Overcoming Resistance to Platinum-Based Drugs in Ovarian Cancer by Salinomycin and Its Derivatives—An In Vitro Study
Abstract
1. Introduction
2. Results
2.1. Derivation of Cisplatin-Resistant Cell Lines
2.2. In Vitro Activity of Cytotoxic Drugs, Salinomycin, and Its Derivatives Against OvCa Cells
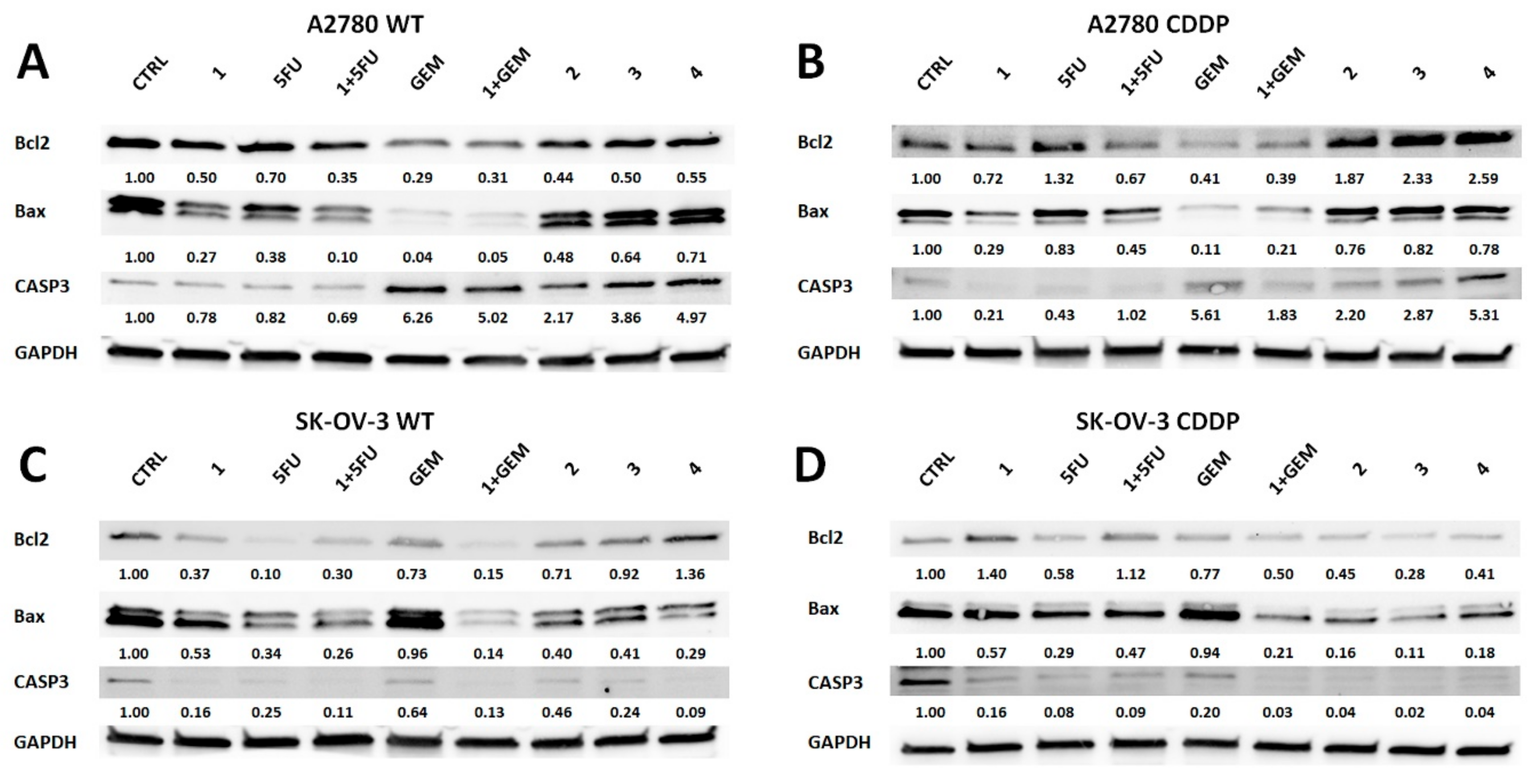
2.3. Salinomycin and Its Derivatives Overcome Cisplatin Resistance and are More Selective Against Cisplatin-Resistant OvCa Cells
3. Discussion
4. Materials and Methods
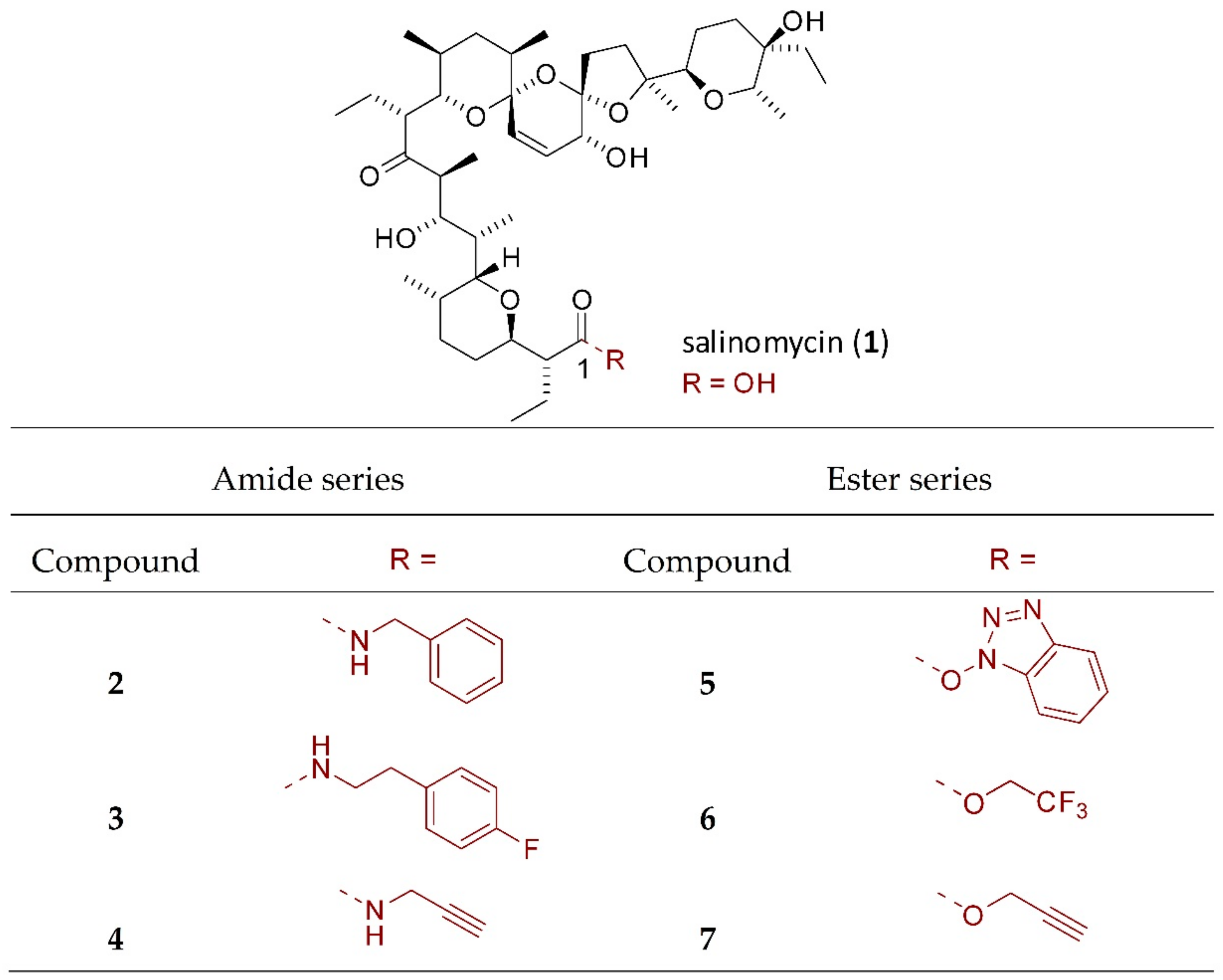
4.1. Chemical Part
4.1.1. Isolation of Salinomycin
4.1.2. Synthesis of Salinomycin Derivatives
4.2. Biological Part
4.2.1. Cell Culture and Derivation of Cisplatin-Resistant Cell Lines
4.2.2. Isolation of RNA and RT-qPCR
4.2.3. Cell Viability Assay
4.2.4. Western Blot Analysis
4.2.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 5FU | 5-fluorouracil |
| ABCB1 | ATP-binding cassette subfamily B member 1 |
| ABCB4 | ATP-binding cassette subfamily B member 4 |
| ABCC2 | ATP-binding cassette subfamily C member 2 |
| ABCG2 | ATP-binding cassette subfamily G member 2 |
| ALL | Acute lymphoblastic leukemia |
| Bax | Bcl2 associated X protein |
| Bcl-2 | B-cell lymphoma 2 |
| CASP3 | Caspase 3 |
| CDDP | Cisplatin |
| CI | Combination index |
| CNT | Concentrative nucleoside transporters |
| CSCs | Cancer stem cells |
| DMEM | Dulbecco modified eagle medium |
| EMT | Epithelial-mesenchymal transition |
| ENT | Equilibrative nucleoside transporters |
| FBS | Fetal bovine serum |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| GEM | Gemcitabine |
| HRP | Horseradish peroxidase |
| MDR | Multidrug resistance |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| NEAA | Non-essential amino acids |
| OvCa | Ovarian cancer |
| P-gp | P-glycoprotein |
| PVDF | Polyvinylidene difluoride |
| RI | Resistance indexes |
| RT-qPCR | Reverse transcriptase quantitative polymerase chain reaction |
| SAR | Structure-activity relationship |
| SI | Selectivity indexes |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef]
- Rosenthal, A.N.; Menon, U.; Jacobs, I.J. Screening for ovarian cancer. Clin. Obstet. Gynecol. 2006, 49, 433–447. [Google Scholar] [CrossRef]
- Marcus, C.S.; Maxwell, G.L.; Darcy, K.M.; Hamilton, C.A.; McGuire, W.P. Current Approaches and Challenges in Managing and Monitoring Treatment Response in Ovarian Cancer. J. Cancer. 2014, 5, 25–30. [Google Scholar]
