholar] [CrossRef]
- Beckwith, A.C. Grain sorghum glutelin. Isolation and characterization. J. Agric. Food Chem. 1972, 20, 761–764. [Google Scholar] [CrossRef]
- McCuistion, K.C.; Selle, P.H.; Liu, S.Y.; Goodband, R.D. Sorghum as a Feed Grain for Animal Production. In Sorghum and Millets: Chemistry, Technology, and Nutritional Attributes, 2nd ed.; Elsevier Inc. in cooperation with AACC International: Amsterdam, The Netherlands, 2019; pp. 355–391. [Google Scholar] [CrossRef]
- Bean, S.R.; Zhu, L.J.; Smith, B.M.; Wilson, J.D.; Ioerger, B.P.; Tilley, M. Starch and protein chemistry and functional properties. In Sorghum and Millets: Chemistry, Technology, and Nutritional Attributes, 2nd ed.; Elsevier Inc. in cooperation with AACC International: Amsterdam, The Netherlands, 2019; pp. 131–170. [Google Scholar] [CrossRef]
- Cremer, J.E.; Bean, S.R.; Tilley, M.M.; Ioerger, B.P.; Ohm, J.B.; Kaufman, R.C.; Wilson, J.D.; Innes, D.J.; Gilding, E.K.; Godwin, I.D. Grain Sorghum Proteomics: Integrated Approach toward Characterization of Endosperm Storage Proteins in Kafirin Allelic Variants. J. Agric. Food Chem. 2014, 62, 9819–9831. [Google Scholar] [CrossRef] [PubMed]
- van der Walt, W.H.; Schussler, L.; Van der Walt, W.H. Fractionation of proteins from low-tannin sorghum grain. J. Agric. Food Chem. 1984, 32, 149–154. [Google Scholar] [CrossRef]
- Bloch, C.; Richardson, M. A new family of small (5 kDa) protein inhibitors of insect α-amylases from seeds or sorghum (Sorghum bicolor (L.) Moench) have sequence homologies with wheat γ-purothionins. FEBS Lett. 1991, 279, 101–104. [Google Scholar] [CrossRef]
- Bloch, C., Jr.; Richardson, M. The amino acid sequences of two 13-kDa alpha-amylase inhibitors from the seeds of Sorghum bicolor (L.) Moench. Protein Seq. Data Anal. 1992, 5, 27–30. [Google Scholar]
- Sagu, S.T.; Huschek, G.; Bonick, J.; Homann, T.; Rawel, H.M. A New Approach of Extraction of alpha-Amylase/trypsin Inhibitors from Wheat (Triticum aestivum L.), Based on Optimization Using Plackett-Burman and Box-Behnken Designs. Molecules 2019, 24, 3589. [Google Scholar] [CrossRef]
- Sagu, S.T.; Zimmermann, L.; Landgraber, E.; Homann, T.; Huschek, G.; Ozpinar, H.; Schweigert, F.J.; Rawel, H.M. Comprehensive Characterization and Relative Quantification of alpha-Amylase/Trypsin Inhibitors from Wheat Cultivars by Targeted HPLC-MS/MS. Foods 2020, 9, 1448. [Google Scholar] [CrossRef]
- Pahr, S.; Constantin, C.; Papadopoulos, N.G.; Giavi, S.; Mäkelä, M.; Pelkonen, A.; Ebner, C.; Mari, A.; Scheiblhofer, S.; Thalhamer, J.; et al. α-Purothionin, a new wheat allergen associated with severe allergy. J. Allergy Clin. Immunol. 2013, 132, 1000–1003. [Google Scholar] [CrossRef]
- Chandrasekher, G.; Raju, D.S.; Pattabiraman, T.N. Natural plant enzyme inhibitors. α-amylase inhibitors in millets. J. Sci. Food Agric. 1981, 32, 9–16. [Google Scholar] [CrossRef]
- Kutty, A.V.M.; Pattabiraman, T.N. Isolation and characterization of an amylase inhibitor from sorghum seeds, specific for human enzymes. J. Agric. Food Chem. 1986, 34, 552–557. [Google Scholar] [CrossRef]
