Assessment of Biodegradation Efficiency of Polychlorinated Biphenyls (PCBs) and Petroleum Hydrocarbons (TPH) in Soil Using Three Individual Bacterial Strains and Their Mixed Culture
Abstract
:1. Introduction
2. Results
2.1. Bacterial Strains Used in this Study
2.2. Assessment of PCB and TPH Biodegradation Effectiveness in Sterile Soils
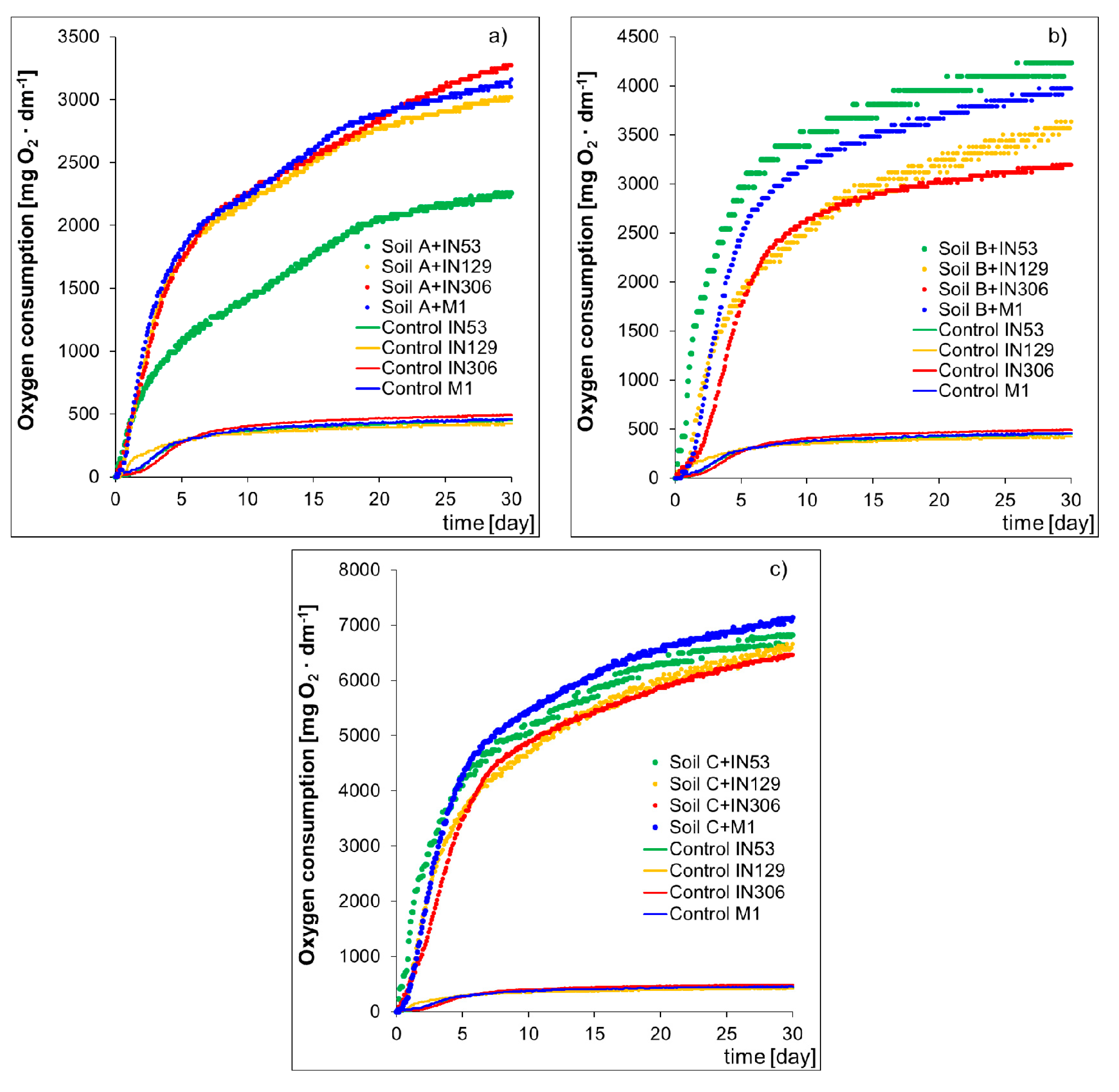
2.2.1. Respirometric Tests
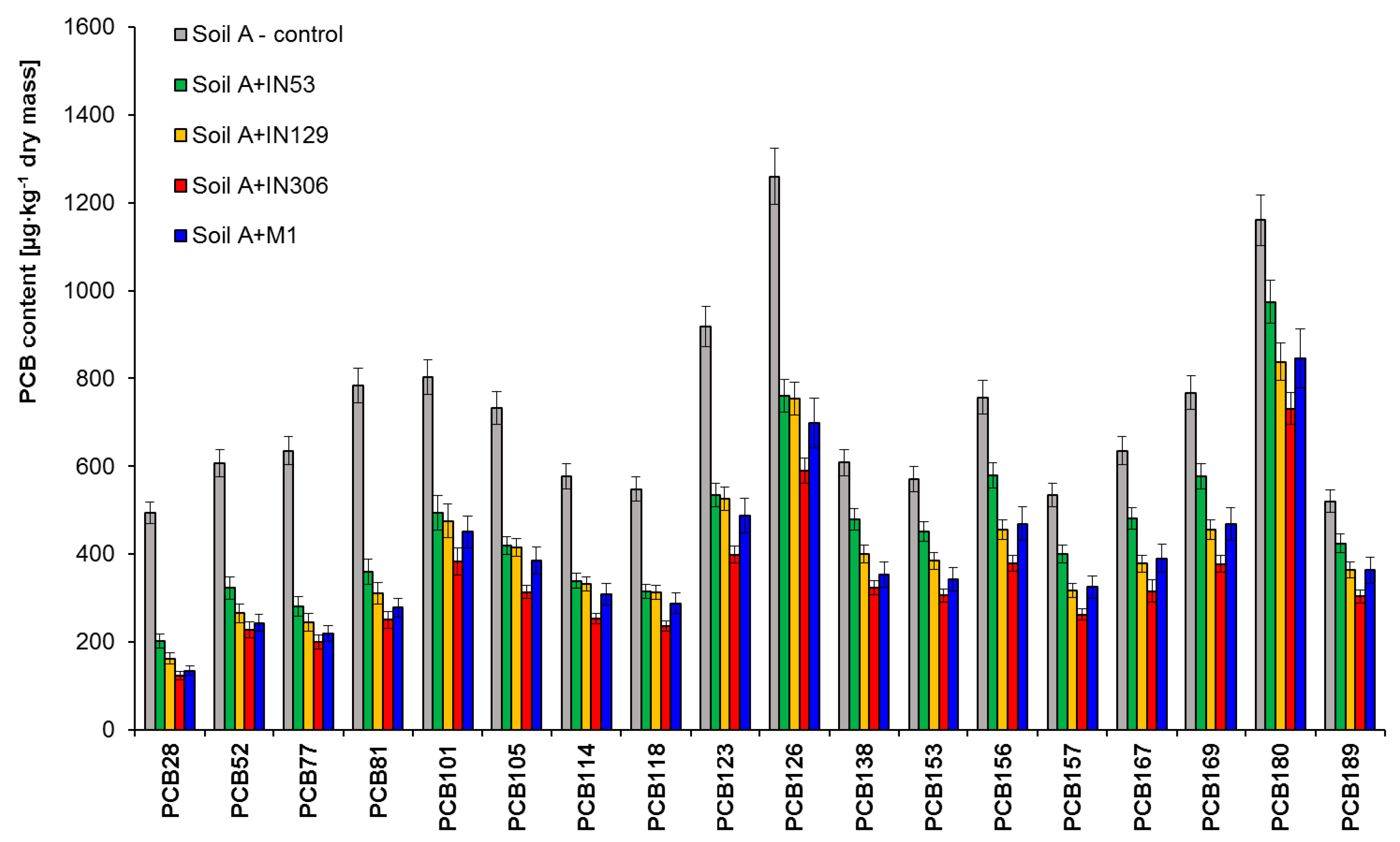
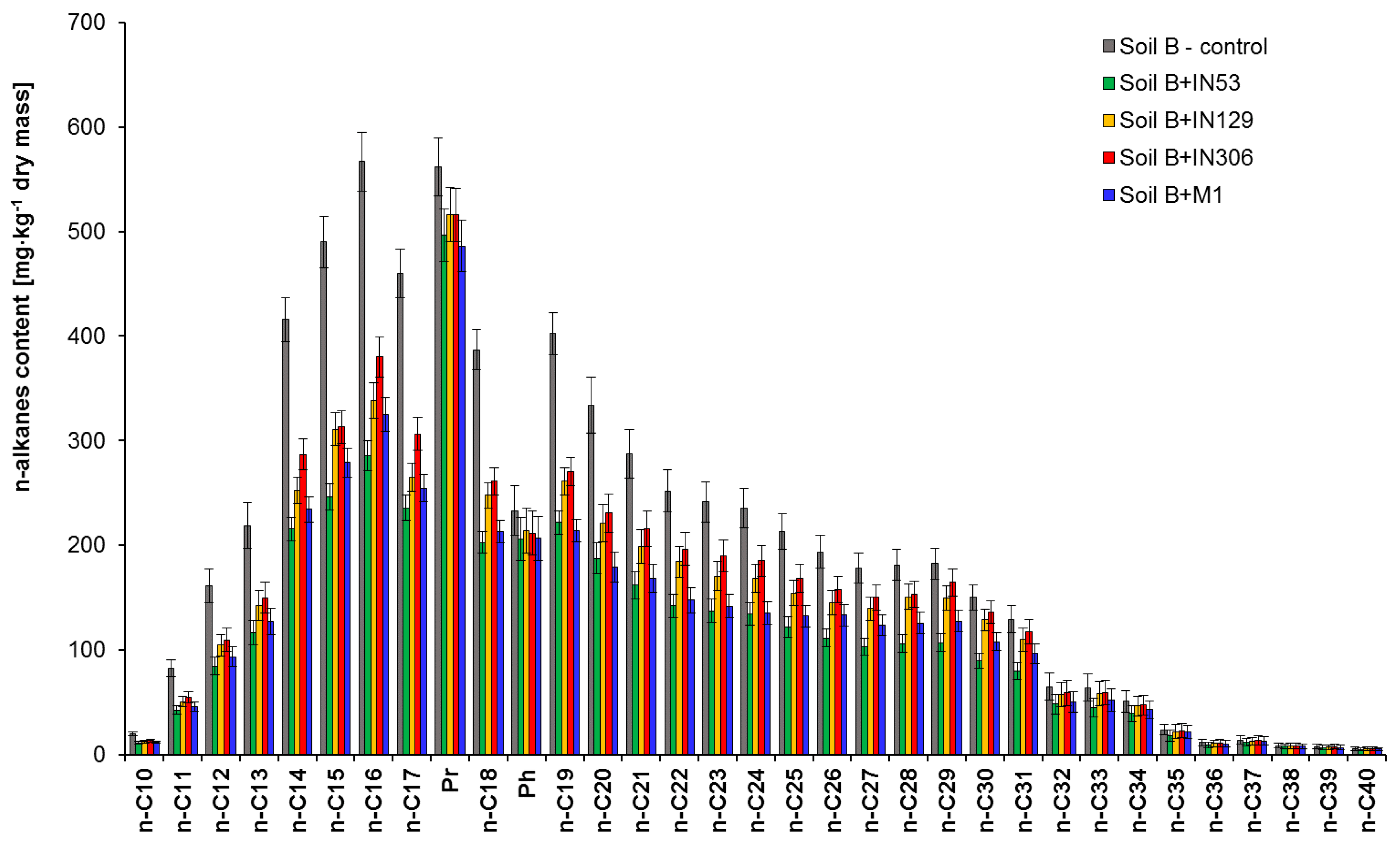
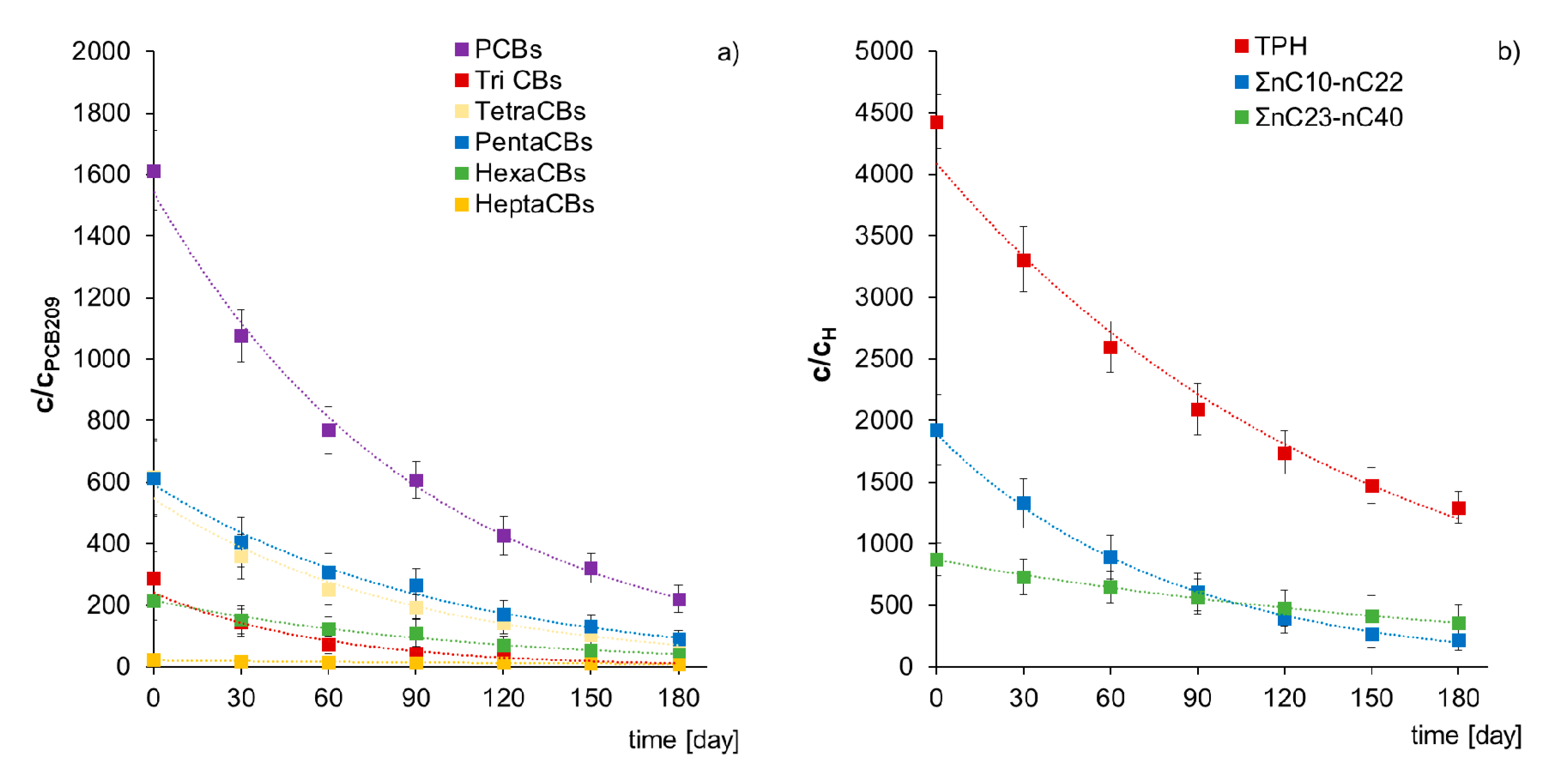
2.2.2. Assessment of PCBs and TPH Biodegradation Based on Chromatographic Analyses
2.3. Assessment of PCBs and TPH Biodegradation in Non-Sterile Soil-the Ex-Situ Prism Method
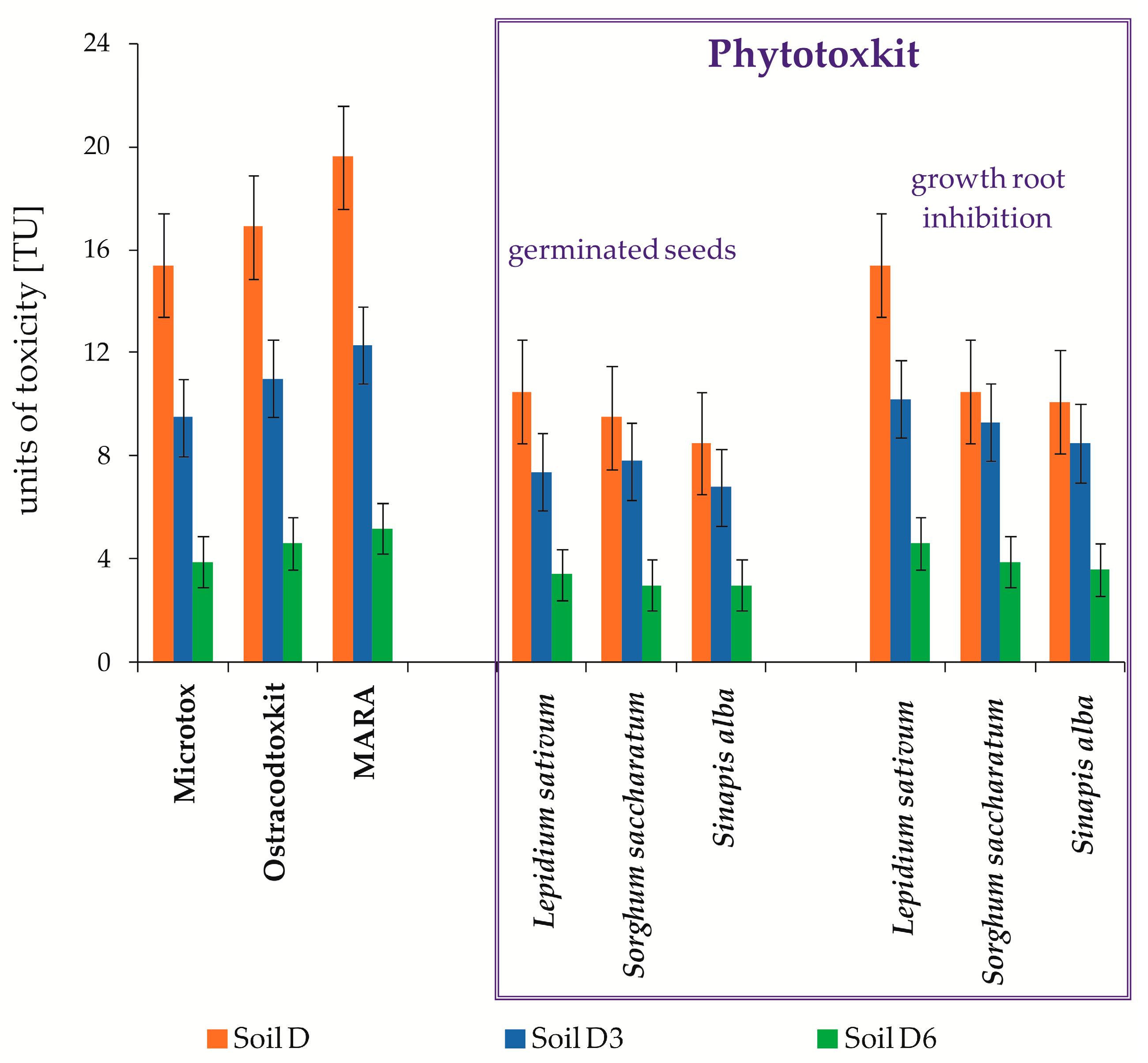
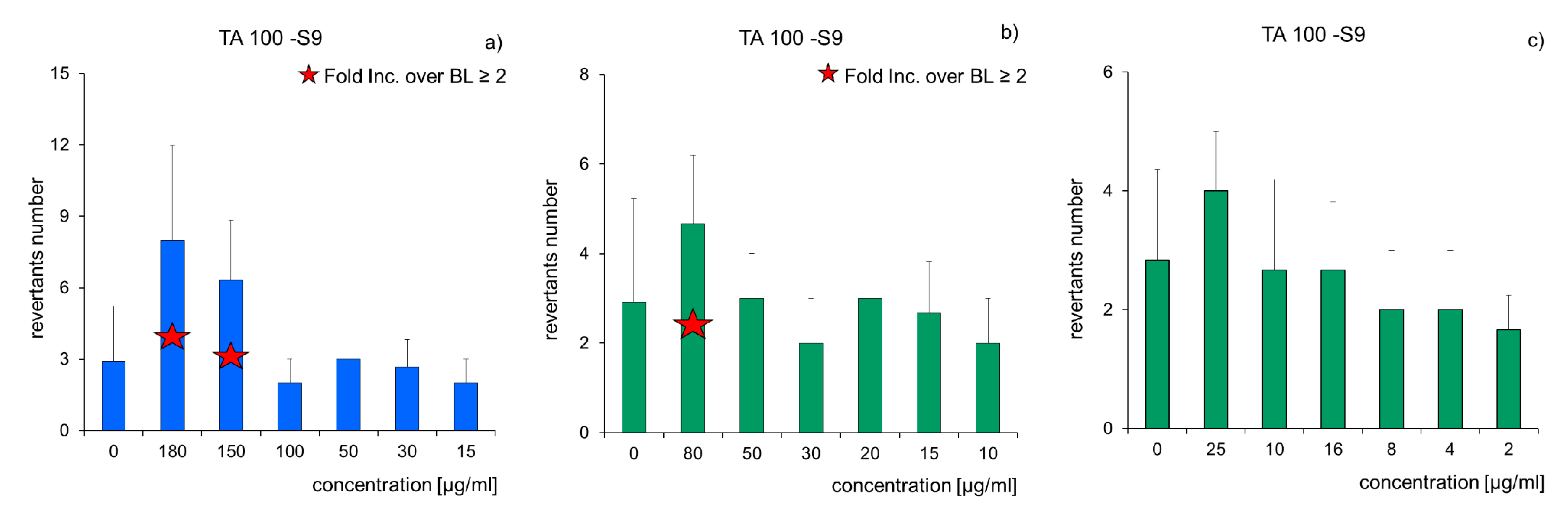
Ecotoxicological Assessment
3. Discussion
4. Materials and Methods
4.1. Soil and Microorganisms
4.2. Experimental Setup
4.2.1. Respirometric Tests
4.2.2. PCBs and TPH Biodegradation in Non-sterile Soil-the Ex-Situ Prism Method
4.3. Chromatographic Analysis
4.3.1. PCB Extraction and Quantification
4.3.2. TPH Extraction and Quantification
4.4. Mathematical Model of PCB and TPH Biodegradation
4.5. Ecotoxicological Analyses
4.6. Data Analysis and Statistical Information
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Field, J.A.; Sierra-Alvarez, R. Microbial transformation and degradation of polychlorinated biphenyls. Environ. Pollut. 2008, 155, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Vasilyeva, G.K.; Strijakova, E.R. Bioremediation of soils and sediments contaminated by polychlorinated biphenyls. Microbiology 2007, 76, 639–653. [Google Scholar] [CrossRef]
- Pieper, D.H.; Seeger, M. Bacterial metabolism of polychlorinated biphenyls. J. Mol. Microbiol. Biotechnol. 2008, 15, 121–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furukawa, K.; Fujihara, H. Microbial degradation of polychlorinated biphenyls: Biochemical and molecular features. J. Biosci. Bioeng. 2008, 105, 433–449. [Google Scholar] [CrossRef]
- Correa, P.A.; Lin, L.; Just, C.L.; Hu, D.; Hornbuckle, K.C.; Schnoor, J.L.; Van Aken, B. The effects of individual PCB congeners on the soil bacterial community structure and the abundance of biphenyl dioxygenase genes. Environ. Int. 2010, 36, 901–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudášová, H.; Lukáčová, L.; Murínová, S.; Puškárová, A.; Pangallo, D.; Dercová, K. Bacterial strains isolated from PCB-contaminated sediments and their use for bioaugmentation strategy in microcosms. J. Basic Microbiol. 2014, 54, 253–260. [Google Scholar] [CrossRef]
- Shaikh, N.S.; Parkin, S.; Luthe, G.; Lehmler, H.-J. The three-dimensional structure of 3,3′,4,4′-tetrachlorobiphenyl, a dioxin-like polychlorinated biphenyl (PCB). Chemosphere 2008, 70, 1694–1698. [Google Scholar] [CrossRef] [Green Version]
- Erickson, M.D.; Kaley, R.G. Applications of polychlorinated biphenyls. Environ. Sci Pollut. Res. 2011, 18, 135–151. [Google Scholar] [CrossRef]
- Borja, J.; Taleon, D.M.; Auresenia, J.; Gallardo, S. Polychlorinated biphenyls and their biodegradation. Process Biochem. 2005, 40, 1999–2013. [Google Scholar] [CrossRef]
- Sharma, J.K.; Gautam, R.K.; Nanekar, S.V.; Weber, R.; Singh, B.K.; Singh, S.K.; Juwarkar, A.A. Advances and perspective in bioremediation of polychlorinated biphenyl-contaminated soils. Environ. Sci Pollut. Res. 2018, 25, 16355–16375. [Google Scholar] [CrossRef]