- Cooke, S.L.; Brenton, J.D. Evolution of platinum resistance in high-grade serous ovarian cancer. Lancet Oncol. 2011, 12, 1169–1174. [Google Scholar] [CrossRef]
- Rocconi, R.P.; Case, A.S.; Straughn, J.M.; Estes, J.M.; Partridge, E.E. Role of chemotherapy for patients with recurrent platinum-resistant advanced epithelial ovarian cancer: A cost-effectiveness analysis. Cancer 2006, 107, 536–543. [Google Scholar] [CrossRef]
- Au, K.K.; Josahkian, J.A.; Francis, J.A.; Squire, J.A.; Koti, M. Current state of biomarkers in ovarian cancer prognosis. Futur. Oncol. 2015, 11, 3187–3195. [Google Scholar] [CrossRef] [PubMed]
- Kurman, R.J.; Shih, I.M. The Dualistic Model of Ovarian Carcinogenesis. Am. J. Pathol. 2016, 186, 733–747. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Q.; Wang, C.; Ngai, S. Ovarian Cancer Stem Cells: A New Target for Cancer. Therapy 2013, 2013, 916819. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, A.B.; Joseph, P.; Kovalenko, O.; Singh, S.; Armstrong, A.; Redline, R.; Resnick, K.; Zanotti, K.; Waggoner, S.; DiFeo, A. Critical role of Wnt/β-catenin signaling in driving epithelial ovarian cancer platinum resistance. Oncotarget 2015, 6, 23720–23734. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe, S.M.; Morgan, S.L.; Wyant, G.A.; Tran, L.T.; Muto, K.W.; Chen, Y.S.; Chin, K.T.; Partridge, J.C.; Poole, B.B.; Cheng, K.H.; et al. Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proc. Natl. Acad. Sci. 2012, 109, E2939–E2948. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, Y.; Shibuya, M.; Sugawara, H.; Kawaguchi, O.; Hirsoe, C. Salinomycin, a new polyether antibiotic. J. Antibiot. (Tokyo). 1974, 27, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.B.; Onder, T.T.; Jiang, G.; Tao, K.; Kuperwasser, C.; Weinberg, R.A.; Lander, E.S. Identification of Selective Inhibitors of Cancer Stem Cells by High-Throughput Screening. Cell 2009, 138, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Naujokat, C.; Steinhart, R. Salinomycin as a Drug for Targeting Human Cancer Stem Cells. J. Biomed. Biotechnol. 2012, 2012, 950658. [Google Scholar] [CrossRef] [PubMed]
- Antoszczak, M. A medicinal chemistry perspective on salinomycin as a potent anticancer and anti-CSCs agent. Eur. J. Med. Chem. 2019, 164, 366–377. [Google Scholar] [CrossRef]