- Bean, S.R.; Ioerger, B.P.; Smith, B.M.; Blackwell, D.L. Sorghum Protein Structure and Chemistry: Implications for Nutrition and Functionality. In Advances in Cereal Science: Implications To Food Processing and Health Promotion; American Chemical Society: Washington, DC, USA, 2011; Volume 1089, pp. 131–147. [Google Scholar] [CrossRef]
- Ciacci, C.; Maiuri, L.; Caporaso, N.; Bucci, C.; Del Giudice, L.; Rita Massardo, D.; Pontieri, P.; Di Fonzo, N.; Bean, S.R.; Ioerger, B.; et al. Celiac disease: In vitro and in vivo safety and palatability of wheat-free sorghum food products. Clin. Nutr. 2007, 26, 799–805. [Google Scholar] [CrossRef] [PubMed]
- de Mesa-Stonestreet, N.J.; Alavi, S.; Bean, S.R. Sorghum proteins: The concentration, isolation, modification, and food applications of kafirins. J. Food Sci. 2010, 75, R90–R104. [Google Scholar] [CrossRef] [PubMed]
- Ratnavathi, C.V. Sorghum Processing and Utilization. Sorghum Biochem. 2016, 311–327. [Google Scholar] [CrossRef]
- Davies, J.M.; Li, H.; Green, M.; Towers, M.; Upham, J.W. Subtropical grass pollen allergens are important for allergic respiratory diseases in subtropical regions. Clin Transl Allergy 2012, 2, 4. [Google Scholar] [CrossRef]
- Bokka, C.S.; Veeramachaneni, G.K.; Thunuguntla, V.B.S.C.; Manda, N.K.; Bondili, J.S. Specific panallergen peptide of Sorghum Polcalcin showing IgE response identified based on in silico and in vivo peptide mapping. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Bonick, J.; Huschek, G.; Rawel, H.M. Determination of wheat, rye and spelt authenticity in bread by targeted peptide biomarkers. J. Food Compos. Anal. 2017, 58, 82–91. [Google Scholar] [CrossRef]
- Martinez-Esteso, M.J.; Norgaard, J.; Brohee, M.; Haraszi, R.; Maquet, A.; O’Connor, G. Defining the wheat gluten peptide fingerprint via a discovery and targeted proteomics approach. J. Proteom. 2016, 147, 156–168. [Google Scholar] [CrossRef]
- Rogniaux, H.; Pavlovic, M.; Lupi, R.; Lollier, V.; Joint, M.; Mameri, H.; Denery, S.; Larré, C. Allergen relative abundance in several wheat varieties as revealed via a targeted quantitative approach using MS. Proteomics 2015, 15, 1736–1745. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [PubMed]
- Altenbach, S.B.; Vensel, W.H.; Dupont, F.M. The spectrum of low molecular weight alpha-amylase/protease inhibitor genes expressed in the US bread wheat cultivar Butte 86. BMC Res. Notes 2011, 4. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Wakabayashi, S.; Matsubara, H. Disulfide Bridges in an α-Amylase Inhibitor from Wheat Kernel. J. Biochem. 1983, 94, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Zevallos, V.F.; Raker, V.; Tenzer, S.; Jimenez-Calvente, C.; Ashfaq-Khan, M.; Rüssel, N.; Pickert, G.; Schild, H.; Steinbrink, K.; Schuppan, D. Nutritional Wheat Amylase-Trypsin Inhibitors Promote Intestinal Inflammation via Activation of Myeloid Cells. Gastroenterology 2017, 152, 1100–1113.e1112. [Google Scholar] [CrossRef] [PubMed]
- Dupont, F.M.; Vensel, W.H.; Tanaka, C.K.; Hurkman, W.J.; Altenbach, S.B. Deciphering the complexities of the wheat flour proteome using quantitative two-dimensional electrophoresis, three proteases and tandem mass spectrometry. Proteome Sci. 2011, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.K.; Leinweber, B.L.; Smith, C.V. Detection of reversible protein thiol modifications in tissues. Anal. Biochem. 2006, 358, 171–184. [Google Scholar] [CrossRef]