- Blanco-Moreno, R.; Sáez, L.P.; Luque-Almagro, V.M.; Roldán, M.D.; Moreno-Vivián, C. Isolation of bacterial strains able to degrade biphenyl, diphenyl ether and the heat transfer fluid used in thermo-solar plants. New Biotechnol. 2017, 35, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Martinez, A.; Hornbuckle, K.C.; Mattes, T.E. Potential for polychlorinated biphenyl biodegradation in sediments from Indiana Harbor and Ship Canal. Int. Biodeterior. Biodegrad. 2014, 89, 50–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukerjee-Dhar, G. Bph genes of the thermophilic PCB degrader, Bacillus sp. JF8: Characterization of the divergent ring-hydroxylating dioxygenase and hydrolase genes upstream of the Mn-dependent BphC. Microbiology 2005, 151, 4139–4151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suyamud, B.; Inthorn, D.; Panyapinyopol, B.; Thiravetyan, P. Biodegradation of bisphenol A by a newly isolated Bacillus megaterium strain ISO-2 from a polycarbonate industrial wastewater. Water Air Soil Pollut. 2018, 229, 348. [Google Scholar] [CrossRef]
- Ponce, B.L.; Latorre, V.K.; González, M.; Seeger, M. Antioxidant compounds improved PCB-degradation by Burkholderia xenovorans strain LB400. Enzym. Microb. Technol. 2011, 49, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Sierra, I.; Valera, J.L.; Marina, M.L.; Laborda, F. Study of the biodegradation process of polychlorinated biphenyls in liquid medium and soil by a new isolated aerobic bacterium (Janibacter sp.). Chemosphere 2003, 53, 609–618. [Google Scholar] [CrossRef]
- Moody, J.; Doerge, D.; Freeman, J.; Cerniglia, C. Degradation of biphenyl by Mycobacterium sp. strain PYR-1. Appl. Microbiol. Biotechnol. 2002, 58, 364–369. [Google Scholar] [CrossRef]
- Murínová, S.; Dercová, K. Potential Use of Newly Isolated Bacterial Strain Ochrobactrum anthropi in bioremediation of polychlorinated biphenyls. Water Air Soil Pollut. 2014, 225, 1980. [Google Scholar] [CrossRef]
- Horváthová, H.; Lászlová, K.; Dercová, K. Bioremediation of PCB-contaminated shallow river sediments: The efficacy of biodegradation using individual bacterial strains and their consortia. Chemosphere 2018, 193, 270–277. [Google Scholar] [CrossRef]
- Zorádová, S.; Dudášová, H.; Lukáčová, L.; Dercová, K.; Čertík, M. The effect of polychlorinated biphenyls (PCBs) on the membrane lipids of Pseudomonas stutzeri. Int. Biodeterior. Biodegrad. 2011, 65, 1019–1023. [Google Scholar] [CrossRef]
- Tao, B.; Zhou, Y.; He, X.; Li, D. Pseudomonas chengduensis sp. nov., isolated from landfill leachate. Int. J. Syst. Evol. Microbiol. 2014, 64, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Ridl, J.; Suman, J.; Fraraccio, S.; Hradilova, M.; Strejcek, M.; Cajthaml, T.; Zubrova, A.; Macek, T.; Strnad, H.; Uhlik, O. Complete genome sequence of Pseudomonas alcaliphila JAB1 (=DSM 26533), a versatile degrader of organic pollutants. Stand Genom. Sci. 2018, 13, 3. [Google Scholar] [CrossRef]
- Chakraborty, J.; Das, S. Characterization of the metabolic pathway and catabolic gene expression in biphenyl degrading marine bacterium Pseudomonas aeruginosa JP-11. Chemosphere 2016, 144, 1706–1714. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, H.; Fujihara, H.; Kimura, N.; Hirose, J.; Watanabe, T.; Futagami, T.; Goto, M.; Shimodaira, J.; Furukawa, K. Insights into the genomic plasticity of Pseudomonas putida KF715, a strain with unique biphenyl-utilizing activity and genome instability properties: Genomic plasticity of Pseudomonas putida KF715. Environ. Microbiol. Rep. 2017, 9, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Weiland-Bräuer, N.; Fischer, M.A.; Schramm, K.-W.; Schmitz, R.A. Polychlorinated biphenyl (PCB)-degrading potential of microbes present in a cryoconite of Jamtalferner Glacier. Front. Microbiol. 2017, 8, 1105. [Google Scholar] [CrossRef] [Green Version]