- Antoszczak, M.; Huczyński, A. Salinomycin and its derivatives–A new class of multiple-targeted “magic bullets”. Eur. J. Med. Chem. 2019, 176, 208–227. [Google Scholar] [CrossRef]
- Kaushik, V.; Yakisich, J.; Kumar, A.; Azad, N.; Iyer, A. Ionophores: Potential Use as Anticancer Drugs and Chemosensitizers. Cancers 2018, 10, 360. [Google Scholar] [CrossRef]
- Versini, A.; Saier, L.; Sindikubwabo, F.; Müller, S.; Cañeque, T.; Rodriguez, R. Chemical biology of salinomycin. Tetrahedron 2018, 74, 5585–5614. [Google Scholar] [CrossRef]
- Antoszczak, M.; Maj, E.; Stefańska, J.; Wietrzyk, J.; Janczak, J.; Brzezinski, B.; Huczyński, A. Synthesis, antiproliferative and antibacterial activity of new amides of salinomycin. Bioorg. Med. Chem. Lett. 2014, 24, 1724–1729. [Google Scholar] [CrossRef]
- Antoszczak, M.; Popiel, K.; Stefańska, J.; Wietrzyk, J.; Maj, E.; Janczak, J.; Michalska, G.; Brzezinski, B.; Huczyński, A. Synthesis, cytotoxicity and antibacterial activity of new esters of polyether antibiotic – salinomycin. Eur. J. Med. Chem. 2014, 76, 435–444. [Google Scholar] [CrossRef]
- Urbaniak, A.; Delgado, M.; Antoszczak, M.; Huczyński, A.; Chambers, T.C. Salinomycin derivatives exhibit activity against primary acute lymphoblastic leukemia (ALL) cells in vitro. Biomed. Pharmacother. 2018, 99, 384–390. [Google Scholar] [CrossRef]
- Huczyński, A.; Rutkowski, J.; Popiel, K.; Maj, E.; Wietrzyk, J.; Stefańska, J.; Majcher, U.; Bartl, F. Synthesis, antiproliferative and antibacterial evaluation of C-ring modified colchicine analogues. Eur. J. Med. Chem. 2015, 90, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Huczyński, A.; Janczak, J.; Antoszczak, M.; Wietrzyk, J.; Maj, E.; Brzezinski, B. Antiproliferative activity of salinomycin and its derivatives. Bioorg. Med. Chem. Lett. 2012, 22, 7146–7150. [Google Scholar] [CrossRef] [PubMed]
- Antonenko, Y.N.; Rokitskaya, T.I.; Huczyński, A. Electrogenic and nonelectrogenic ion fluxes across lipid and mitochondrial membranes mediated by monensin and monensin ethyl ester. Biochim. Biophys. Acta - Biomembr. 2015, 1848, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Harker, W.G.; Slade, D.L.; Dalton, W.S.; Meltzer, P.S.; Trent, J.M. Multidrug resistance in mitoxantrone-selected HL-60 leukemia cells in the absence of P-glycoprotein overexpression. Cancer Res. 1989, 49, 4542–4549. [Google Scholar] [PubMed]