- Suttapitugsakul, S.; Xiao, H.; Smeekens, J.; Wu, R. Evaluation and optimization of reduction and alkylation methods to maximize peptide identification with MS-based proteomics. Mol. Biosyst. 2017, 13, 2574–2582. [Google Scholar] [CrossRef]
- Sebastiano, R.; Citterio, A.; Lapadula, M.; Righetti, P.G. A new deuterated alkylating agent for quantitative proteomics. Rapid Commun. Mass Spectrom. 2003, 17, 2380–2386. [Google Scholar] [CrossRef]
- Hill, B.G.; Reily, C.; Oh, J.-Y.; Johnson, M.S.; Landar, A. Methods for the determination and quantification of the reactive thiol proteome. Free Radic. Biol. Med. 2009, 47, 675–683. [Google Scholar] [CrossRef]
- Zhou, W.; Yang, S.; Wang, P.G. Matrix effects and application of matrix effect factor. Bioanalysis 2017, 9, 1839–1844. [Google Scholar] [CrossRef]
- Matuszewski, B.K.; Constanzer, M.L.; Chavez-Eng, C.M. Strategies for the assessment of matrix effect in quantitative bioanalytical methods based on HPLC-MS/MS. Anal. Chem. 2003, 75, 3019–3030. [Google Scholar] [CrossRef]
- Smith, R.H.; Bhaskaran, S. Sorghum [Sorghum bicolor (L.) Moench]; Springer: Berlin/Heidelberg, Germany, 1986; Volume 2, pp. 220–233. [Google Scholar] [CrossRef]
- Berenguer, M.J.; Faci, J.M. Sorghum (Sorghum Bicolor L. Moench) yield compensation processes under different plant densities and variable water supply. Eur. J. Agron. 2001, 15, 43–55. [Google Scholar] [CrossRef]
- Dahlberg, J.; Berenji, J.; Sikora, V.; Latković, D. Assessing sorghum (Sorghum bicolor (L.) Moench) germplasm for new traits: Food, fuels & unique uses. Maydica 2011, 56, 8. [Google Scholar]
- de Wet, J.M.J. Special Paper: Systematics and Evolution of Sorghum Sect. Sorghum (Gramineae). Am. J. Bot. 1978, 65, 477–484. [Google Scholar] [CrossRef]
- Sander, I.; Rozynek, P.; Rihs, H.P.; van Kampen, V.; Chew, F.T.; Lee, W.S.; Kotschy-Lang, N.; Merget, R.; Brüning, T.; Raulf-Heimsoth, M. Multiple wheat flour allergens and cross-reactive carbohydrate determinants bind IgE in baker’s asthma. Allergy 2011, 66, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Monge, R.; Gomez, L.; Barber, D.; Lopez-Otin, C.; Armentia, A.; Salcedo, G. Wheat and barley allergens associated with baker’s asthma. Glycosylated subunits of the α-amylase-inhibitor family have enhanced IgE-binding capacity. Biochem. J. 1992, 281, 401–405. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Shrivastava, A.; Gupta, V. Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron. Young Sci. 2011, 2, 21. [Google Scholar] [CrossRef]


| No. | Accession Number | Accession Name | Scientific Name | Race | Country of Origin | Region | Total Proteins * (g/100 g) | Extracted Proteins ** (mg/mL) |
|---|---|---|---|---|---|---|---|---|
| 1 | B.Sc | S35 | Sorghum bicolor (L.) Moench | Madjeru | Cameroon | Africa | 7.9 | 2.137 ± 0.139 a |
| 2 | R.Sc | Damougari | Sorghum bicolor (L.) Moench | Safari | Cameroon | Africa | 8.5 | 2.170 ± 0.132 a,c |