- Chung, S.-Y.; Maeda, M.; Song, E.; Horikoshij, K.; Kudo, T. A Gram-positive polychlorinated biphenyl-degrading bacterium, Rhodococcus erythropolis strain TA421, isolated from a termite ecosystem. Biosci. Biotechnol. Biochem. 1994, 58, 2111–2113. [Google Scholar] [CrossRef]
- Taguchi, K.; Motoyama, M.; Kudo, T. Multiplicity of 2,3-dihydroxybiphenyl dioxygenase genes in the gram-positive polychlorinated biphenyl degrading bacterium Rhodococcus rhodochrous K37. Biosci. Biotechnol. Biochem. 2004, 68, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Murínová, S.; Dercová, K.; Dudášová, H. Degradation of polychlorinated biphenyls (PCBs) by four bacterial isolates obtained from the PCB-contaminated soil and PCB-contaminated sediment. Int. Biodeterior. Biodegrad. 2014, 91, 52–59. [Google Scholar] [CrossRef]
- Pham, T.T.M.; Pino Rodriguez, N.J.; Hijri, M.; Sylvestre, M. Optimizing polychlorinated biphenyl degradation by flavonoid-induced cells of the Rhizobacterium Rhodococcus erythropolis U23A. PLoS ONE 2015, 10, e0126033. [Google Scholar] [CrossRef] [Green Version]
- Shumkova, E.S.; Olsson, B.E.; Kudryavtseva, A.V.; Plotnikova, E.G. Draft genome sequence of Rhodococcus ruber strain P25, an active polychlorinated biphenyl degrader. Genome Announc. 2015, 3. [Google Scholar] [CrossRef] [Green Version]
- Atago, Y.; Shimodaira, J.; Araki, N.; Bin Othman, N.; Zakaria, Z.; Fukuda, M.; Futami, J.; Hara, H. Identification of novel extracellular protein for PCB/biphenyl metabolism in Rhodococcus jostii RHA1. Biosci. Biotechnol. Biochem. 2016, 80, 1012–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Yu, M.; Shen, A. Complete genome sequence of the polychlorinated biphenyl degrader Rhodococcus sp. WB1. Genome Announc. 2016, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Hu, J.; Xu, K.; Tang, X.; Xu, X.; Shen, C. Biodegradation and chemotaxis of polychlorinated biphenyls, biphenyls, and their metabolites by Rhodococcus sp. Biodegradation 2018, 29, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Todoroki, T.; Shimoda, K.; Terada, A.; Hosomi, M. Behavior of PCDDs/PCDFs in remediation of PCBs-contaminated sediments by thermal desorption. Chemosphere 2010, 80, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Teng, Y.; Luo, Y.; Dick, R.P. Biodegradation of 3,3′,4,4′-tetrachlorobiphenyl by Sinorhizobium meliloti NM. Bioresour. Technol. 2016, 201, 261–268. [Google Scholar] [CrossRef]
- Jing, R.; Fusi, S.; Kjellerup, B.V. Remediation of polychlorinated biphenyls (PCBs) in contaminated soils and sediment: State of knowledge and perspectives. Front. Environ. Sci. 2018, 6, 1–17. [Google Scholar] [CrossRef]
- Pathiraja, G.; Egodawatta, P.; Goonetilleke, A.; Teo, V.S.J. Effective degradation of polychlorinated biphenyls by a facultative anaerobic bacterial consortium using alternating anaerobic aerobic treatments. Sci. Total Environ. 2019, 659, 507–514. [Google Scholar] [CrossRef]
- Cervantes-González, E.; Guevara-García, M.A.; García-Mena, J.; Ovando-Medina, V.M. Microbial diversity assessment of polychlorinated biphenyl–contaminated soils and the biostimulation and bioaugmentation processes. Environ. Monit. Assess 2019, 191, 118. [Google Scholar] [CrossRef]