- Badisa, R.B.; Darling-Reed, S.F.; Joseph, P.; Cooperwood, J.S.; Latinwo, L.M.; Goodman, C.B. Selective cytotoxic activities of two novel synthetic drugs on human breast carcinoma MCF-7 cells. Anticancer Res. 2009, 29, 2993–2996. [Google Scholar]
- Chou, T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef]
- Ichite, N.; Chougule, M.B.; Jackson, T.; Fulzele, S.V.; Safe, S.; Singh, M. Enhancement of Docetaxel Anticancer Activity by a Novel Diindolylmethane Compound in Human Non-Small Cell Lung Cancer. Clin. Cancer Res. 2009, 15, 543–552. [Google Scholar] [CrossRef]
- Dewangan, J.; Srivastava, S.; Rath, S.K. Salinomycin: A new paradigm in cancer therapy. Tumor Biol. 2017, 39, 101042831769503. [Google Scholar] [CrossRef]
- Piperno, A.; Marrazzo, A.; Scala, A.; Rescifina, A. Chemistry and biology of salinomycin and its analogues. salinomycin and its analogues. In Targets In Heterocyclic Systems; Attanasi, O.A., Merino, P., Spinelli, D., Eds.; Società Chimica Italiana: Rome, Italy, 2015; Volume 19, pp. 177–213. [Google Scholar]
- Kocieński, P.J.; Brown, R.C.D.; Pommier, A.; Procter, M.; Schmidt, B. Synthesis of salinomycin. J. Chem. Soc. Perkin Trans. 1998, 1, 9–40. [Google Scholar] [CrossRef]
- Genovese, I.; Ilari, A.; Assaraf, Y.G.; Fazi, F.; Colotti, G. Not only P-glycoprotein: Amplification of the ABCB1- containing chromosome region 7q21 confers multidrug resistance upon cancer cells by coordinated overexpression of an assortment of resistance-related proteins. Drug Resist. Updat. 2017, 32, 23–46. [Google Scholar] [CrossRef]
- Comsa, E.; Nguyen, K.; Loghin, F.; Boumendjel, A.; Peuchmaur, M.; Andrieu, T.; Falson, P. Ovarian cancer cells cisplatin sensitization agents selected by mass cytometry target ABCC2 inhibition. Future Med. Chem. 2018, 10, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Rubiś, B.; Hołysz, H.; Barczak, W.; Gryczka, R.; Łaciński, M.; Jagielski, P.; Czernikiewicz, A.; Półrolniczak, A.; Wojewoda, A.; Perz, K.; et al. Study of ABCB1 polymorphism frequency in breast cancer patients from Poland. Pharmacol. Reports 2012, 64, 1560–1566. [Google Scholar] [CrossRef]
