Microbial Transformation of Prenylquercetins by Mucor hiemalis
Abstract
1. Introduction
2. Results and Discussions
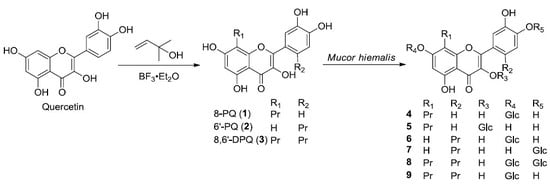
2.1. Preparation and Microbial Biotransformation of Prenylquercetins
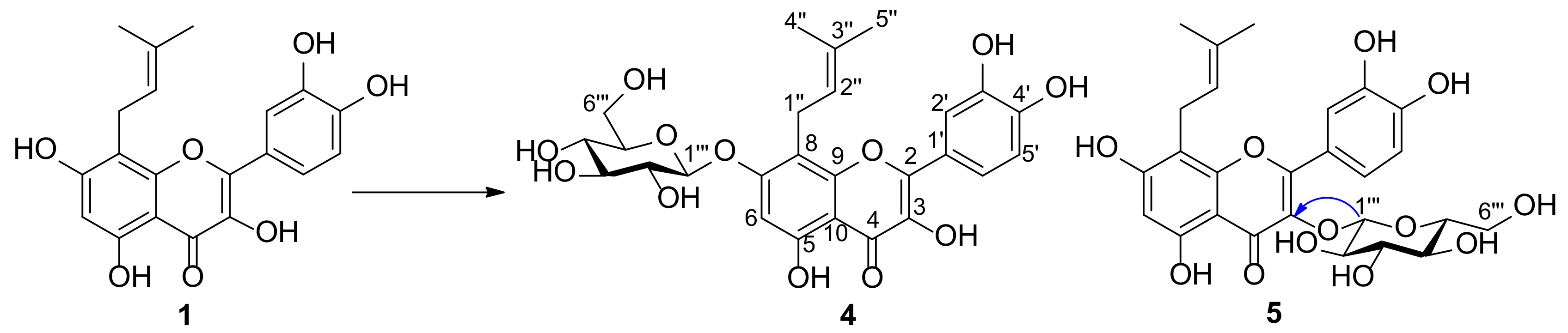
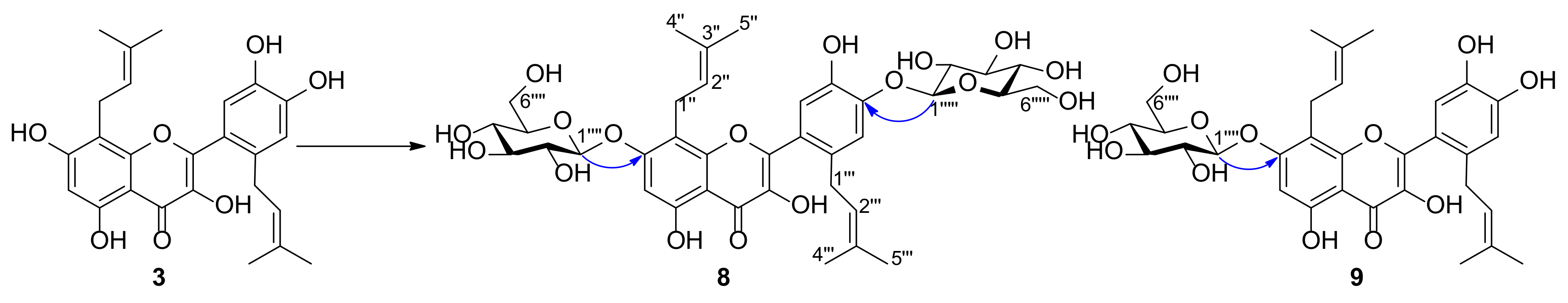
2.2. Structure Elucidation of Metabolites of Prenylquercetins
2.3. Water Solubility of Prenylquercetins and Their Metabolites
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Chemicals and Ingredients
3.3. Microorganisms and Culture Media
3.4. Procedure for Microbial Transformation
3.5. Scale-up Fermentation and Isolation of Metabolites
3.6. Spectroscopic Data of Metabolites
3.7. Acid Hydrolysis
3.8. Water Solubility Test
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Erlund, I. Review of the flavonoids quercetin, hesperetin, and naringenin. Dietary sources, bioactivities, bioavailability, and epidemiology. Nutr. Res. 2004, 24, 851–874. [Google Scholar] [CrossRef]
- Lo Piero, A.R. The state of the art in biosynthesis of anthocyanins and its regulation in pigmented sweet oranges [(Citrus sinensis) L. Osbeck]. J. Agric. Food Chem. 2015, 63, 4031–4041. [Google Scholar] [CrossRef]
- Ross, J.A.; Kasum, C.M. Dietary flavonoids: Bioavailability, metabolic effects, and safety. Annu. Rev. Nutr. 2002, 22, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Havsteen, B.H. The biochemistry and medical significance of the flavonoids. Pharmacol. Therapeut. 2002, 96, 67–202. [Google Scholar] [CrossRef]
- Williams, C.A.; Harborne, J.B. The leaf flavonoids of the Zingiberales. Biochem. Syst. Ecol. 1977, 5, 221–229. [Google Scholar] [CrossRef]
- Mpalantinos, M.A.; Soares De Moura, R.; Parente, J.P.; Kuster, R.M. Biologically active flavonoids and kava pyrones from the aqueous extract of Alpinia Zerumbet. Phytother. Res. 1998, 12, 442–444. [Google Scholar] [CrossRef]
- David, A.V.A.; Arulmoli, R.; Parasuraman, S. Overviews of biological importance of quercetin: A bioactive flavonoid. Pharmacogn. Rev. 2016, 10, 84–89. [Google Scholar]
- Jelinek, L.; Karabin, M.; Kincl, T.; Hudcova, T.; Kotlikova, B.; Dostalek, P. Xanthohumol: Possible Isolation and Beer Enrichment. Chem. Listy 2013, 107, 209–213. [Google Scholar]
- Yang, X.; Jiang, Y.; Yang, J.; He, J.; Sun, J.; Chen, F.; Zhang, M.; Yang, B. Prenylated flavonoids, promising nutraceuticals with impressive biological activities. Trends Food Sci. Technol. 2015, 44, 93–104. [Google Scholar] [CrossRef]
- Mukai, R.; Fujikura, Y.; Murota, K.; Uehara, M.; Minekawa, S.; Matsui, N.; Kawamura, T.; Nemoto, H.; Terao, J. Prenylation enhances quercetin uptake and reduces efflux in Caco-2 cells and enhances tissue accumulation in mice fed long-term. J. Nutr. 2013, 143, 1558–1564. [Google Scholar] [CrossRef]
- Zheng, Z.-P.; Cheng, K.-W.; Chao, J.; Wu, J.; Wang, M. Tyrosinase inhibitors from paper mulberry (Broussonetia papyrifera). Food Chem. 2008, 106, 529–535. [Google Scholar] [CrossRef]