- Steliga, T.; Jakubowicz, P.; Kapusta, P. Changes in toxicity during in situ bioremediation of weathered drill wastes contaminated with petroleum hydrocarbons. Bioresour. Technol. 2012, 125, 1–10. [Google Scholar] [CrossRef]
- Silva-Castro, G.A.; Rodriguez-Calvo, A.; Laguna, J.; González-López, J.; Calvo, C. Autochthonous microbial responses and hydrocarbons degradation in polluted soil during biostimulating treatments under different soil moisture. Assay in pilot plant. Int. Biodeterior. Biodegrad. 2016, 108, 91–98. [Google Scholar] [CrossRef]
- Roy, A.; Dutta, A.; Pal, S.; Gupta, A.; Sarkar, J.; Chatterjee, A.; Saha, A.; Sarkar, P.; Sar, P.; Kazy, S.K. Biostimulation and bioaugmentation of native microbial community accelerated bioremediation of oil refinery sludge. Bioresour. Technol. 2018, 253, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, P.; Roy, A.; Pal, S.; Mohapatra, B.; Kazy, S.K.; Maiti, M.K.; Sar, P. Enrichment and characterization of hydrocarbon-degrading bacteria from petroleum refinery waste as potent bioaugmentation agent for in situ bioremediation. Bioresour. Technol. 2017, 242, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, M.; da Fonseca, M.M.R.; de Carvalho, C.C.C.R. Bioaugmentation and biostimulation strategies to improve the effectiveness of bioremediation processes. Biodegradation 2011, 22, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Havel, J.; Reineke, W. Degradation of Aroclor 1221 and survival of strains in soil microcosms. Appl Microbiol. Biotechnol. 1992, 38, 129–134. [Google Scholar] [CrossRef]
- Egorova, D.O.; Demakov, V.A.; Plotnikova, E.G. Bioaugmentation of a polychlorobiphenyl contaminated soil with two aerobic bacterial strains. J. Hazard. Mater. 2013, 261, 378–386. [Google Scholar] [CrossRef]
- Wu, M.; Li, W.; Dick, W.A.; Ye, X.; Chen, K.; Kost, D.; Chen, L. Bioremediation of hydrocarbon degradation in a petroleum-contaminated soil and microbial population and activity determination. Chemosphere 2017, 169, 124–130. [Google Scholar] [CrossRef]
- Lászlová, K.; Dudášová, H.; Olejníková, P.; Horváthová, G.; Velická, Z.; Horváthová, H.; Dercová, K. The application of biosurfactants in bioremediation of the aged sediment contaminated with polychlorinated biphenyls. Water Air Soil Pollut. 2018, 229, 219. [Google Scholar] [CrossRef]
- Passatore, L.; Rossetti, S.; Juwarkar, A.A.; Massacci, A. Phytoremediation and bioremediation of polychlorinated biphenyls (PCBs): State of knowledge and research perspectives. J. Hazard. Mater. 2014, 278, 189–202. [Google Scholar] [CrossRef]
- Steliga, T.; Kapusta, P.; Jakubowicz, P. Effectiveness of bioremediation processes of hydrocarbon pollutants in weathered drill wastes. Water Air Soil Pollut. 2009, 202, 211–228. [Google Scholar] [CrossRef]
- Steliga, T.; Jakubowicz, P.; Kapusta, P.; Kluk, D. Badania biodegradacji odpadów wiertniczych zanieczyszczonych substancjami ropopochodnymi (ang. Study on biodegradation of drilling wastes contaminated with petroleum hydrocarbons). Przemysł Chem. 2018, 97, 1666–1675. [Google Scholar] [CrossRef]