- Duan, Z.; Brakora, K.A.; Seiden, M.V. Inhibition of ABCB1 (MDR1 ) and ABCB4 ( MDR3 ) expression by small interfering RNA and reversal of paclitaxel resistance in human ovarian cancer cells. Mol. Cancer Ther. 2004, 3, 833–838. [Google Scholar] [PubMed]
- Luqmani, Y.A. Mechanisms of drug resistance in cancer chemotherapy. Med. Princ. Pract. 2005, 14 (Suppl. 1), 35–48. [Google Scholar] [CrossRef]
- Januchowski, R.; Wojtowicz, K.; Sujka-kordowska, P.; Andrzejewska, M.; Zabel, M. MDR Gene Expression Analysis of Six Drug-Resistant Ovarian Cancer Cell Lines. Biomed. Res. Int. 2013, 2013, 241763. [Google Scholar] [CrossRef] [PubMed]
- Eckford, P.D.; Sharom, F.J. ABC Efflux Pump-Based Resistance to Chemotherapy Drugs. Chem. Rev. 2009, 2989–3011. [Google Scholar] [CrossRef] [PubMed]
- Boesch, M.; Zeimet, A.G.; Rumpold, H.; Gastl, G.; Sopper, S.; Wolf, D. Drug Transporter-Mediated Protection of Cancer Stem Cells From Ionophore Antibiotics. Stem Cells Transl. Med. 2015, 4, 1028–1032. [Google Scholar] [CrossRef]
- Riccioni, R.; Dupuis, M.L.; Bernabei, M.; Petrucci, E.; Pasquini, L.; Mariani, G.; Cianfriglia, M.; Testa, U. The cancer stem cell selective inhibitor salinomycin is a p-glycoprotein inhibitor. Blood Cells Mol. Dis. 2010, 45, 86–92. [Google Scholar] [CrossRef]
- Antoszczak, M.; Urbaniak, A.; Delgado, M.; Maj, E.; Borgström, B.; Wietrzyk, J.; Huczyński, A.; Yuan, Y.; Chambers, T.C.; Strand, D. Biological activity of doubly modified salinomycin analogs – Evaluation in vitro and ex vivo. Eur. J. Med. Chem. 2018, 156, 510–523. [Google Scholar] [CrossRef]
- Li, R.; Dong, T.; Hu, C.; Lu, J.; Dai, J.; Liu, P. Salinomycin repressed the epithelial-mesenchymal transition of epithelial ovarian cancer cells via downregulating Wnt/β-catenin pathway. Onco. Targets. Ther. 2017, 10, 1317–1325. [Google Scholar] [CrossRef]
- Chung, H.; Kim, Y.H.; Kwon, M.; Shin, S.J.; Kwon, S.H.; Cha, S.D.; Cho, C.H. The effect of salinomycin on ovarian cancer stem-like cells. Obstet. Gynecol. Sci. 2016, 59, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Shin, S.; Chung, H.; Kwon, S.; Cha, S.; Lee, J.; Cho, C.; Lee, J. Salinomycin reduces stemness and induces apoptosis on human ovarian cancer stem cell. J. Gynecol. Oncol. 2017, 28, e14. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, B.; Lee, H.G.; Kwon, S.; Cha, S.; Shin, S.; Lee, G.; Bae, I.; Cho, C. Salinomycin inhibits Akt/NF-κB and induces apoptosis in cisplatin resistant ovarian cancer cells. Cancer Epidemiol. 