| 3 | SOR 557 | n.a. | Sorghum bicolor (L.) Moench | Durra | Egypt | Africa | 13.5 | 2.684 ± 0.145 b |
| 4 | SOR 1055 | HAMRA | Sorghum bicolor (L.) Moench | Kafir-Durra | Yemen | Africa | 15.4 | 2.553 ± 0.013 b |
| 5 | SOR 555 | H × 60 | Sorghum bicolor (L.) Moench | Kafir-Bicolor | Tansania | Africa | 13.6 | 2.506 ± 0.066 a,b |
| 6 | SOR 777 | n.a. | Sorghum halepense (L.) Pers. | n.a. | Columbia | America | 14.4 | 4.046 ± 0.106 c |
| 7 | SOR 1099 | BATAN73 620 | Sorghum bicolor (L.) Moench | Guinea | Mexico | America | 17.0 | 2.049 ± 0.026 a |
| 8 | SOR 1236 | WRAY | Sorghum bicolor (L.) Moench | Caudatum | USA | America | 12.2 | 2.506 ± 0.198 a,b |
| 9 | SOR 226 | Jan-Da-Li | Sorghum bicolor (L.) Moench | Durra | China | Asia | 12.8 | 3.719 ± 0.238 c |
| 10 | SOR 618 | n.a. | Sorghum bicolor (L.) Moench | Bicolor | n.a. | Asia | 12.2 | 4.140 ± 0.066 c |
| 11 | SOR 759 | n.a. | Sorghum bicolor (L.) Moench | Bicolor | Korea, PRK | Asia | 11.3 | 2.343 ± 0.059 a,b |
| 12 | SOR 844 | n.a. | Sorghum bicolor (L.) Moench | Guinea | Iraq | Asia | 12.1 | 0.588 ± 0.047 d |
| 13 | SOR 654 | Blackhull | Sorghum bicolor (L.) Moench | Durra-Bicolor | United Kingdom | Europe | 12.1 | 3.383 ± 0.238 c |
| 14 | SOR 20 | n.a. | Sorghum bicolor (L.) Moench | Durra | Denmark | Europe | 14.7 | 2.474 ± 0.125 a,b |
| 15 | SOR 958 | Tech 8 | Sorghum bicolor (L.) Moench | Bicolor | Germany | Europe | 14.0 | 3.048 ± 0.026 b,c |
| Validation Criterion | IS-H2O (GWGG) | β-Lg (GLDIQK) | P81367 (ELAAVPSR) | P81368 (TYMVR) |
|---|---|---|---|---|
| IS recovery (%) | 100.0 ± 0.5 | 104.1 ± 0.6 | 99.4 ± 0.9 | 99.4 ± 0.9 |
| Repeatability (% RSD) | 1.98 | 0.61 | 1.50 | 0.41 |
| Reproducibility (% RSD) | 1.42 | 1.03 | 2.32 | 1.17 |
| Linearity () | 0.9973 | 0.9992 | 0.9995 | 0.9968 |
| LoD (ng protein) | 0.075 * | 0.001 | 0.048 | 0.346 |
| LoQ (ng protein) | 0.250 * | 0.004 | 0.161 | 1.155 |
| Entry | Protein Name | Short Name | MW (Da) | pI | Length (AA) |
|---|---|---|---|---|---|
| P81367 | Alpha-amylase inhibitor 4 | IAA4_SORBI | 12,449.45 | 8.96 | 118 |
| P81368 | Alpha-amylase inhibitor 5 | IAA5_SORBI | 12,776.11 | 8.55 | 116 |
Sample Availability: Not available. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sagu, S.T.; Landgräber, E.; Rackiewicz, M.; Huschek, G.; Rawel, H. Relative Abundance of Alpha-Amylase/Trypsin Inhibitors in Selected Sorghum Cultivars. Molecules 2020, 25, 5982. https://doi.org/10.3390/molecules25245982
Sagu ST, Landgräber E, Rackiewicz M, Huschek G, Rawel H. Relative Abundance of Alpha-Amylase/Trypsin Inhibitors in Selected Sorghum Cultivars. Molecules. 2020; 25(24):5982. https://doi.org/10.3390/molecules25245982
Chicago/Turabian StyleSagu, Sorel Tchewonpi, Eva Landgräber, Michal Rackiewicz, Gerd Huschek, and Harshadrai Rawel. 2020. "Relative Abundance of Alpha-Amylase/Trypsin Inhibitors in Selected Sorghum Cultivars" Molecules 25, no. 24: 5982. https://doi.org/10.3390/molecules25245982
APA StyleSagu, S. T., Landgräber, E., Rackiewicz, M., Huschek, G., & Rawel, H. (2020). Relative Abundance of Alpha-Amylase/Trypsin Inhibitors in Selected Sorghum Cultivars. Molecules, 25(24), 5982. https://doi.org/10.3390/molecules25245982