- Brzeszcz, J.; Steliga, T.; Kapusta, P.; Turkiewicz, A.; Kaszycki, P. r-strategist versus K-strategist for the application in bioremediation of hydrocarbon-contaminated soils. Int. Biodeterior. Biodegrad. 2016, 106, 41–52. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, W.; Strunk, O.; Westram, R.; Richter, L.; Meier, H.; Yadhukumar; Buchner, A.; Lai, T.; Steppi, S.; Jobb, G.; et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004, 32, 1363–1371. [Google Scholar] [CrossRef] [Green Version]
- Elangovan, S.; Pandian, S.B.S.; Geetha, S.J.; Joshi, S.J. Polychlorinated biphenyls (PCBs): Environmental fate, challenges and bioremediation. In Microbial Metabolism of Xenobiotic Compounds; Arora, P.K., Ed.; Springer: Singapore, 2019; pp. 165–188. [Google Scholar] [CrossRef]
- Tu, C.; Teng, Y.; Luo, Y.; Li, X.; Sun, X.; Li, Z.; Liu, W.; Christie, P. Potential for biodegradation of polychlorinated biphenyls (PCBs) by Sinorhizobium meliloti. J. Hazard. Mater. 2011, 186, 1438–1444. [Google Scholar] [CrossRef]
- Horváthová, H.; Lászlová, K.; Dercová, K. Bioremediation vs. nanoremediation: Degradation of polychlorinated biphenyls (PCBS) using integrated remediation approaches. Water Air Soil Pollut. 2019, 230, 204. [Google Scholar] [CrossRef]
- Nabavi, B.; Nikaeen, M.; Amin, M.; Hatamzadeh, M. Isolation and identification of aerobic polychlorinated biphenyls degrading bacteria. Int. J. Environ. Health Eng. 2013, 2, 47. [Google Scholar] [CrossRef]
- McLeod, M.P.; Warren, R.L.; Hsiao, W.W.L.; Araki, N.; Myhre, M.; Fernandes, C.; Miyazawa, D.; Wong, W.; Lillquist, A.L.; Wang, D.; et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA 2006, 103, 15582–15587. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, J.L.M.; Kachel, C.A.; Aiello, M.R.; Quensen, J.F.; Maltseva, O.V.; Tsoi, T.V.; Tiedje, J.M. Degradation of Aroclor 1242 dechlorination products in sediments by Burkholderia xenovorans LB400(ohb) and Rhodococcus sp. Strain RHA1(fcb). Appl. Environ. Microbiol. 2006, 72, 2476–2482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohmori, T.; Morita, H.; Tanaka, M.; Miyauchi, K.; Kasai, D.; Furukawa, K.; Miyashita, K.; Ogawa, N.; Masai, E.; Fukuda, M. Development of a strain for efficient degradation of polychlorinated biphenyls by patchwork assembly of degradation pathways. J. Biosci. Bioeng. 2011, 111, 437–442. [Google Scholar] [CrossRef] [PubMed]
- McKay, D.B.; Prucha, M.; Reineke, W.; Timmis, K.N.; Pieper, D.H. Substrate specificity and expression of three 2,3-dihydroxybiphenyl 1,2-dioxygenases from Rhodococcus globerulus Strain P6. J. Bacteriol. 2003, 185, 2944–2951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Sun, Y.; Qian, S. Biodegradation of seven polychlorinated biphenyls by a newly isolated aerobic bacterium (Rhodococcus sp. R04). J. Ind. Microbiol. Biotechnol. 2004, 31, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Zhang, Y.; Tang, X.; Luo, W. PCB biodegradation and bphA1 gene expression induced by salicylic acid and biphenyl with Pseudomonas fluorescence P2W and Ralstonia eutropha H850. Pol. J. Environ. Stud. 2014, 23, 1591–1598. [Google Scholar]
- Zhang, H.; Jiang, X.; Lu, L.; Xiao, W. Biodegradation of polychlorinated biphenyls (PCBs) by the novel identified cyanobacterium Anabaena PD-1. PLoS ONE 2015, 10, e0131450. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, K. Molecular genetics and evolutionary relationship of PCB-degrading bacteria. Biodegradation 1994, 5, 289–300. [Google Scholar] [CrossRef]
- Abbasian, F.; Lockington, R.; Megharaj, M.; Naidu, R. A review on the genetics of aliphatic and aromatic hydrocarbon degradation. Appl. Biochem. Biotechnol. 2016, 178, 224–250. [Google Scholar] [CrossRef] [PubMed]