2013, 37, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Préfontaine, M.; Donovan, J.T.; Powell, J.L.; Buley, L. Treatment of Refractory Ovarian Cancer with 5-Fluorouracil and Leucovorin. Gynecol. Oncol. 1996, 61, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Burnett, A.F.; Barter, J.F.; Potkul, R.K.; Jarvis, T.; Barnes, W.A. Ineffectiveness of continuous 5-fluorouracil as salvage therapy for ovarian cancer. Am. J. Clin. Oncol. 1994, 17, 490–493. [Google Scholar] [CrossRef]
- Braly, P.S.; Berek, J.S.; Blessing, J.A.; Homesley, H.D.; Averette, H. Intraperitoneal Administration of Cisplatin and 5-Fluorouracil in Residual Ovarian Cancer: A Phase II Gynecologic Oncology Group Trial. Gynecol. Oncol. 1995, 56, 164–168. [Google Scholar] [CrossRef]
- Wang, F.; Dai, W.; Wang, Y.; Shen, M.; Chen, K.; Cheng, P.; Zhang, Y.; Wang, C.; Li, J.; Zheng, Y.; et al. The synergistic in vitro and in vivo antitumor effect of combination therapy with salinomycin and 5-fluorouracil against hepatocellular carcinoma. PLoS ONE 2014, 9, e97414. [Google Scholar] [CrossRef]
- Klose, J.; Eissele, J.; Volz, C.; Schmitt, S.; Ritter, A.; Ying, S.; Schmidt, T.; Heger, U.; Schneider, M.; Ulrich, A. Salinomycin inhibits metastatic colorectal cancer growth and interferes with Wnt/β-catenin signaling in CD133+ human colorectal cancer cells. BMC Cancer 2016, 16, 896. [Google Scholar] [CrossRef]
- Berg, T.; Nøttrup, T.J.; Roed, H. Gemcitabine for recurrent ovarian cancer—A systematic review and meta-analysis. Gynecol Oncol. 2019, 155, 530–537. [Google Scholar] [CrossRef]
- Zhang, G.N.; Liang, Y.; Zhou, L.J.; Chen, S.P.; Chen, G.; Zhang, T.P.; Kang, T.; Zhao, Y.P. Combination of salinomycin and gemcitabine eliminates pancreatic cancer cells. Cancer Lett. 2011, 313, 137–144. [Google Scholar] [CrossRef]
- Hagmann, W.; Jesnowski, R.; Löhr, J.M. Interdependence of gemcitabine treatment, ransporter expression, and resistance in human pancreatic carcinoma cells. Neoplasia. 2010, 12, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Anglada, M.; Pérez-Torras, S. Emerging Roles of Nucleoside Transporters. Front Pharmacol. 2018, 9, 606. [Google Scholar] [CrossRef] [PubMed]