- Chen, R.M.; Hu, L.H.; An, T.Y.; Li, J.; Shen, Q. Natural PTP1B inhibitors from Broussonetia papyrifera. Bioorg. Med. Chem. Lett. 2002, 12, 3387–3390. [Google Scholar] [CrossRef]
- Fan, Y.-H.; Ye, R.; Xu, H.-Y.; Feng, X.-H.; Ma, C.-M. Structures and in vitro antihepatic fibrosis activities of prenylated dihydrostilbenes and flavonoids from Glycyrrhiza uralensis Leaves. J. Food Sci. 2019, 84, 1124–1230. [Google Scholar] [CrossRef] [PubMed]
- Tronina, T.; Strugała, P.; Popłoński, J.; Włoch, A.; Sordon, S.; Bartmańska, A.; Huszcza, E. The influence of glycosylation of natural and synthetic prenylated flavonoids on binding to human serum albumin and inhibition of cyclooxygenases COX-1 and COX-2. Molecules 2017, 22, 1230. [Google Scholar] [CrossRef]
- Kren, V.; Martínkovâ, I. Glycosides in medicine: “the role of glycosidic residue in biological activity”. Curr. Med. Chem. 2001, 8, 1303–1328. [Google Scholar] [CrossRef]
- Chebil, L.; Humeau, C.; Anthoni, J.; Dehez, F.; Engasser, J.-M.; Ghoul, M. Solubility of flavonoids in organic solvents. J. Chem. Eng. Data 2007, 52, 1552–1556. [Google Scholar] [CrossRef]
- Murota, K.; Matsuda, N.; Kashino, Y.; Fujikura, Y.; Nakamura, T.; Kato, Y.; Shimiu, R.; Okuyama, S.; Tanaka, H.; Koda, T.; et al. α-Oligoglucosylation of a sugar moiety enhances the bioavailability of quercetin glucosides in humans. Arch. Biochem. Biophys. 2010, 501, 91–97. [Google Scholar] [CrossRef]
- Plaza, M.; Pozzo, T.; Liu, J.; Ara, K.Z.G.; Turner, C.; Karlsson, E.N. Substituent effects on in vitro antioxidizing properties, stability, and solubility in flavonoids. J. Agric. Food Chem. 2014, 62, 3321–3333. [Google Scholar] [CrossRef]
- Brglez Mojzer, E.; Knez Hrnčič, M.; Škerget, M.; Knez, Ž.; Bren, U. Polyphenols: Extraction methods, antioxidative action, bioavailability and anticarcinogenic effects. Molecules 2016, 21, 901. [Google Scholar] [CrossRef]
- Cao, H.; Xu, H.; Zhu, G.; Liu, S. Isoquercetin ameliorated hypoxia/reoxygenation-induced H9C2 cardiomyocyte apoptosis via a mitochondrial-dependent pathway. Biomed. Pharmacother. 2017, 95, 938–943. [Google Scholar] [CrossRef]
- Tronina, T.; Bartmańska, A.; Milczarek, M.; Wietrzyk, J.; Popłoński, J.; Rój, E.; Huszcza, E. Antioxidant and antiproliferative activity of glycosides obtained by biotransformation of xanthohumol. Bioorg. Med. Chem. Lett. 2013, 23, 1957–1960. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Liang, W.-F.; Li, Z.-W.; Feng, J.; Wang, Q.; Qiao, X.; Ye, M. Efficient and selective glucosylation of prenylated phenolic compounds by Mucor hiemalis. RSC Adv. 2016, 6, 20791–20799. [Google Scholar] [CrossRef]
- Zhu, X.; Schmidt, R.R. New principles for glycoside-bond formation. Angew. Chem. Int. Ed. Engl. 2009, 48, 1900–1934. [Google Scholar] [CrossRef] [PubMed]
- Christensen, H.M.; Oscarson, S.; Jensen, H.H. Common side reaction of the glycosyl donor in chemical glycosylation. Carbohydr. Res. 2015, 408, 51–95. [Google Scholar] [CrossRef]