- Nwankwegu, A.S.; Orji, M.U.; Onwosi, C.O. Studies on organic and in-organic biostimulants in bioremediation of diesel-contaminated arable soil. Chemosphere 2016, 162, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Dercová, K.; Čičmanová, J.; Lovecká, P.; Demnerová, K.; Macková, M.; Hucko, P.; Kušnír, P. Isolation and identification of PCB-degrading microorganisms from contaminated sediments. Int. Biodeterior. Biodegrad. 2008, 62, 219–225. [Google Scholar] [CrossRef]
- Guarino, C.; Zuzolo, D.; Marziano, M.; Conte, B.; Baiamonte, G.; Morra, L.; Benotti, D.; Gresia, D.; Stacul, E.R.; Cicchella, D.; et al. Investigation and assessment for an effective approach to the reclamation of polycyclic aromatic hydrocarbon (PAHs) contaminated site: SIN Bagnoli, Italy. Sci. Rep. 2019, 9, 11522. [Google Scholar] [CrossRef] [Green Version]
- Adams, R.H.; Kanga-Leyva, K.; Guzmán-Osorio, F.J.; Escalante- Espinosa, E. Comparison of moisture management methods for the bioremediation of hydrocarbon contaminated soil. Afr. J. Biotechnol. 2011, 19, 394–404. [Google Scholar] [CrossRef]
- Foucault, Y.; Durand, M.-J.; Tack, K.; Schreck, E.; Geret, F.; Leveque, T.; Pradere, P.; Goix, S.; Dumat, C. Use of ecotoxicity test and ecoscores to improve the management of polluted soils: Case of a secondary lead smelter plant. J. Hazard. Mater. 2013, 246–247, 291–299. [Google Scholar] [CrossRef] [Green Version]
- Lima, T.M.S.; Procópio, L.C.; Brandão, F.D.; Leão, B.A.; Tótola, M.R.; Borges, A.C. Evaluation of bacterial surfactant toxicity towards petroleum degrading microorganisms. Bioresour. Technol. 2011, 102, 2957–2964. [Google Scholar] [CrossRef] [PubMed]
- Oleszczuk, P.; Jośko, I.; Kuśmierz, M.; Futa, B.; Wielgosz, E.; Ligęza, S.; Pranagal, J. Microbiological, biochemical and ecotoxicological evaluation of soils in the area of biochar production in relation to polycyclic aromatic hydrocarbon content. Geoderma 2014, 213, 502–511. [Google Scholar] [CrossRef]
- Niyommaneerat, W.; Nakajima, F.; Tobino, T.; Yamamoto, K. Development of a chronic sediment toxicity test using the benthic ostracod Heterocypris incongruens and their application to toxicity assessments of urban road dust. Ecotoxicol. Environ. Saf. 2017, 143, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Steliga, T. The use of biotests in estimation of bioremediation processes in weathered drilling wastes. Arch. Environ. Prot. 2011, 2, 61–70. [Google Scholar]
- Baran, A.; Tarnawski, M. Phytotoxkit/Phytotestkit and Microtox® as tools for toxicity assessment of sediments. Ecotoxicol. Environ. Saf. 2013, 98, 19–27. [Google Scholar] [CrossRef]
- Fai, P.B.; Grant, A. An assessment of the potential of the microbial assay for risk assessment (MARA) for ecotoxicological testing. Ecotoxicology 2010, 19, 1626–1633. [Google Scholar] [CrossRef]
- Mamindy-Pajany, Y.; Hamer, B.; Roméo, M.; Géret, F.; Galgani, F.; Durmiši, E.; Hurel, C.; Marmier, N. The toxicity of composted sediments from Mediterranean ports evaluated by several bioassays. Chemosphere 2011, 82, 362–369. [Google Scholar] [CrossRef] [Green Version]
- Gabrielson, J.; Kühn, I.; Colque-Navarro, P.; Hart, M.; Iversen, A.; McKenzie, D.; Möllby, R. Microplate-based microbial assay for risk assessment and (eco)toxic fingerprinting of chemicals. Anal. Chim. Acta 2003, 485, 121–130. [Google Scholar] [CrossRef]