- Mackey, J.R.; Yao, S.Y.; Smith, K.M.; Karpinski, E.; Baldwin, S.A.; Cass, C.E.; Young, J.D. Gemcitabine transport in xenopus oocytes expressing recombinant plasma membrane mammalian nucleoside transporters. J Natl Cancer Inst. 1999, 91, 1876–1881. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.W.; Marrache, S.; Cummins, S.; Bhutia, Y.D.; Mody, H.; Hooks, S.B.; Dhar, S.; Govindarajan, R. Defective hCNT1 transport contributes to gemcitabine chemoresistance in ovarian cancer subtypes: Overcoming transport defects using a nanoparticle approach. Cancer Lett. 2015, 359, 233–240. [Google Scholar] [CrossRef]
- Parajuli, B.; Shin, S.J.; Kwon, S.H.; Cha, S.D.; Chung, R.; Park, W.J.; Lee, H.G.; Cho, C.H. Salinomycin induces apoptosis via death receptor-5 up-regulation in cisplatin-resistant ovarian cancer cells. Anticancer Res. 2013, 33, 1457–1462. [Google Scholar]
- Kaplan, F.; Teksen, F. Apoptotic effects of salinomycin on human ovarian cancer cell line (OVCAR-3). Tumour Biol. 2016, 37, 3897–3903. [Google Scholar] [CrossRef]
- Lach, M.S.; Kulcenty, K.; Jankowska, K.; Trzeciak, T.; Richter, M.; Suchorska, W.M. Effect of cellular mass on chondrogenic differentiation during embryoid body formation. Mol. Med. Rep. 2018, 18, 2705–2714. [Google Scholar] [CrossRef]
- Blaszczak, W.; Lach, M.; Barczak, W.; Suchorska, W. Fucoidan Exerts Anticancer Effects Against Head and Neck Squamous Cell Carcinoma In Vitro. Molecules 2018, 23, 3302. [Google Scholar] [CrossRef]
- Chou, T.C. Theoretical Basis, Experimental Design, and Computerized Simulation of Synergism and Antagonism in Drug Combination Studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef]
Sample Availability: Samples of the all compounds are available from the authors. |


| Compound | A2780 | A2780 CDDP | SK-OV-3 | SK-OV-3 CDDP | MRC-5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IC50 (µM) | CI 95% | IC50 (µM) | CI 95% | IC50 (µM) | CI 95% | IC50 (µM) | CI 95% | IC50 (µM) | CI 95% | ||
| SAL | 1 | 0.11 | 0.08‒0.13 | 0.29 | 0.27‒0.29 | 4.01 | 3.13‒5.11 | 3.72 | 3.24‒4.25 | 10.15 | 5.14–20.01 |
| salinomycin amides | 2 | 27.05 | 24.25‒30.18 | 24.90 | 19.98‒31.04 | 183.45 | 156.43‒215.24 | 140.60 | 119.40‒165.71 | 213.33 | 186.19–244.52 |
| 3 | 8.49 | 6.74‒10.68 | 6.48 | 4.71‒8.89 | 79.08 | 71.25‒87.76 | 69.40 | 61.88‒77.84 | 66.19 | 57.31–83.54 | |
| 4 | 14.38 | 11.99‒17.23 | 12.54 | 9.58‒16.40 | 91.17 | 81.42‒102.07 | 58.53 | 53.22‒64.37 | 98.96 | 88.13–111.13 | |