- Semeniuchenko, V.; Garazd, Y.; Garazd, M.; Shokol, T.; Groth, U.; Khilya, V. Highly efficient glucosylation of flavonoids. Monatsh. Chem. 2009, 140, 1503–1512. [Google Scholar] [CrossRef]
- Sordon, S.; Popłoński, J.; Huszcza, E. Microbial glycosylation of flavonoids. Pol. J. Microbiol. 2016, 65, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Sordon, S.; Popłoński, J.; Tronina, T.; Huszcza, E. Regioselective O-glycosylation of flavonoids by fungi Beauveria bassiana, Absidia coerulea and Absidia glauca. Bioorg. Chem. 2019, 93, 102750. [Google Scholar] [CrossRef] [PubMed]
- Klingel, T.; Hadamjetz, M.; Fischer, A.; Wefers, D. Glycosylation of flavonoids and flavonoid glycosides by mutant dextransucrase from Lactobacillus reuteri TMW 1.106. Carbohydr. Res. 2019, 483, 107741. [Google Scholar] [CrossRef]
- Xiao, Y.; Lee, I.-S. Microbial metabolism of prenylated apigenin derivatives by Mucor hiemalis. Phytochem. Lett. 2016, 16, 197–202. [Google Scholar] [CrossRef]
- Xiao, Y.; Lee, I.-S. Microbial transformation of quercetin and its prenylated derivatives. Nat. Prod. Res. 2018, 32, 902–908. [Google Scholar] [CrossRef]
- Kawamura, T.; Hayashi, M.; Mukai, R.; Terao, J.; Nemoto, H. An efficient method for C8-prenylation of flavonols and flavanones. Synthesis 2012, 44, 1308–1314. [Google Scholar]
- Kajjout, M.; Rolando, C. Regiospecific synthesis of quercetin O-β-d-glucosylated and O-β-d-glucuronidated isomers. Tetrahedron 2011, 67, 4731–4741. [Google Scholar] [CrossRef]
- Clark, A.M.; Hufford, C.D. Use of microorganisms for the study of drug metabolism: An update. Med. Res. Rev. 1991, 11, 473–501. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Lee, I.-S. A new flavonol glycoside from the aerial parts of Epimedium koreanum Nakai. Nat. Prod. Res. 2017, 31, 320–325. [Google Scholar] [CrossRef]
- Kaminaga, Y.; Nagatsu, A.; Akiyama, T.; Sugimoto, N.; Yamazaki, T.; Maitani, T.; Mizukami, H. Production of unnatural glucosides of curcumin with drastically enhanced water solubility by cell suspension cultures of Catharanthus roseus. FEBS Lett. 2003, 555, 311–316. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Huh, J.-Y.; Nam, S.-H.; Kim, D.; Lee, S.-B. Synthesis of quercetin-3-O-glucoside from rutin by Penicillium decumbens naringinase. J. Food Sci. 2013, 78, C411–C415. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, F.; Xiao, Y.; Lee, I.-S. Microbial Transformation of Prenylquercetins by Mucor hiemalis. Molecules 2020, 25, 528. https://doi.org/10.3390/molecules25030528
Han F, Xiao Y, Lee I-S. Microbial Transformation of Prenylquercetins by Mucor hiemalis. Molecules. 2020; 25(3):528. https://doi.org/10.3390/molecules25030528
Chicago/Turabian StyleHan, Fubo, Yina Xiao, and Ik-Soo Lee. 2020. "Microbial Transformation of Prenylquercetins by Mucor hiemalis" Molecules 25, no. 3: 528. https://doi.org/10.3390/molecules25030528
APA StyleHan, F., Xiao, Y., & Lee, I.-S. (2020). Microbial Transformation of Prenylquercetins by Mucor hiemalis. Molecules, 25(3), 528. https://doi.org/10.3390/molecules25030528