- Gabrielson, J. Assessing the Toxic Impact of Chemicals Using Bacteria; Microbiology and Tumӧbiologist Centrum (MTC)/Microbiology and Tumor Biology Center (MTC); Karolinska University Press: Stockholm, Sweden, 2004; ISBN 978917140134. [Google Scholar]
- Paskuliakova, A.; McGowan, T.; Tonry, S.; Touzet, N. Phycoremediation of landfill leachate with the chlorophyte Chlamydomonas sp. SW15aRL and evaluation of toxicity pre and post treatment. Ecotoxicol. Environ. Saf. 2018, 147, 622–630. [Google Scholar] [CrossRef]
- Wadhia, K.; Dando, T.; Clive Thompson, K. Intra-laboratory evaluation of Microbial Assay for Risk Assessment (MARA) for potential application in the implementation of the Water Framework Directive (WFD). J. Environ. Monit. 2007, 9, 953. [Google Scholar] [CrossRef]
- Kamber, M.; Fluckiger-Isler, S.; Engelhardt, G.; Jaeckh, R.; Zeiger, E. Comparison of the Ames II and traditional Ames test responses with respect to mutagenicity, strain specificities, need for metabolism and correlation with rodent carcinogenicity. Mutagenesis 2009, 24, 359–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijay, U.; Gupta, S.; Mathur, P.; Suravajhala, P.; Bhatnagar, P. Microbial mutagenicity assay: Ames Test. Biol. Protoc. 2018, 8, 32763. [Google Scholar] [CrossRef]
- Wang, D.; Lin, J.; Lin, J.; Wang, W.; Li, S. Biodegradation of petroleum hydrocarbons by Bacillus subtilis BL-27, a strain with weak hydrophobicity. Molecules 2019, 24, 3021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wrenn, B.A.; Venosa, A.D. Selective enumeration of aromatic and aliphatic hydrocarbon degrading bacteria by a most-probable-number procedure. Can. J. Microbiol. 1996, 42, 252–258. [Google Scholar] [CrossRef]
- Malińska, K. Application of a modified OxiTop® respirometer for laboratory composting studies. Arch. Environ. Prot. 2016, 42, 56–62. [Google Scholar] [CrossRef] [Green Version]
- Steliga, T.; Wojtowicz, K. Wykorzystanie testów respirometrycznych do oceny efektywności biodegradacji osadów z instalacji kopalnianych. Naft. Gaz 2019, 75, 29–37. [Google Scholar] [CrossRef]
- Steliga, T.; Uliasz, M. Spent drilling muds management and natural environment protection. Gospod. Surowcami Min. 2014, 30, 135–155. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Huang, D.; Shi, Y.; Han, F.; Wang, Y.; Ye, H.; Tang, Y.; Yu, H. Method for the analysis of 7 indictor polychlorinated biphenyls (PCBs) and 13 organochlorine pesticide residues in sediment by gas chromatography (GC). IOP Conf. Ser. Earth Environ. Sci. 2019, 237, 022053. [Google Scholar] [CrossRef]
- Wojtowicz, K.; Jakubowicz, P. Opracowanie metodyki oznaczania polichlorowanych bifenyli w próbkach gleb. Naft. Gaz 2019, 75, 420–429. [Google Scholar] [CrossRef]
- Chaîneau, C.H.; Yepremian, C.; Vidalie, J.F.; Ducreux, J.; Ballerini, D. Bioremediation of a crude oil-polluted soil: Biodegradation, leaching and toxicity assessments. Water Air Soil Pollut. 2003, 144, 419–440. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
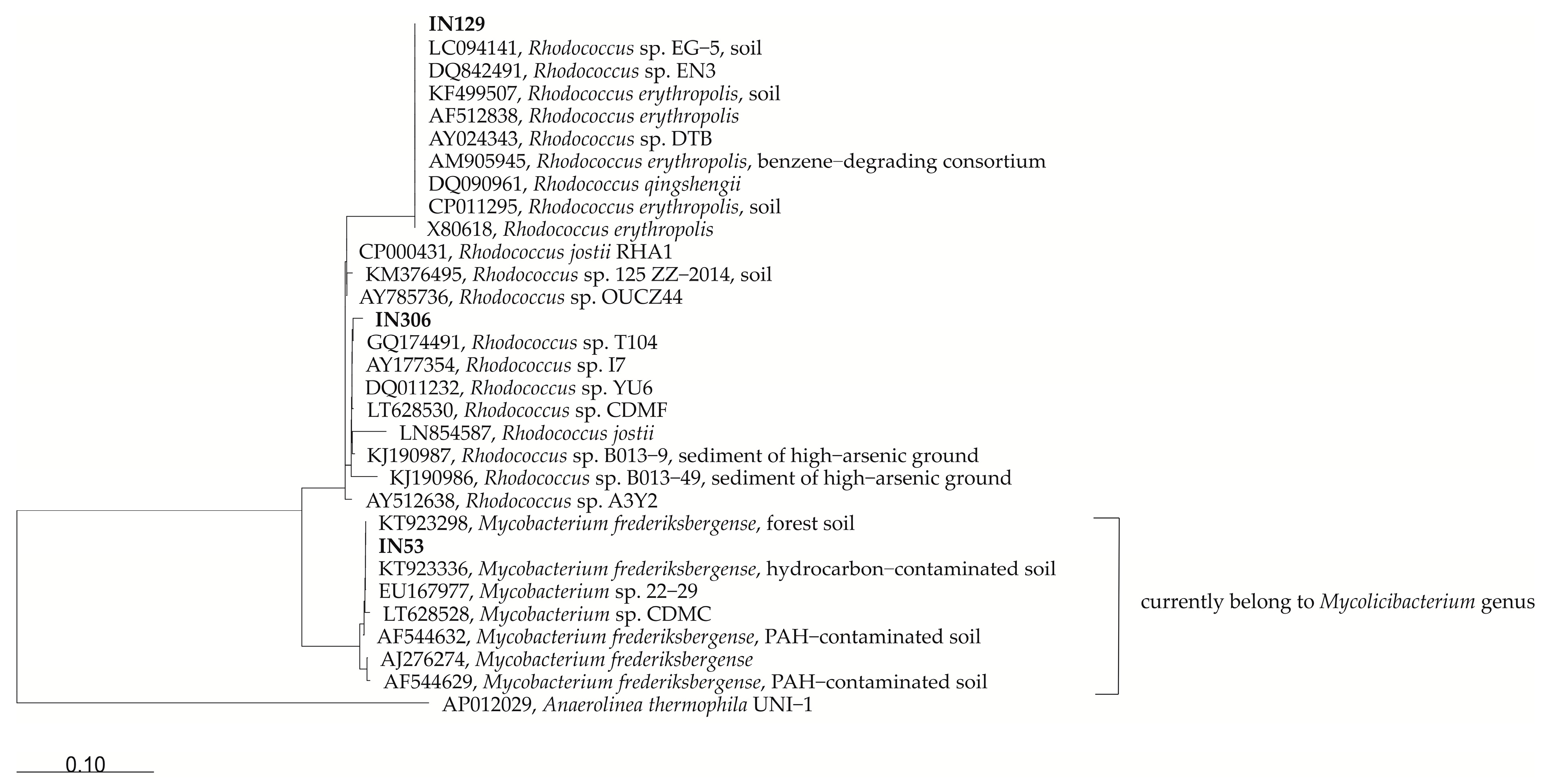


| Strain | The Closest Relative Based on 16S rRNA Accession Number, (% of Identity) * | The Closest Relative for Which the Genome Sequence Is Available in NCBI GenBank, Accession Number, (% of Identity) | Gene Encoding for the Enzymes Degrading Hydrocarbons |
|---|---|---|---|
| Mycolicibacterium frederiksbergense IN53 JN572675 | Mycolicibacterium frederiksbergense DSM 44346 (typical strain) NR_025393.1 99.58% | Mycobacterium sp. YC-RL4, CP015596, (99.24%) | Alkane 1-monooxygenase (2 copies), pentachlorophenol monooxygenase, 2,3-dihydroxybiphenyl 1,2-dioxygenase (2 copies), ring-hydroxylating dioxygenase alpha subunit |