| salinomycin esters | 5 | 25.50 | 20.44‒31.79 | 7.74 | 6.04‒9.94 | 205.88 | 188.71‒224.65 | 103.41 | 82.63‒129.38 | 58.81 | 47.05–73.53 |
| 6 | 13.41 | 10.58‒17.01 | 50.16 | 39.26‒64.08 | 125.33 | 117.36‒133.85 | 91.58 | 83.78‒100.11 | 110.46 | 98.87–123.41 | |
| 7 | 37.31 | 27.95‒49.56 | 45.74 | 35.49‒57.05 | 179.72 | 146.77‒219.90 | 94.56 | 85.65‒104.41 | 160.96 | 96.41–268.69 | |
| 1:1 molar mixtures | 1 + 5FU | 0.16 | 0.14‒0.19 | 0.14 | 0.14‒0.15 | 3.65 | 2.96‒4.49 | 3.45 | 2.91‒4.11 | 1.93 | 0.83–4.49 |
| 1 + GEM | 0.018 | 0.01‒0.03 | 0.024 | 0.01‒0.04 | 1.17 | 1.07‒1.29 | 1.07 | 1.07‒1.29 | 1.93 | 1.09–3.43 | |
| reference anticancer drugs | 5FU | 3.62 | 2.15‒6.00 | 1.62 | 1.23‒2.08 | 20.23 | 8.85‒46.23 | 36.38 | 16.85‒78.46 | 13.38 | 7.23–24.75 |
| GEM | 0.007 | 0.006‒0.007 | 0.02 | 0.01‒0.02 | 0.002 | 0.00007‒0.01 | 0.003 | 0.00003‒0.02 | 0.04 | 0.01–0.012 | |
| CDDP | 0.47 | 0.40‒0.50 | 8.33 | 7.40‒9.37 | 4.03 | 3.48‒4.61 | 6.72 | 5.96‒7.56 | 15.7 | 12.9 –19.27 | |
| Combination of Compounds | A2780 | A2780 CDDP | SK-OV-3 | SK-OV-3 CDDP |
|---|---|---|---|---|
| 1+5FU | 1.57 | 0.56 | 1.09 | 1.02 |
| 1+GEM | 2.86 | 1.60 | 691 | 360 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michalak, M.; Lach, M.S.; Antoszczak, M.; Huczyński, A.; Suchorska, W.M. Overcoming Resistance to Platinum-Based Drugs in Ovarian Cancer by Salinomycin and Its Derivatives—An In Vitro Study. Molecules 2020, 25, 537. https://doi.org/10.3390/molecules25030537
Michalak M, Lach MS, Antoszczak M, Huczyński A, Suchorska WM. Overcoming Resistance to Platinum-Based Drugs in Ovarian Cancer by Salinomycin and Its Derivatives—An In Vitro Study. Molecules. 2020; 25(3):537. https://doi.org/10.3390/molecules25030537
Chicago/Turabian StyleMichalak, Marcin, Michał Stefan Lach, Michał Antoszczak, Adam Huczyński, and Wiktoria Maria Suchorska. 2020. "Overcoming Resistance to Platinum-Based Drugs in Ovarian Cancer by Salinomycin and Its Derivatives—An In Vitro Study" Molecules 25, no. 3: 537. https://doi.org/10.3390/molecules25030537
APA StyleMichalak, M., Lach, M. S., Antoszczak, M., Huczyński, A., & Suchorska, W. M. (2020). Overcoming Resistance to Platinum-Based Drugs in Ovarian Cancer by Salinomycin and Its Derivatives—An In Vitro Study. Molecules, 25(3), 537. https://doi.org/10.3390/molecules25030537