| Rhodococcus erythropolis IN129 KT923311 | Rhodococcus erythropolis IN104 KT923338 100% | Rhodococcus erythropolis X5, CP044284, (99.93%) | Alkane 1-monooxygenase (5copies), pentachlorophenol monooxygenase, phenol/toluene hydroxylase, biphenyl 2,3-dioxygenase, 2,3-dihydroxybiphenyl 1,2-dioxygenase (3 copies), ring-hydroxylating dioxygenase alpha subunit |
| Rhodococcus sp. IN306 KX058399 | Rhodococcus sp. S2-17 KY765341 99.30% | Rhodococcus jostii RHA1, CP000431, (98.75%) | Alkane 1-monooxygenase, pentachlorophenol monooxygenase, phenol/toluene hydroxylase, biphenyl 2,3-dioxygenase, 2,3-dihydrodihydroxybiphenyl 2,3-dehydrogenase, 2,3-dihydroxybiphenyl 1,2-dioxygenase (5 copies), |
| Trophic Level | Life Form | Test Type | Test Reaction | Reference | |
|---|---|---|---|---|---|
| Consumers | Heterocypris incongruens | Ostracodtoxkit(F)TM (MicroBioTest Inc., Gent, Belgium) | growth inhibition, mortality | [74,75] | |
| Producers | Sorghum saccharatum Sinapis alba Lepidium sativum | PhytotoxkitTM test (MicroBioTests Inc., Belgium), | growth inhibition, germinated seeds | [76,77,78] | |
| Decomposers | Vibrio fischeri | Microtox® Solid Phase Test (SDI, New Castle, DE, USA) | luminescence inhibitionTU = 100/EC50 | [70,71,72,73] | |
| Decomposers | Set of 11 bacteria strains | MARA environmental risk test | growth inhibition | [77,79,80,81,82] | |
| oil contaminants were extracted (SPE) and vaporised, deposit was dissolved in dimethylsulfooxygen (DMSO) | |||||
| Microbacterium sp., Brevundimonas diminuta, Citrobacter freudii, Comamonas testosterone, Enterococcus casseliflavus, Delftia acidovorans, Kurthia gibsoni, Staphylococcus warnerii, Pseudomonas aurantiaca, Serratia rubidaea, Pichia anomala) | |||||
| Decomposers | Salmonella typhimurium (TA-98, TA-100) | AMES (Muta-ChromoPlateTM Kit) (EBPI, Mississauga, ON, Canada). | revertants number | [39,83,84] | |
| oil contaminants were extracted (SPE) and vaporised, deposit was dissolved in DMSO | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steliga, T.; Wojtowicz, K.; Kapusta, P.; Brzeszcz, J. Assessment of Biodegradation Efficiency of Polychlorinated Biphenyls (PCBs) and Petroleum Hydrocarbons (TPH) in Soil Using Three Individual Bacterial Strains and Their Mixed Culture. Molecules 2020, 25, 709. https://doi.org/10.3390/molecules25030709
Steliga T, Wojtowicz K, Kapusta P, Brzeszcz J. Assessment of Biodegradation Efficiency of Polychlorinated Biphenyls (PCBs) and Petroleum Hydrocarbons (TPH) in Soil Using Three Individual Bacterial Strains and Their Mixed Culture. Molecules. 2020; 25(3):709. https://doi.org/10.3390/molecules25030709
Chicago/Turabian StyleSteliga, Teresa, Katarzyna Wojtowicz, Piotr Kapusta, and Joanna Brzeszcz. 2020. "Assessment of Biodegradation Efficiency of Polychlorinated Biphenyls (PCBs) and Petroleum Hydrocarbons (TPH) in Soil Using Three Individual Bacterial Strains and Their Mixed Culture" Molecules 25, no. 3: 709. https://doi.org/10.3390/molecules25030709
APA StyleSteliga, T., Wojtowicz, K., Kapusta, P., & Brzeszcz, J. (2020). Assessment of Biodegradation Efficiency of Polychlorinated Biphenyls (PCBs) and Petroleum Hydrocarbons (TPH) in Soil Using Three Individual Bacterial Strains and Their Mixed Culture. Molecules, 25(3), 709. https://doi.org/10.3390/molecules25030709






